Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Almerinda Di Venere | -- | 1939 | 2024-03-04 15:03:24 | | | |
| 2 | Rita Xu | -8 word(s) | 1931 | 2024-03-05 02:35:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Erba, F.; Mei, G.; Minicozzi, V.; Sabatucci, A.; Di Venere, A.; Maccarrone, M. Conformational Dynamics of Lipoxygenases. Encyclopedia. Available online: https://encyclopedia.pub/entry/55834 (accessed on 07 February 2026).
Erba F, Mei G, Minicozzi V, Sabatucci A, Di Venere A, Maccarrone M. Conformational Dynamics of Lipoxygenases. Encyclopedia. Available at: https://encyclopedia.pub/entry/55834. Accessed February 07, 2026.
Erba, Fulvio, Giampiero Mei, Velia Minicozzi, Annalaura Sabatucci, Almerinda Di Venere, Mauro Maccarrone. "Conformational Dynamics of Lipoxygenases" Encyclopedia, https://encyclopedia.pub/entry/55834 (accessed February 07, 2026).
Erba, F., Mei, G., Minicozzi, V., Sabatucci, A., Di Venere, A., & Maccarrone, M. (2024, March 04). Conformational Dynamics of Lipoxygenases. In Encyclopedia. https://encyclopedia.pub/entry/55834
Erba, Fulvio, et al. "Conformational Dynamics of Lipoxygenases." Encyclopedia. Web. 04 March, 2024.
Copy Citation
Lipoxygenases (LOXs) are a family of enzymes that includes different fatty acid oxygenases with a common tridimensional structure. The main functions of LOXs are the production of signaling compounds and the structural modifications of biological membranes.
lipoxygenase
membrane binding
conformational flexibility
1. Introduction
Lipoxygenases (LOXs) are dioxygenases that play a key role in the metabolism of polyunsaturated fatty acids (PUFAs)—among which is arachidonic (eicosatetraenoic) acid—in a large variety of living cells [1] ranging from microorganisms [2] to plants [3] and mammals [4]. Such a ubiquitous distribution is suggestive of relevant biological roles conferred to LOXs by natural evolution [5][6]. The reaction catalyzed by LOXs is the oxygenation of PUFAs, i.e., the insertion of molecular oxygen (O2) in their acyl chain with the generation of hydroperoxyl (HOO-) groups at different positions. Insertion of O2 is specific for each LOX isoform that is indeed named with the number of the carbon atom where O2 has been bound. LOXs products initiate crucial biosynthetic pathways in living organisms. In mammals, 5- and 15-LOXs induce the synthesis of important signaling molecules [1][7], such as leukotrienes (5-LOX) and lipoxins (15-LOX), that play a crucial role in inflammation and immunity [8]. They are also involved in the development of pathological atherosclerosis [9], as a direct consequence of their ability to bind low density lipoproteins [10]. Plant lipoxygenases (8-, 13-LOXs) are instead involved in germination and growth, as well as in pathogen resistance [3]. Additionally, some LOXs can modify the structure of lipid bilayers to reach specific metabolic goals: for example, in plants, oxidation of membranes by LOXs drives leaf senescence or lipid mobilization during the germination phase [3]; in animals, 15-LOX of reticulocytes attack the mitochondria envelope and promote the maturation of red [11][12].
The first biochemical characterization of a (plant) LOX dates back to the 1970s [13][14] but, despite the widespread presence of LOXs in the seeds of legumes [15], the first tridimensional structure of a soybean 15-LOX (also known as LOX-1) was obtained only 20 years later [16][17]. The main 3D features of soybean 15-LOX are: (i) the existence of two rather distinct structural domains, and (ii) the presence of a non-heme catalytic iron located in the C-terminal domain of the polypeptide chain [16][17]. For several years, only soybean 15-LOX 3D structure was available, and thus was used as a template to model human 5-, 12-, and 15-LOXs [18]. A more reliable model for mammalian (and in particular human) LOXs was possible when rabbit reticulocyte 15-LOX was crystallized [19] and its preliminary structure was determined [20] and then refined [21]. The further characterization of human [22][23], porcine [24], and coral [25] LOXs has provided evidence that a common 3D architecture does exist in animal LOXs.
Starting in the 1970s, the attention on LOXs has increased considerably, as it appears from the number of papers published since then (Figure 1A). Human 5-LOX and 15-LOX are clearly the most studied members (Figure 1A) due to their major impact on health and disease conditions [26][27][28][29][30]. Of note, only a small number of studies have interrogated the interaction of LOXs with bio-membranes (Figure 1B), despite two fundamental questions arising from this event: (i) How do soluble enzymes (such as LOXs) search and find their substrates in a rather peculiar environment like lipid bilayers? (ii) How does the interaction with bio-membranes modulate the activity of LOXs? The first issue is obviously not specific for LOXs, yet these enzymes may represent a paradigmatic example to shed light on other membrane-interacting proteins [31].
