
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Nawal Merjaneh | -- | 1614 | 2024-02-27 14:14:02 | | | |
| 2 | Fanny Huang | -1 word(s) | 1613 | 2024-03-05 09:18:49 | | |
Video Upload Options
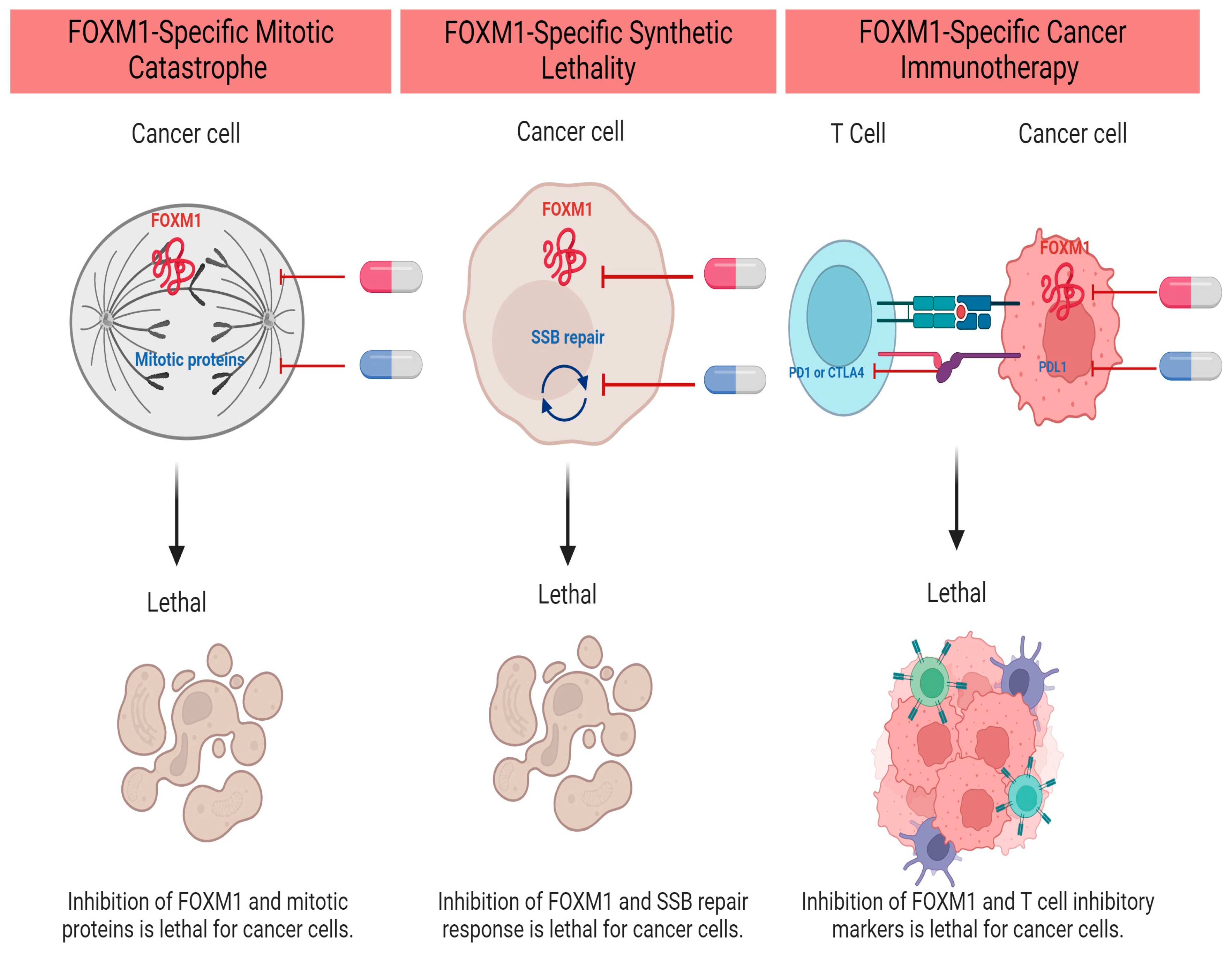
Forkhead box M1 (FOXM1) is a transcription factor in the forkhead (FOX) family, which is required for cellular proliferation in normal and neoplastic cells. FOXM1 is highly expressed in many different cancers, and its expression is associated with a higher tumor stage and worse patient-related outcomes. Abnormally high expression of FOXM1 in cancers compared to normal tissue makes FOXM1 an attractive target for pharmacological inhibition. FOXM1-inhibiting agents and specific FOXM1-targeted small-molecule inhibitors have been developed in the lab and some of them have shown promising efficacy and safety profiles in mouse models. While the future goal is to translate FOXM1 inhibitors to clinical trials, potential synergistic drug combinations can maximize anti-tumor efficacy while minimizing off-target side effects.
