
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Imane Rezig | -- | 1763 | 2024-03-06 20:08:12 | | | |
| 2 | Fanny Huang | Meta information modification | 1763 | 2024-03-07 04:11:38 | | |
Video Upload Options
Cytokinesis, as the last stage of the cell division cycle, is a tightly controlled process amongst all eukaryotes, with defective division leading to severe cellular consequences and implicated in serious human diseases and conditions such as cancer. Both mammalian cells and the fission yeast Schizosaccharomyces pombe use binary fission to divide into two equally sized daughter cells. Similar to mammalian cells, in S. pombe, cytokinetic division is driven by the assembly of an actomyosin contractile ring (ACR) at the cell equator between the two cell tips. The ACR is composed of a complex network of membrane scaffold proteins, actin filaments, myosin motors and other cytokinesis regulators. The contraction of the ACR leads to the formation of a cleavage furrow which is severed by the endosomal sorting complex required for transport (ESCRT) proteins, leading to the final cell separation during the last stage of cytokinesis, abscission.
1. The Use of Fission Yeast to Study Eukaryotic Cytokinesis
2. Actin–Myosin Contractile Ring (ACR) Assembly in Fission Yeast
2.1. Positioning of the Cell Division Plane
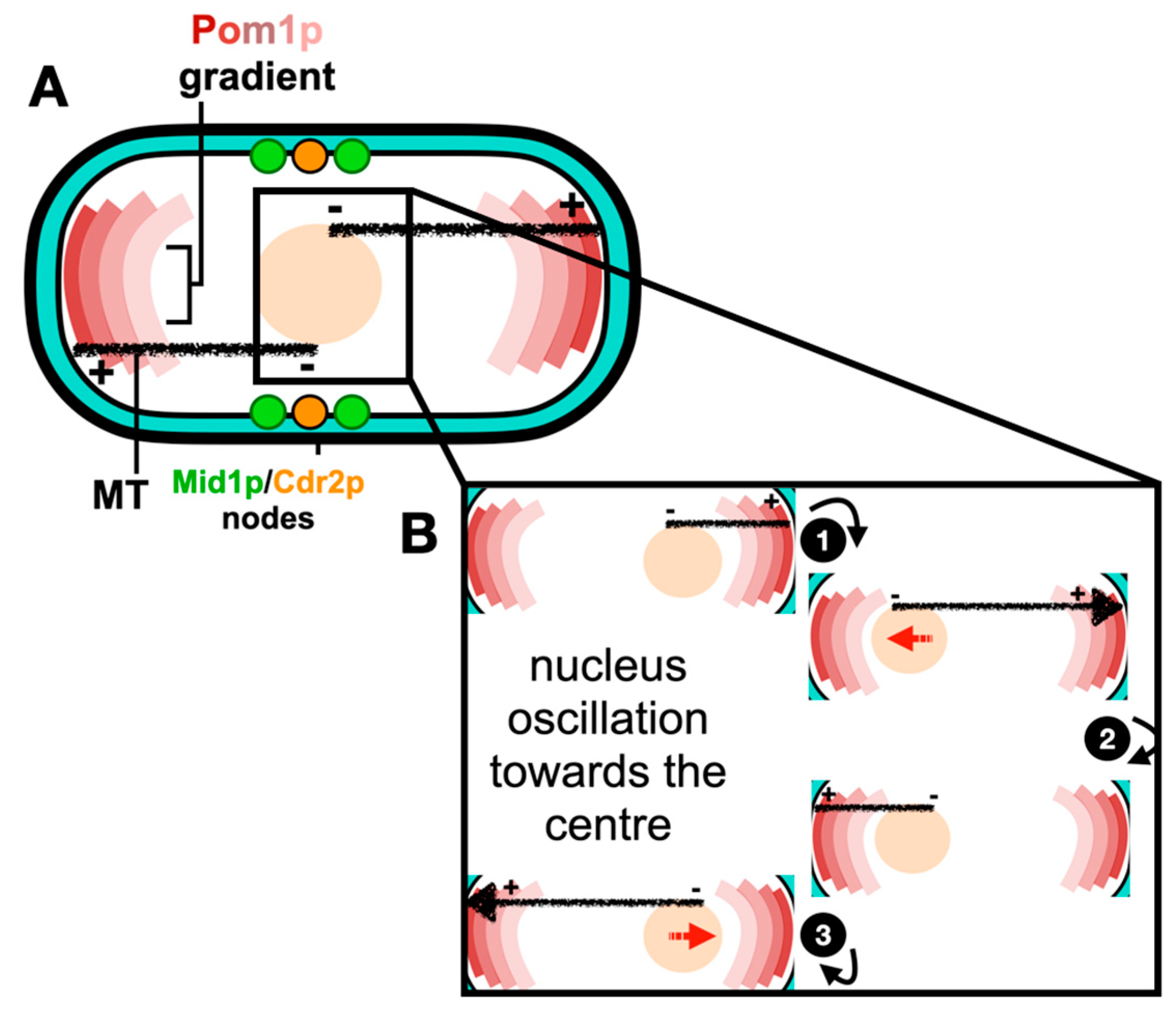
2.2. Molecular Organization of Nodes within the ACR
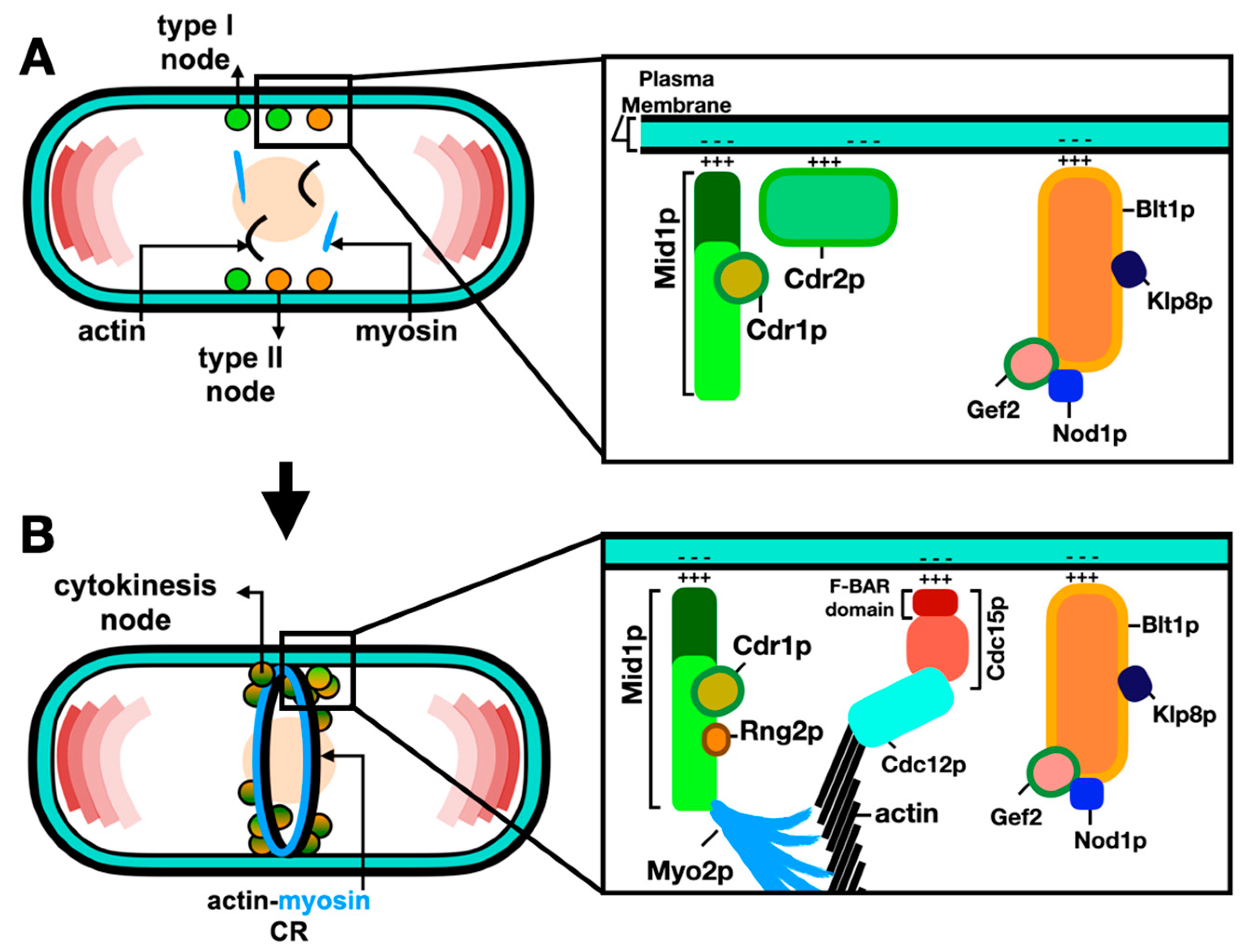
2.3. Anchorage of the ACR to the Plasma Membrane
References
- Pollard, T.D.; Wu, J.-Q. Understanding cytokinesis: Lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 2010, 11, 149–155.
- Hartwell, L.H.; Culotti, J.; Reid, B. Genetic Control of the Cell-Division Cycle in Yeast, I. Detection of Mutants. Proc. Natl. Acad. Sci. USA 1970, 66, 352–359.
- Hartwell, L.H.; Mortimer, R.K.; Culotti, J.; Culotti, M. Genetic control of the cell division cycle in yeast: V. genetic analysis of cdc mutants. Genetics 1973, 74, 267–286.