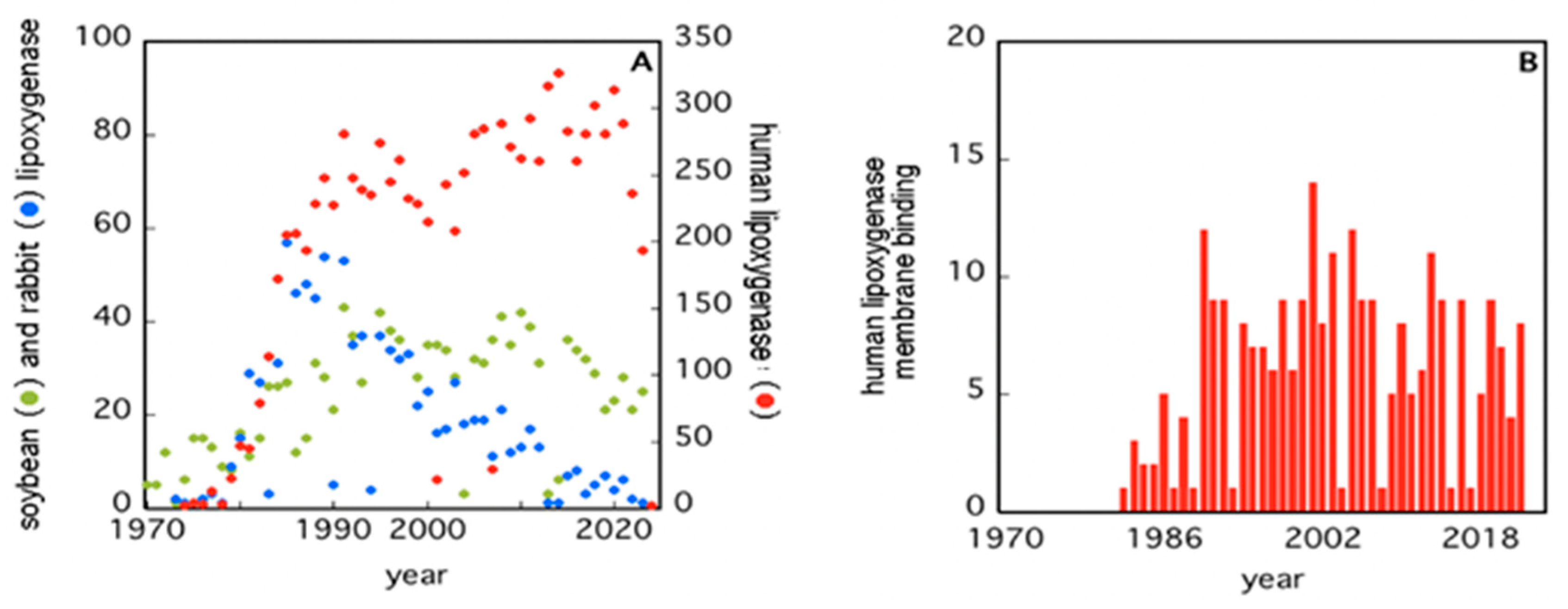
Figure 1. Number of papers retrieved from PubMed when using the keywords “soybean”, “rabbit”, “human”, and “lipoxygenases” (Panel (A)), or “human lipoxygenases membrane binding” (Panel (B)).
The second issue seems to be of great relevance for LOXs, especially for human 5-LOX, whose activity leads to the synthesis of bioactive compounds—leukotrienes—from arachidonic acid [26][29]. Human 5-LOX is, indeed, the only member of the LOXs family that is present both in the cytoplasm and in the nucleus of a cell and that is able to bind different types of membranes (plasma membranes, nuclear membranes) both directly or through the specific 5-LOX activating protein, FLAP [27].
2. Insights into the Architecture of LOXs
Despite the huge amount of data available on phylogenetics, biological activity, design and action of inhibitors, and in vivo localization of LOXs, these proteins remain quite elusive from the structural point of view. In fact, a limited number of 3D structures are as yet available because of crystallization problems due to unstable segments in the protein sequence. The first two characterized LOXs are a plant enzyme, namely soybean 15-LOX (the first ever to be crystallized), and its mammalian counterpart, 12/15-LOX from rabbit reticulocyte. These two proteins share a limited sequence homology (<24%, Figure 2) and have different molecular weights (94,000 and 77,000, respectively); nonetheless, they display the same 3D organization. In particular, X-ray measurements [16][20] revealed the presence of two different domains, namely an N-terminal β-barrel PLAT (Polycystin-1, Lipoxygenase, Alpha-Toxin) domain, and a larger C-terminal domain that is mainly characterized by α-helices (Figure 3 and Figure 4). The two domains are connected by a short flexible linker and play different functional roles: the C-terminal contains the catalytic site [16][20], while the N-terminal has regulatory functions and, for instance, influences the membrane binding ability of mammalian enzymes [32]. This general structural organization is also highly conserved in other human variants, such as 5-, 12-, and 15-LOX [22][23][33], and in coral 8-LOX [25], despite the degree of sequence identity with both plant and rabbit LOXs being on the average quite low (Figure 2).