1. Introduction
2. FOXM1 Inhibitors in Combination with Cytotoxic Chemotherapy
2.1. Combination with Alkylating Agents
2.2. Combination with Topoisomerase II Inhibitors
2.3. Combination with Mitotic Spindle Inhibitors
2.4. Combination with Antimetabolites
References
- Clark, K.L.; Halay, E.D.; Lai, E.; Burley, S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 1993, 364, 412–420.
- Clevidence, D.E.; Overdier, D.G.; Tao, W.; Qian, X.; Pani, L.; Lai, E.; Costa, R.H. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc. Natl. Acad. Sci. USA 1993, 90, 3948–3952.
- Kaestner, K.H.; Lee, K.H.; Schlondorff, J.; Hiemisch, H.; Monaghan, A.P.; Schutz, G. Six members of the mouse forkhead gene family are developmentally regulated. Proc. Natl. Acad. Sci. USA 1993, 90, 7628–7631.
- Kalin, T.V.; Ustiyan, V.; Kalinichenko, V.V. Multiple faces of FoxM1 transcription factor: Lessons from transgenic mouse models. Cell Cycle 2011, 10, 396–405.
- Kim, I.M.; Ramakrishna, S.; Gusarova, G.A.; Yoder, H.M.; Costa, R.H.; Kalinichenko, V.V. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J. Biol. Chem. 2005, 280, 22278–22286.
- Kalin, T.V.; Wang, I.C.; Meliton, L.; Zhang, Y.; Wert, S.E.; Ren, X.; Snyder, J.; Bell, S.M.; Graf, L., Jr.; Whitsett, J.A.; et al. Forkhead Box m1 transcription factor is required for perinatal lung function. Proc. Natl. Acad. Sci. USA 2008, 105, 19330–19335.
- Ustiyan, V.; Zhang, Y.; Perl, A.K.; Whitsett, J.A.; Kalin, T.V.; Kalinichenko, V.V. β-catenin and Kras/Foxm1 signaling pathway are critical to restrict Sox9 in basal cells during pulmonary branching morphogenesis. Dev. Dyn. 2016, 245, 590–604.
- Wang, I.C.; Meliton, L.; Ren, X.; Zhang, Y.; Balli, D.; Snyder, J.; Whitsett, J.A.; Kalinichenko, V.V.; Kalin, T.V. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS ONE 2009, 4, e6609.
- Bolte, C.; Zhang, Y.; Wang, I.C.; Kalin, T.V.; Molkentin, J.D.; Kalinichenko, V.V. Expression of Foxm1 transcription factor in cardiomyocytes is required for myocardial development. PLoS ONE 2011, 6, e22217.
- Bolte, C.; Zhang, Y.; York, A.; Kalin, T.V.; Schultz Jel, J.; Molkentin, J.D.; Kalinichenko, V.V. Postnatal ablation of Foxm1 from cardiomyocytes causes late onset cardiac hypertrophy and fibrosis without exacerbating pressure overload-induced cardiac remodeling. PLoS ONE 2012, 7, e48713.