- Nurse, P. Genetic control of cell size at cell division in yeast. Nature 1975, 256, 547–551.
- Nurse, P.; Thuriaux, P.; Nasmyth, K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1976, 146, 167–178.
- Chang, F.; Nurse, P. Finishing the cell cycle: Control of mitosis and cytokinesis in fission yeast. Trends Genet. 1993, 9, 333–335.
- Chang, F.; Nurse, P. How fission yeast fission in the middle. Cell 1996, 84, 191–194.
- Nurse, P. Fission yeast cell cycle mutants and the logic of eukaryotic cell cycle control. Mol. Biol. Cell 2020, 31, 2871–2873.
- Chang, F. Forces that shape fission yeast cells. Mol. Biol. Cell 2017, 28, 1819–1824.
- Piel, M.; Tran, P.T. Cell shape and cell division in fission yeast. Curr. Biol. 2009, 19, R823–R827.
- Tran, P.T.; Marsh, L.; Doye, V.; Inoue, S.; Chang, F. A mechanism for nuclear positioning in fission yeast based upon microtubule pushing. J. Cell Biol. 2001, 153, 397–411.
- Daga, R.R.; Chang, F. Dynamic positioning of the fission yeast cell division plane. Proc. Natl. Acad. Sci. USA 2005, 102, 8228–8232.
- Daga, R.R.; Yonetani, A.; Chang, F. Asymmetric Microtubule Pushing Forces in Nuclear Centering. Curr. Biol. 2006, 16, 1544–1550.
- Gallardo, P.; Barrales, R.R.; Daga, R.R.; Salas-Pino, S. Nuclear Mechanics in the Fission Yeast. Cells 2019, 8, 1285.
- Martin, S.G.; McDonald, W.H.; Yates, J.R., 3rd; Chang, F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell. 2005, 8, 479–491.
- Hersch, M.; Hachet, O.; Dalessi, S.; Ullal, P.; Bhatia, P.; Bergmann, S.; Martin, S.G. Pom1 gradient buffering through intermolecular auto-phosphorylation. Mol. Syst. Biol. 2015, 11, 818.
- Moseley, J.B.; Mayeux, A.; Paoletti, A.; Nurse, P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009, 7248, 857–860.
- Gerganova, V.; Floderer, C.; Archetti, A.; Michon, L.; Carlini, L.; Reichler, T.; Manley, S.; Martin, S.G. Author response: Multi-phosphorylation reaction and clustering tune Pom1 gradient mid-cell levels according to cell size. Elife 2019, 3, e45983.
- Pan, K.Z.; Saunders, T.E.; Flor-Parra, I.; Howard, M.; Chang, F. Cortical regulation of cell size by a sizer cdr2p. Elife 2014, 3, e02040.
- Almonacid, M.; Celton-Morizur, S.; Jakubowski, J.L.; Dingli, F.; Loew, D.; Mayeux, A.; Chen, J.-S.; Gould, K.L.; Clifford, D.M.; Paoletti, A. Temporal control of contractile ring assembly by Plo1 regulation of Myosin II recruitment by Mid1/Anillin. Curr. Biol. 2011, 21, 473–479.
- Paoletti, A.; Chang, F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission Yeast. Mol. Biol. Cell 2000, 11, 2757–2773.
- Rezig, I.M.; Yaduma, W.G.; Gould, G.W.; McInerny, C.J. Anillin/Mid1p interacts with the ESCRT-associated protein Vps4p and mitotic kinases to regulate cytokinesis in fission yeast. Cell Cycle 2021, 20, 1845–1860.
- Saha, S.; Pollard, T.D.; Akamatsu, M.; Lin, Y.; Bewersdorf, J.; Lidke, M.E.D.; Sherlekar, A.; Rikhy, R.; Montell, M.E.D.; Goss, J.W.; et al. Anillin-related protein Mid1p coordinates the assembly of the cytokinetic contractile ring in fission yeast. Mol. Biol. Cell 2012, 23, 3982–3992.
- Chatterjee, M.; Pollard, T.D. The functionally important N-terminal half of fission yeast mid1p anillin is intrinsically disordered and undergoes phase separation. Biochemistry 2019, 58, 3031–3041.
- Steever, A.; Pringle, J.; Bähler, J.; Wang, Y.-L.; Gould, K.; McCollum, D.; Wheatley, S. Role of polo kinase and mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 1998, 143, 1603–1616.
- Rezig, I.M.; Yaduma, W.G.; Gould, G.W.; McInerny, C.J. The role of anillin/Mid1p during medial division and cytokinesis: From fission yeast to cancer cells. Cell Cycle 2022, 22, 633–644.