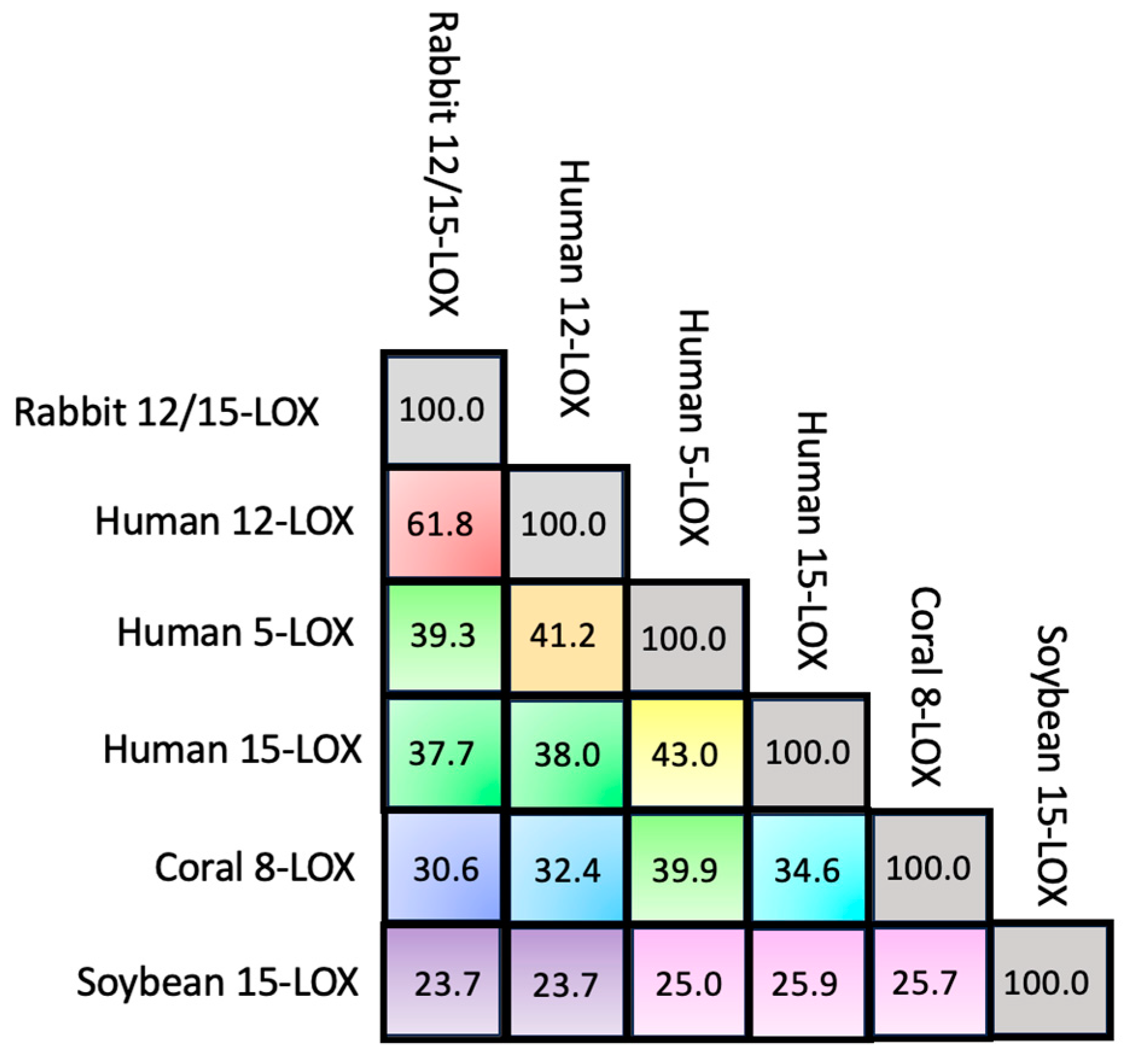
Figure 2. Percentage of sequence identity among the six LOX isoforms.
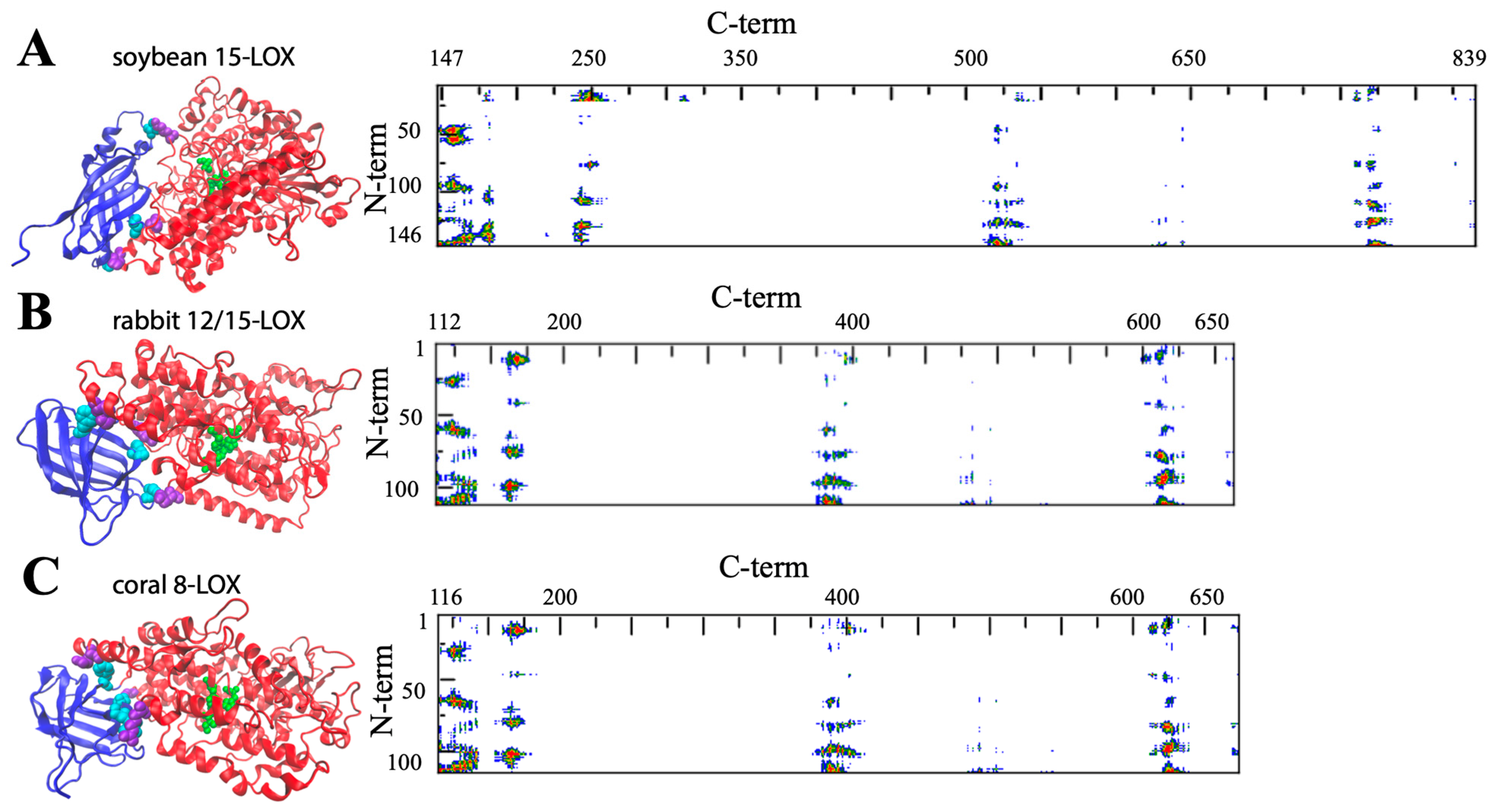
Figure 3. General structure of soybean 15-LOX (A), rabbit 12/15-LOX (B), and coral 8-LOX (C). The models are shown in a secondary structure rendering. Fe ligands are in green. Some representative contact residues between the two domains are shown in cyan and violet for the N- and C-terminals, respectively (A: V22, G54, F144, P157, K252, V520; B: S13, F62, Y98, H128, E169, Y614; C: P102, W106, F114, Q132, R167, E647). On the right side, the contact maps of the two domains are reported (red, yellow, green, and blue spots correspond to 7, 10, 13, and 16 Å inter-residues distances).