- Gao, F.; Bian, F.; Ma, X.; Kalinichenko, V.V.; Das, S.K. Control of regional decidualization in implantation: Role of FoxM1 downstream of Hoxa10 and cyclin D3. Sci. Rep. 2015, 5, 13863.
- Wang, I.C.; Ustiyan, V.; Zhang, Y.; Cai, Y.; Kalin, T.V.; Kalinichenko, V.V. Foxm1 transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras(G12D.). Oncogene 2014, 33, 5391–5396.
- Ustiyan, V.; Wert, S.E.; Ikegami, M.; Wang, I.C.; Kalin, T.V.; Whitsett, J.A.; Kalinichenko, V.V. Foxm1 transcription factor is critical for proliferation and differentiation of Clara cells during development of conducting airways. Dev. Biol. 2012, 370, 198–212.
- Bolte, C.; Ustiyan, V.; Ren, X.; Dunn, A.W.; Pradhan, A.; Wang, G.; Kolesnichenko, O.A.; Deng, Z.; Zhang, Y.; Shi, D.; et al. Nanoparticle Delivery of Proangiogenic Transcription Factors into the Neonatal Circulation Inhibits Alveolar Simplification Caused by Hyperoxia. Am. J. Respir. Crit. Care Med. 2020, 202, 100–111.
- Cheng, X.H.; Black, M.; Ustiyan, V.; Le, T.; Fulford, L.; Sridharan, A.; Medvedovic, M.; Kalinichenko, V.V.; Whitsett, J.A.; Kalin, T.V. SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS Genet. 2014, 10, e1004656.
- Wang, I.C.; Snyder, J.; Zhang, Y.; Lander, J.; Nakafuku, Y.; Lin, J.; Chen, G.; Kalin, T.V.; Whitsett, J.A.; Kalinichenko, V.V. Foxm1 mediates cross talk between Kras/mitogen-activated protein kinase and canonical Wnt pathways during development of respiratory epithelium. Mol. Cell Biol. 2012, 32, 3838–3850.
- Wang, I.C.; Zhang, Y.; Snyder, J.; Sutherland, M.J.; Burhans, M.S.; Shannon, J.M.; Park, H.J.; Whitsett, J.A.; Kalinichenko, V.V. Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes Clara cell hyperplasia. Dev. Biol. 2010, 347, 301–314.
- Ribeiro, R.; Macedo, J.C.; Costa, M.; Ustiyan, V.; Shindyapina, A.V.; Tyshkovskiy, A.; Gomes, R.N.; Castro, J.P.; Kalin, T.V.; Vasques-Nóvoa, F.; et al. In vivo cyclic induction of the FOXM1 transcription factor delays natural and progeroid aging phenotypes and extends healthspan. Nat. Aging 2022, 2, 397–411.
- Black, M.; Arumugam, P.; Shukla, S.; Pradhan, A.; Ustiyan, V.; Milewski, D.; Kalinichenko, V.V.; Kalin, T.V. FOXM1 nuclear transcription factor translocates into mitochondria and inhibits oxidative phosphorylation. Mol. Biol. Cell 2020, 31, 1411–1424.
- Goda, C.; Balli, D.; Black, M.; Milewski, D.; Le, T.; Ustiyan, V.; Ren, X.; Kalinichenko, V.V.; Kalin, T.V. Loss of FOXM1 in macrophages promotes pulmonary fibrosis by activating p38 MAPK signaling pathway. PLoS Genet. 2020, 16, e1008692.
- Hasegawa, T.; Kikuta, J.; Sudo, T.; Matsuura, Y.; Matsui, T.; Simmons, S.; Ebina, K.; Hirao, M.; Okuzaki, D.; Yoshida, Y.; et al. Identification of a novel arthritis-associated osteoclast precursor macrophage regulated by FoxM1. Nat. Immunol. 2019, 20, 1631–1643.