- Akamatsu, M.; Berro, J.; Pu, K.-M.; Tebbs, I.R.; Pollard, T.D. Cytokinetic nodes in fission yeast arise from two distinct types of nodes that merge during interphase. J. Cell Biol. 2014, 204, 977–988.
- Laplante, C.; Huang, F.; Tebbs, I.R.; Bewersdorf, J.; Pollard, T.D. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc. Natl. Acad. Sci. USA 2016, 113, E5876–E5885.
- Zhu, Y.-H.; Ye, Y.; Wu, Z.; Wu, J.-Q. Cooperation between Rho-GEF Gef2 and its binding partner Nod1 in the regulation of fission yeast cytokinesis. Mol. Biol. Cell 2013, 24, 3187–3204.
- Akamatsu, M.; Lin, Y.; Bewersdorf, J.; Pollard, T.D.; Saha, S.; Wang, M.E.Y.-L.; Wills, R.C.; Goulden, B.D.; Hammond, G.R.V.; Kozminski, M.E.K.G.; et al. Analysis of interphase node proteins in fission yeast by quantitative and superresolution fluorescence microscopy. Mol. Biol. Cell 2017, 28, 3203–3214.
- Malla, M.; Pollard, T.D.; Chen, Q. Counting actin in contractile rings reveals novel contributions of cofilin and type II myosins to fission yeast cytokinesis. Mol. Biol. Cell 2022, 33, ar51.
- McDonald, N.A.; Lind, A.L.; Smith, S.E.; Li, R.; Gould, K.L. Nanoscale architecture of the Schizosaccharomyces pombe contractile ring. Elife 2017, 15, e28865.
- Sayyad, W.A.; Pollard, T.D. The number of cytokinesis nodes in mitotic fission yeast scales with cell size. Elife 2022, 12, e76249.
- Vavylonis, D.; Wu, J.-Q.; Hao, S.; O’Shaughnessy, B.; Pollard, T.D. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 2008, 319, 97–100.
- Zimmermann, D.; Homa, K.E.; Hocky, G.M.; Pollard, L.W.; De La Cruz, E.M.; Voth, G.A.; Trybus, K.M.; Kovar, D.R. Mechanoregulated inhibition of formin facilitates contractile actomyosin ring assembly. Nat. Commun. 2017, 8, 703.
- Swulius, M.T.; Nguyen, L.T.; Ladinsky, M.S.; Ortega, D.R.; Aich, S.; Mishra, M.; Jensen, G.J. Structure of the fission yeast actomyosin ring during constriction. Proc. Natl. Acad. Sci. USA 2018, 115, E1455–E1464.
- Bellingham-Johnstun, K.; Anders, E.C.; Ravi, J.; Bruinsma, C.; Laplante, C. Molecular organization of cytokinesis node predicts the constriction rate of the contractile ring. J. Cell Biol. 2021, 220, e202008032.
- Snider, C.E.; Chandra, M.; McDonald, N.A.; Willet, A.H.; Collier, S.E.; Ohi, M.D.; Jackson, L.P.; Gould, K.L. Opposite surfaces of the Cdc15 F-BAR domain create a membrane platform that coordinates cytoskeletal and signaling components for cytokinesis. Cell Rep. 2020, 33, 108526.
- Bhattacharjee, R.; Mangione, M.C.; Wos, M.; Chen, J.-S.; Snider, C.E.; Roberts-Galbraith, R.H.; McDonald, N.A.; Presti, L.L.; Martin, S.G.; Gould, K.L. DYRK kinase Pom1 drives F-BAR protein Cdc15 from the membrane to promote medial division. Mol. Biol. Cell 2020, 31, 917–929.
- Magliozzi, J.O.; Sears, J.; Cressey, L.; Brady, M.; Opalko, H.E.; Kettenbach, A.N.; Moseley, J.B. Fission yeast Pak1 phosphorylates anillin-like Mid1 for spatial control of cytokinesis. J. Cell Biol. 2020, 219, e201908017.
- Snider, C.E.; Noor, W.N.I.W.M.; Nguyen, N.T.H.; Gould, K.L.; Suetsugu, S. The state of F-BAR domains as membrane-bound oligomeric platforms. Trends Cell Biol. 2021, 31, 644–655.
- Bhattacharjee, R.; Hall, A.R.; Mangione, M.C.; Igarashi, M.G.; Roberts-Galbraith, R.H.; Chen, J.-S.; Vavylonis, D.; Gould, K.L. Author response: Multiple polarity kinases inhibit phase separation of F-BAR protein Cdc15 and antagonize cytokinetic ring assembly in fission yeast. Elife 2023, 7, e83062.
- Moshtohry, M.; Bellingham-Johnstun, K.; Elting, M.W.; Laplante, C. Laser ablation reveals the impact of Cdc15p on the stiffness of the contractile ring. Mol. Biol. Cell 2022, 33, br9.