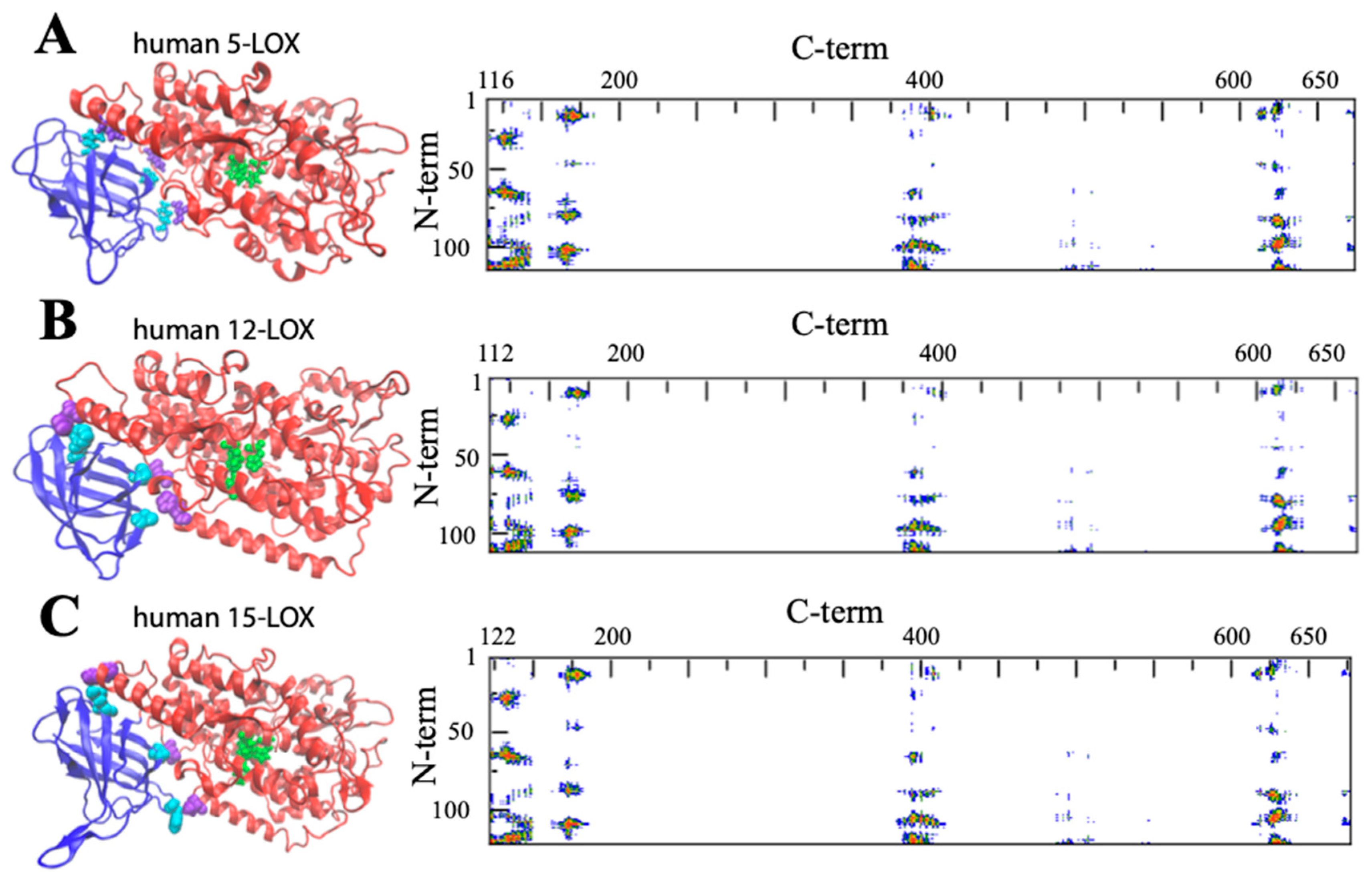
Figure 4. General structure of human 5-LOX (A), human12-LOX (B), and human 15-LOX (C). Graphic rendering and contact map color codes are the same used in Figure 3. Representative contact residues between the two domains are shown in cyan and violet for the N- and C-terminals, respectively (A: Q13, L67, Y101, H131, D171, H625; B: A12, F62, Y98, H128, E168, Y614; C: F14, L65, Y107, H131, T178, D625). The models reproduced in A, B, and C have been obtained from the available PDB files: (3o8y, 8ghb, and 4nre, respectively). Missing atoms in the 5-LOX structure were added using the Chimera interface to Modeller.