- Kurahashi, T.; Yoshida, Y.; Ogura, S.; Egawa, M.; Furuta, K.; Hikita, H.; Kodama, T.; Sakamori, R.; Kiso, S.; Kamada, Y.; et al. Forkhead Box M1 Transcription Factor Drives Liver Inflammation Linking to Hepatocarcinogenesis in Mice. Cell Mol. Gastroenterol. Hepatol. 2020, 9, 425–446.
- Xia, H.; Ren, X.; Bolte, C.S.; Ustiyan, V.; Zhang, Y.; Shah, T.A.; Kalin, T.V.; Whitsett, J.A.; Kalinichenko, V.V. Foxm1 regulates resolution of hyperoxic lung injury in newborns. Am. J. Respir. Cell Mol. Biol. 2015, 52, 611–621.
- Balli, D.; Ren, X.; Chou, F.S.; Cross, E.; Zhang, Y.; Kalinichenko, V.V.; Kalin, T.V. Foxm1 transcription factor is required for macrophage migration during lung inflammation and tumor formation. Oncogene 2012, 31, 3875–3888.
- Ren, X.; Zhang, Y.; Snyder, J.; Cross, E.R.; Shah, T.A.; Kalin, T.V.; Kalinichenko, V.V. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol. Cell Biol. 2010, 30, 5381–5393.
- Barger, C.J.; Branick, C.; Chee, L.; Karpf, A.R. Pan-Cancer Analyses Reveal Genomic Features of FOXM1 Overexpression in Cancer. Cancers 2019, 11, 251.
- Korver, W.; Roose, J.; Heinen, K.; Weghuis, D.O.; de Bruijn, D.; van Kessel, A.G.; Clevers, H. The human TRIDENT/HFH-11/FKHL16 gene: Structure, localization, and promoter characterization. Genomics 1997, 46, 435–442.
- Jiang, M.; Li, M.; Fu, X.; Huang, Y.; Qian, H.; Sun, R.; Mao, Y.; Xie, Y.; Li, Y. Simultaneously detection of genomic and expression alterations in prostate cancer using cDNA microarray. Prostate 2008, 68, 1496–1509.
- Lensch, R.; Gotz, C.; Andres, C.; Bex, A.; Lehmann, J.; Zwergel, T.; Unteregger, G.; Kamradt, J.; Stoeckle, M.; Wullich, B. Comprehensive genotypic analysis of human prostate cancer cell lines and sublines derived from metastases after orthotopic implantation in nude mice. Int. J. Oncol. 2002, 21, 695–706.
- Spirin, K.S.; Simpson, J.F.; Takeuchi, S.; Kawamata, N.; Miller, C.W.; Koeffler, H.P. p27/Kip1 mutation found in breast cancer. Cancer Res. 1996, 56, 2400–2404.
- Singh, B.; Gogineni, S.K.; Sacks, P.G.; Shaha, A.R.; Shah, J.P.; Stoffel, A.; Rao, P.H. Molecular cytogenetic characterization of head and neck squamous cell carcinoma and refinement of 3q amplification. Cancer Res. 2001, 61, 4506–4513.
- Rodriguez, S.; Khabir, A.; Keryer, C.; Perrot, C.; Drira, M.; Ghorbel, A.; Jlidi, R.; Bernheim, A.; Valent, A.; Busson, P. Conventional and array-based comparative genomic hybridization analysis of nasopharyngeal carcinomas from the Mediterranean area. Cancer Genet. Cytogenet. 2005, 157, 140–147.
- Heselmeyer, K.; Macville, M.; Schrock, E.; Blegen, H.; Hellstrom, A.C.; Shah, K.; Auer, G.; Ried, T. Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes. Chromosomes Cancer 1997, 19, 233–240.
- Kelleher, F.C.; O’Sullivan, H. FOXM1 in sarcoma: Role in cell cycle, pluripotency genes and stem cell pathways. Oncotarget 2016, 7, 42792–42804.
- Liao, G.-B.; Li, X.-Z.; Zeng, S.; Liu, C.; Yang, S.-M.; Yang, L.; Hu, C.-J.; Bai, J.-Y. Regulation of the master regulator FOXM1 in cancer. Cell Commun. Signal 2018, 16, 57.