The composition of the two N- and C-terminal domains is significantly different in all the above-mentioned LOX structures. The smaller β-barrel N-terminal is rather dense, mainly containing tightly packed hydrophobic residues; the C-terminal core is instead characterized by large cavities suitable for oxygen transit and substrate [34][35][36][37]. The two domains are separated by a large gap, yet they are not fully independent. In Figure 3 and Figure 4, the analysis of the domain–domain interface is reported in terms of “contact maps”, which indicate the closest points of contact between the two protein sections. Plant and mammal lipoxygenases share a similar architecture, in which a few (≈4) main groups of contacts characterize the interaction between the two domains (Figure 3 and Figure 4). However, the positions of such contacts along the polypeptide chain of soybean 15-LOX are not the same reported for mammalian enzymes or coral 8-LOX, due to the larger size of the plant enzyme. Indeed, except for soybean 15-LOX, the other 5 enzymes exhibit the most relevant contacts in similar locations, namely at positions 15, 25, 60, and 100 in the N-terminal domain, and approximately at positions 160, 390, and 620 in the C-terminal domain (Figure 3 and Figure 4). A more detailed analysis of these regions reveals a high homology score among the five LOX polypeptide chains (Figure 5), suggesting that such amino acids might play an important role in the protein dynamics. In particular, it could be speculated that these regions communicate movements of the N-terminal to the C-terminal and vice versa, in analogy to the “hot spots” found in the contact networks of oligomeric protein inter-subunit surfaces [38]. Furthermore, it should be stressed that many of the conserved or semi-conserved residues are histidines and aromatic amino acids (Figure 5), whose large side chain fills the gap at the domain–domain interface.
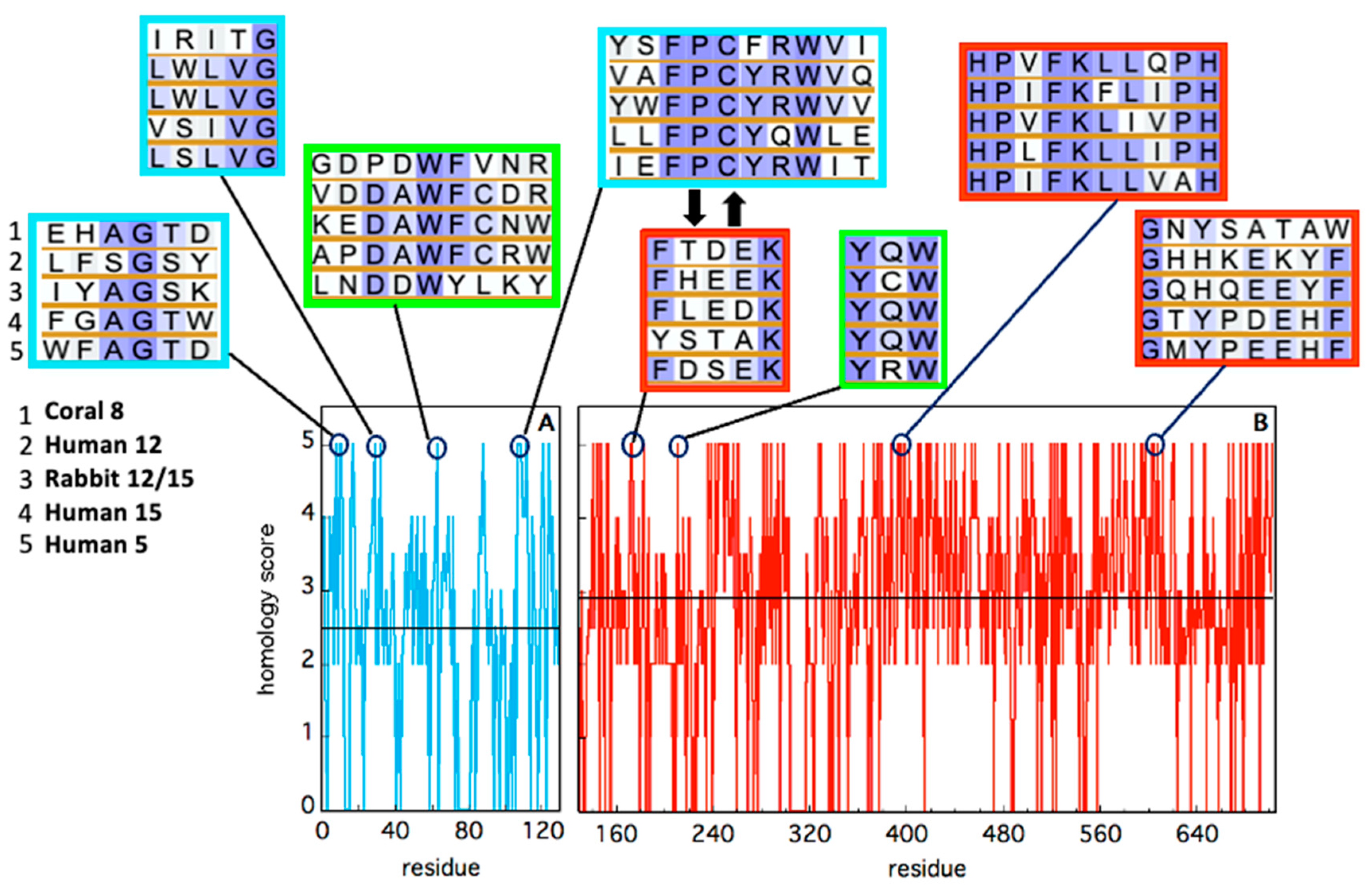
Figure 5. Sequence homology of the five animal LOX isoforms, in the N-terminal (cyan) and C-terminal (red) domains. Alignment has been obtained using BLAST. Identity has been quantified by assigning an “homology score” using the following scheme: XXXXX = 5; XXXXZ = 4; XXXZZ = 3.5; XXXZB = 3; XXZZB = 2.5; XXZBO = 2.0; XZBOU = 0. The average score in each domain is reported as a black horizontal line. The local sequence segments containing residues involved in relevant domain-domain contacts (i.e., the red spots in Figure 3 and Figure 4) are reported in the cyan (N-terminal) and red (C-terminal) rectangular boxes. The two thick black arrows indicate two interacting segments of particular relevance, as discussed in the text. Putative conserved residues involved in protein–membrane interaction are shown in the two green rectangles.
3. Structural Flexibility of LOXs
One important feature of a protein structure is the intrinsic plasticity due to the presence of inner empty cavities, water molecules, and inter-domains movements. LOXs accomplish, at the same time, quite different tasks, hosting the acyl chain of polyunsaturated fatty acids and binding membranes, two functions that require a certain degree of elasticity. Small angle X-ray scattering experiments suggested that rabbit 12/15-LOX [39] and human platelet 12-LOX [40] display a certain degree of conformational flexibility due to both local and global structural changes. Temperature- and pressure-dependent dynamic fluorescence data [41][42] led to a similar conclusion. Local flexibility was probed by the conformational changes induced by an eicosatetraynoic acid (ETYA) inhibitor, and was thus associated with the active site in the C-terminal domain [42]. These data are compatible with the mobility of a few α-helix segments that characterize the opening of the catalytic pocket in rabbit 12/15-LOX [21]. Instead, global flexibility arises from interdomain movements [39][40], as also suggested by molecular dynamics simulations [43]. The few key contacts that characterize the domain–domain interface (Figure 3 and Figure 4) play a major role in regulating protein plasticity, especially if they are characterized by a specific aromatic side chain. For instance, Y98 is a highly conserved residue (Figure 5) that, in rabbit 12/15-LOX and in human 12-LOX, occupies a relevant position at the domain–domain interface (Figure 3B and Figure 4B). Its substitution with smaller amino acids (for instance phenylalanine or alanine) does not influence protein secondary and tertiary structures, but has a strong impact on enzyme catalysis and stability by modulating domain association and substrate binding [44]. Flexibility is fundamental for interdomain communication also in human 15-LOX [45] and coral 11-LOX [46]. In fact, it was proposed that the N-terminal domain could exert an allosteric regulation of LOX catalytic activity through residues at the domain-domain interface [46]. A highly conserved tryptophan in the FPCYRW segment (Figure 5) seems to be the best candidate to accomplish such a task, due to its strong interaction with the group of amino acids located in the C-terminal domain between positions 160 and 170 (Figure 3 and Figure 4). It should be noted that, within the same region, a lysine and a phenylalanine—a tyrosine in human 15-LOX—are always present in animal isoforms (Figure 5). Therefore, it is tempting to speculate that the proposed communication mechanism between the two protein domains [46] could be a common feature of all animal LOXs. Finally, the comparison between soybean and rabbit LOXs demonstrated that a higher flexibility of the mammal enzyme facilitates its membrane binding in both in vitro and ex vivo measurements [42], indicating that the global flexibility of (some) LOXs can directly modulate their interaction with lipid bilayers.
References
- Brash, A.R. Lipoxygenases: Occurrence, functions, catalysis, and acquisition of substrate. J. Biol. Chem. 1999, 274, 23679–23682.
- Chrisnasari, R.; Hennebelle, M.; Vincken, J.P.; van Berkel, W.J.H.; Ewing, T.A. Bacterial lipoxygenases: Biochemical characteristics, molecular structure and potential applications. Biotechnol. Adv. 2022, 61, 108046.