- Nandi, D.; Cheema, P.S.; Jaiswal, N.; Nag, A. FoxM1: Repurposing an oncogene as a biomarker. Semin. Cancer Biol. 2018, 52, 74–84.
- Kalinichenko, V.V.; Kalin, T.V. Is there potential to target FOXM1 for ‘undruggable’ lung cancers? Expert. Opin. Ther. Targets 2015, 19, 865–867.
- Halasi, M.; Hitchinson, B.; Shah, B.N.; Váraljai, R.; Khan, I.; Benevolenskaya, E.V.; Gaponenko, V.; Arbiser, J.L.; Gartel, A.L. Honokiol is a FOXM1 antagonist. Cell Death Dis. 2018, 9, 84.
- Zhang, J.R.; Lu, F.; Lu, T.; Dong, W.H.; Li, P.; Liu, N.; Ma, D.X.; Ji, C.Y. Inactivation of FoxM1 transcription factor contributes to curcumin-induced inhibition of survival, angiogenesis, and chemosensitivity in acute myeloid leukemia cells. J. Mol. Med. 2014, 92, 1319–1330.
- Wu, Y.H.; Chiu, W.T.; Young, M.J.; Chang, T.H.; Huang, Y.F.; Chou, C.Y. Solanum Incanum Extract Downregulates Aldehyde Dehydrogenase 1-Mediated Stemness and Inhibits Tumor Formation in Ovarian Cancer Cells. J. Cancer 2015, 6, 1011–1019.
- Tian, T.; Li, J.; Li, B.; Wang, Y.; Li, M.; Ma, D.; Wang, X. Genistein exhibits anti-cancer effects via down-regulating FoxM1 in H446 small-cell lung cancer cells. Tumour Biol. 2014, 35, 4137–4145.
- Dong, G.Z.; Jeong, J.H.; Lee, Y.I.; Lee, S.Y.; Zhao, H.Y.; Jeon, R.; Lee, H.J.; Ryu, J.H. Diarylheptanoids suppress proliferation of pancreatic cancer PANC-1 cells through modulating shh-Gli-FoxM1 pathway. Arch. Pharm. Res. 2017, 40, 509–517.
- Bhat, U.G.; Halasi, M.; Gartel, A.L. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS ONE 2009, 4, e5592.
- Bhat, U.G.; Halasi, M.; Gartel, A.L. FoxM1 is a general target for proteasome inhibitors. PLoS ONE 2009, 4, e6593.
- Borhani, S.; Gartel, A.L. FOXM1: A potential therapeutic target in human solid cancers. Expert. Opin. Ther. Targets 2020, 24, 205–217.
- Sun, L.; Ren, X.; Wang, I.-C.; Pradhan, A.; Zhang, Y.; Flood, H.M.; Han, B.; Whitsett, J.A.; Kalin, T.V.; Kalinichenko, V.V. The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Sci. Signal. 2017, 10, eaai8583.
- Shukla, S.; Milewski, D.; Pradhan, A.; Rama, N.; Rice, K.; Le, T.; Flick, M.J.; Vaz, S.; Zhao, X.; Setchell, K.D.; et al. The FOXM1 Inhibitor RCM-1 Decreases Carcinogenesis and Nuclear β-Catenin. Mol. Cancer Ther. 2019, 18, 1217–1229.
- Chesnokov, M.S.; Halasi, M.; Borhani, S.; Arbieva, Z.; Shah, B.N.; Oerlemans, R.; Khan, I.; Camacho, C.J.; Gartel, A.L. Novel FOXM1 inhibitor identified via gene network analysis induces autophagic FOXM1 degradation to overcome chemoresistance of human cancer cells. Cell Death Dis. 2021, 12, 704.