- Viswanath, K.K.; Varakumar, P.; Pamuru, R.R.; Basha, S.J.; Mehta, S.; Rao, A.D. Plant Lipoxygenases and Their Role in Plant Physiology. J. Plant Biol. 2020, 63, 83–95.
- Skrzypczak-Jankun, E.; Jankun, J.; Al-Senaidy, A. Human lipoxygenase: Developments in its structure, function, relevance to diseases and challenges in drug development. Curr. Med. Chem. 2012, 19, 5122–5127.
- Kuhn, H.; Banthiya, S.; van Leyen, K. Mammalian lipoxygenases and their biological relevance. Biochim. Biophys. Acta 2015, 1851, 308–330.
- Horn, T.; Adel, S.; Schumann, R.; Sur, S.; Kakularam, K.R.; Polamarasetty, A.; Redanna, P.; Kuhn, H.; Heydeck, D. Evolutionary aspects of lipoxygenases and genetic diversity of human leukotriene signaling. Prog. Lipid Res. 2015, 57, 13–39.
- Joo, Y.C.; Oh, D.K. Lipoxygenases: Potential starting biocatalysts for the synthesis of signaling compounds. Biotechnol. Adv. 2012, 30, 1524–1532.
- Samuelsson, B.; Dahlén, S.E.; Lindgren, J.A.; Rouzer, C.A.; Serhan, C.N. Leukotrienes and lipoxins: Structures, biosynthesis, and biological effects. Science 1987, 237, 1171–1176.
- Feinmark, S.J.; Cornicelli, J.A. Is there a role for 15-lipoxygenase in atherogenesis? Biochem. Pharmacol. 1997, 54, 953–959.
- Belkner, J.; Wiesner, R.; Rathman, J.; Barnett, J.; Sigal, E.; Kühn, H. Oxygenation of lipoproteins by mammalian lipoxygenases. Eur. J. Biochem. 1993, 213, 251–261.
- Rapoport, S.M.; Schewe, T.; Wiesner, R.; Halangk, W.; Ludwig, P.; Janicke-Höhne, M.; Tannert, C.; Hiebsch, C.; Klatt, D. The lipoxygenase of reticulocytes. Purification, characterization and biological dynamics of the lipoxygenase; its identity with the respiratory inhibitors of the reticulocyte. Eur. J. Biochem. 1979, 96, 545–561.
- Rapoport, S.M.; Schewe, T. The maturational breakdown of mitochondria in reticulocytes. Biochim. Biophys. Acta 1986, 864, 471–495.
- Roza, M.; Francke, A. Soyabean lipoxygenase: An iron-containing enzyme. Biochim. Biophys. Acta 1973, 327, 24–31.
- Chan, H.W. Soya-bean lipoxygenase: An iron-containing dioxygenase. Biochim. Biophys. Acta 1973, 327, 32–35.
- Siedow, J.N. Plant lipoxygenase: Structure and function. Annu. Rev. Plant Biol. 1991, 42, 145–188.
- Boyington, J.C.; Gaffney, B.J.; Amzel, L.M. The three-dimensional structure of an arachidonic acid 15-lipoxygenase. Science 1993, 260, 1482–1486.
- Boyington, J.C.; Gaffney, B.J.; Amzel, L.M. Crystallization and preliminary x-ray analysis of soybean lipoxygenase-1, a non-heme iron-containing dioxygenase. J. Biol. Chem. 1990, 265, 12771–12773.
- Prigge, S.T.; Boyington, J.C.; Gaffney, B.J.; Amzel, L.M. Structure conservation in lipoxygenases: Structural analysis of soybean lipoxygenase-1 and modeling of human lipoxygenases. Proteins 1996, 24, 275–291.
- Sloane, D.L.; Browner, M.F.; Dauter, Z.; Wilson, K.; Fletterick, R.J.; Sigal, E. Purification and crystallization of 15-lipoxygenase from rabbit reticulocytes. Biochem. Biophys. Res. Commun. 1990, 173, 507–513.
- Gillmor, S.A.; Villaseñor, A.; Fletterick, R.; Sigal, E.; Browner, M.F. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nat. Struct. Biol. 1997, 4, 1003–1009.
- Choi, J.; Chon, J.K.; Kim, S.; Shin, W. Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data. Proteins 2008, 70, 1023–1032.
- Gilbert, N.C.; Bartlett, S.G.; Waight, M.T.; Neau, D.B.; Boeglin, W.E.; Brash, A.R.; Newcomer, M.E. The structure of human 5-lipoxygenase. Science 2011, 331, 217–219.
- Kobe, M.J.; Neau, D.B.; Mitchell, C.E.; Bartlett, S.G.; Newcomer, M.E. The structure of human 15-lipoxygenase-2 with a substrate mimic. J. Biol. Chem. 2014, 289, 8562–8569.
- Xu, S.; Mueser, T.C.; Marnett, L.J.; Funk, M.O., Jr. Crystal structure of 12-lipoxygenase catalytic-domain-inhibitor complex identifies a substrate-binding channel for catalysis. Structure 2012, 20, 1490–1497.
- Oldham, M.L.; Brash, A.R.; Newcomer, M.E. Insights from the X-ray crystal structure of coral 8R-lipoxygenase: Calcium activation via a C2-like domain and a structural basis of product chirality. J. Biol. Chem. 2005, 280, 39545–39552.
- Brock, T.G. Regulating leukotriene synthesis: The role of nuclear 5-lipoxygenase. J. Cell. Biochem. 2005, 96, 1203–1211.
- Werz, O.; Steinhilber, D. Therapeutic options for 5-lipoxygenase inhibitors. Pharmacol. Ther. 2006, 112, 701–718.
- Werz, O.; Steinhilber, D. Development of 5-lipoxygenase inhibitors--lessons from cellular enzyme regulation. Biochem. Pharmacol. 2005, 70, 327–333.
- Rådmark, O.; Werz, O.; Steinhilber, D.; Samuelsson, B. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim. Biophys. Acta 2015, 1851, 331–339.
- Gallegos, E.M.; Reed, T.D.; Mathes, F.A.; Guevara, N.V.; Neau, D.B.; Huang, W.; Newcomer, M.E.; Gilbert, N.C. Helical remodeling augments 5-lipoxygenase activity in the synthesis of proinflammatory mediators. J. Biol. Chem. 2022, 298, 102282.
- Epand, R.M. Recognition of polyunsaturated acyl chains by enzymes acting on membrane lipids. Biochim. Biophys. Acta 2012, 1818, 957–962.
- Walther, M.; Hofheinz, K.; Vogel, R.; Roffeis, J.; Kühn, H. The N-terminal β-barrel domain of mammalian lipoxygenases including mouse 5-lipoxygenase is not essential for catalytic activity and membrane binding but exhibits regulatory functions. Arch. Biochem. Biophys. 2011, 516, 1–9.
- Mobbs, J.I.; Black, K.A.; Tran, M.; Burger, W.A.C.; Venugopal, H.; Holman, T.R.; Holinstat, M.; Thal, D.M.; Glukhova, A. Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors. Blood 2023, 142, 1233–1242.
- Gilbert, N.C.; Gerstmeier, J.; Schexnaydre, E.E.; Börner, F.; Garscha, U.; Neau, D.B.; Werz, O.; Newcomer, M.E. Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products. Nat. Chem. Biol. 2020, 16, 783–790.
- Gaffney, B.J. Lipoxygenases: Structural principles and spectroscopy. Annu. Rev. Biophys. Biomol. Struct. 1996, 25, 431–459.
- Prigge, S.T.; Boyington, J.C.; Faig, M.; Doctor, K.S.; Gaffney, B.J.; Amzel, L.M. Structure and mechanism of lipoxygenases. Biochimie 1997, 79, 629–636.
- Ivanov, I.; Heydeck, D.; Hofheinz, K.; Roffeis, J.; O’Donnell, V.B.; Kuhn, H.; Walther, M. Molecular enzymology of lipoxygenases. Arch. Biochem. Biophys. 2010, 503, 161–174.
- Di Paola, L.; Mei, G.; Di Venere, A.; Giuliani, A. Exploring the stability of dimers through protein structure topology. Curr. Protein Pept. Sci. 2016, 17, 30–36.
- Hammel, M.; Walther, M.; Prassl, R.; Kuhn, H. Structural flexibility of the N-terminal beta-barrel domain of 15-lipoxygenase-1 probed by small angle X-ray scattering. Functional consequences for activity regulation and membrane binding. J. Mol. Biol. 2004, 343, 917–929.
- Shang, W.; Ivanov, I.; Svergun, D.I.; Borbulevych, O.Y.; Aleem, A.M.; Stehling, S.; Jankun, J.; Kühn, H.; Skrzypczak-Jankun, E. Probing dimerization and structural flexibility of mammalian lipoxygenases by small-angle X-ray scattering. J. Mol. Biol. 2011, 409, 654–668.
- Mei, G.; Di Venere, A.; Nicolai, E.; Angelucci, C.B.; Ivanov, I.; Sabatucci, A.; Dainese, E.; Kuhn, H.; Maccarrone, M. Structural properties of plant and mammalian lipoxygenases. Temperature-dependent conformational alterations and membrane binding ability. Biochemistry 2008, 47, 9234–9242.
- Di Venere, A.; Nicolai, E.; Ivanov, I.; Dainese, E.; Adel, S.; Angelucci, B.C.; Kuhn, H.; Maccarrone, M.; Mei, G. Probing conformational changes in lipoxygenases upon membrane binding: Fine-tuning by the active site inhibitor ETYA. Biochim. Biophys. Acta 2014, 1841, 1–10.
- Moin, S.T.; Hofer, T.S.; Sattar, R.; Ul-Haq, Z. Molecular dynamics simulation of mammalian 15S-lipoxygenase with AMBER force field. Eur. Biophys. J. 2011, 40, 715–726.
- Ivanov, I.; Di Venere, A.; Horn, T.; Scheerer, P.; Nicolai, E.; Stehling, S.; Richter, C.; Skrzypczak-Jankun, E.; Mei, G.; Maccarrone, M.; et al. Tight association of N-terminal and catalytic subunits of rabbit 12/15-lipoxygenase is important for protein stability and catalytic activity. Biochim. Biophys. Acta 2011, 1811, 1001–1010.
- Droege, K.D.; Keithly, M.E.; Sanders, C.R.; Armstrong, R.N.; Thompson, M.K. Structural Dynamics of 15-Lipoxygenase-2 via Hydrogen-Deuterium Exchange. Biochemistry 2017, 56, 5065–5074.
- Eek, P.; Järving, R.; Järving, I.; Gilbert, N.C.; Newcomer, M.E.; Samel, N. Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca2+ regulation. J. Biol. Chem. 2012, 287, 22377–22386.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
413
Revisions:
2 times
(View History)
Update Date:
05 Mar 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