- Khan, I.; Kaempf, A.; Raghuwanshi, S.; Chesnokov, M.; Zhang, X.; Wang, Z.; Domling, A.; Tyner, J.W.; Camacho, C.; Gartel, A.L. Favorable outcomes of NPM1mut AML patients are due to transcriptional inactivation of FOXM1, presenting a new target to overcome chemoresistance. Blood Cancer J. 2023, 13, 128.
- Donovan, J.; Deng, Z.; Bian, F.; Shukla, S.; Gomez-Arroyo, J.; Shi, D.; Kalinichenko, V.V.; Kalin, T.V. Improving anti-tumor efficacy of low-dose Vincristine in rhabdomyosarcoma via the combination therapy with FOXM1 inhibitor RCM1. Front. Oncol. 2023, 13, 1112859.
- Gormally, M.V.; Dexheimer, T.S.; Marsico, G.; Sanders, D.A.; Lowe, C.; Matak-Vinković, D.; Michael, S.; Jadhav, A.; Rai, G.; Maloney, D.J.; et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat. Commun. 2014, 5, 5165.
- Zhang, Z.; Xue, S.-T.; Gao, Y.; Li, Y.; Zhou, Z.; Wang, J.; Li, Z.; Liu, Z. Small molecule targeting FOXM1 DNA binding domain exhibits anti-tumor activity in ovarian cancer. Cell Death Discov. 2022, 8, 280.
- Nestal de Moraes, G.; Bella, L.; Zona, S.; Burton, M.J.; Lam, E.W. Insights into a Critical Role of the FOXO3a-FOXM1 Axis in DNA Damage Response and Genotoxic Drug Resistance. Curr. Drug Targets 2016, 17, 164–177.
- Yao, S.; Fan, L.Y.; Lam, E.W. The FOXO3-FOXM1 axis: A key cancer drug target and a modulator of cancer drug resistance. Semin. Cancer Biol. 2018, 50, 77–89.
- Laoukili, J.; Kooistra, M.R.; Brás, A.; Kauw, J.; Kerkhoven, R.M.; Morrison, A.; Clevers, H.; Medema, R.H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005, 7, 126–136.
- Replogle, J.M.; Zhou, W.; Amaro, A.E.; McFarland, J.M.; Villalobos-Ortiz, M.; Ryan, J.; Letai, A.; Yilmaz, O.; Sheltzer, J.; Lippard, S.J.; et al. Aneuploidy increases resistance to chemotherapeutics by antagonizing cell division. Proc. Natl. Acad. Sci. USA 2020, 117, 30566–30576.
- Ippolito, M.R.; Martis, V.; Hong, C.; Wardenaar, R.; Zerbib, J.; Spierings, D.C.J.; Ben-David, U.; Foijer, F.; Santaguida, S. Aneuploidy-driven genome instability triggers resistance to chemotherapy. bioRxiv 2020.
- Chiu, W.T.; Huang, Y.F.; Tsai, H.Y.; Chen, C.C.; Chang, C.H.; Huang, S.C.; Hsu, K.F.; Chou, C.Y. FOXM1 confers to epithelial-mesenchymal transition, stemness and chemoresistance in epithelial ovarian carcinoma cells. Oncotarget 2015, 6, 2349–2365.
- Choi, H.S.; Kim, Y.K.; Hwang, K.G.; Yun, P.Y. Increased FOXM1 Expression by Cisplatin Inhibits Paclitaxel-Related Apoptosis in Cisplatin-Resistant Human Oral Squamous Cell Carcinoma (OSCC) Cell Lines. Int. J. Mol. Sci. 2020, 21, 8897.
- Zhou, J.; Wang, Y.; Wang, Y.; Yin, X.; He, Y.; Chen, L.; Wang, W.; Liu, T.; Di, W. FOXM1 modulates cisplatin sensitivity by regulating EXO1 in ovarian cancer. PLoS ONE 2014, 9, e96989.
- Tang, J.H.; Yang, L.; Chen, J.X.; Li, Q.R.; Zhu, L.R.; Xu, Q.F.; Huang, G.H.; Zhang, Z.X.; Xiang, Y.; Du, L.; et al. Bortezomib inhibits growth and sensitizes glioma to temozolomide (TMZ) via down-regulating the FOXM1-Survivin axis. Cancer Commun. 2019, 39, 81.
- Zhang, N.; Wu, X.; Yang, L.; Xiao, F.; Zhang, H.; Zhou, A.; Huang, Z.; Huang, S. FoxM1 inhibition sensitizes resistant glioblastoma cells to temozolomide by downregulating the expression of DNA-repair gene Rad51. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 5961–5971.
- Peng, W.X.; Han, X.; Zhang, C.L.; Ge, L.; Du, F.Y.; Jin, J.; Gong, A.H. FoxM1-mediated RFC5 expression promotes temozolomide resistance. Cell Biol. Toxicol. 2017, 33, 527–537.
- Yang, F.; Kemp, C.J.; Henikoff, S. Anthracyclines induce double-strand DNA breaks at active gene promoters. Mutat. Res. 2015, 773, 9–15.
- Khongkow, M.; Olmos, Y.; Gong, C.; Gomes, A.R.; Monteiro, L.J.; Yagüe, E.; Cavaco, T.B.; Khongkow, P.; Man, E.P.; Laohasinnarong, S.; et al. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis 2013, 34, 1476–1486.
- Millour, J.; de Olano, N.; Horimoto, Y.; Monteiro, L.J.; Langer, J.K.; Aligue, R.; Hajji, N.; Lam, E.W. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol. Cancer Ther. 2011, 10, 1046–1058.
- Khongkow, P.; Karunarathna, U.; Khongkow, M.; Gong, C.; Gomes, A.R.; Yagüe, E.; Monteiro, L.J.; Kongsema, M.; Zona, S.; Man, E.P.; et al. FOXM1 targets NBS1 to regulate DNA damage-induced senescence and epirubicin resistance. Oncogene 2014, 33, 4144–4155.
- Nestal de Moraes, G.; Delbue, D.; Silva, K.L.; Robaina, M.C.; Khongkow, P.; Gomes, A.R.; Zona, S.; Crocamo, S.; Mencalha, A.L.; Magalhães, L.M.; et al. FOXM1 targets XIAP and Survivin to modulate breast cancer survival and chemoresistance. Cell Signal 2015, 27, 2496–2505.
- Ghandhariyoun, N.; Jaafari, M.R.; Nikoofal-Sahlabadi, S.; Taghdisi, S.M.; Moosavian, S.A. Reducing Doxorubicin resistance in breast cancer by liposomal FOXM1 aptamer: In vitro and in vivo. Life Sci. 2020, 262, 118520.
- Wang, J.Y.; Jia, X.H.; Xing, H.Y.; Li, Y.J.; Fan, W.W.; Li, N.; Xie, S.Y. Inhibition of Forkhead box protein M1 by thiostrepton increases chemosensitivity to doxorubicin in T-cell acute lymphoblastic leukemia. Mol. Med. Rep. 2015, 12, 1457–1464.
- Huang, C.; Zhang, X.; Jiang, L.; Zhang, L.; Xiang, M.; Ren, H. FoxM1 Induced Paclitaxel Resistance via Activation of the FoxM1/PHB1/RAF-MEK-ERK Pathway and Enhancement of the ABCA2 Transporter. Mol. Ther. Oncolytics 2019, 14, 196–212.
- Westhoff, G.L.; Chen, Y.; Teng, N.N.H. Targeting FOXM1 Improves Cytotoxicity of Paclitaxel and Cisplatinum in Platinum-Resistant Ovarian Cancer. Int. J. Gynecol. Cancer 2017, 27, 1602–1609.
- Varghese, V.; Magnani, L.; Harada-Shoji, N.; Mauri, F.; Szydlo, R.M.; Yao, S.; Lam, E.W.; Kenny, L.M. FOXM1 modulates 5-FU resistance in colorectal cancer through regulating TYMS expression. Sci. Rep. 2019, 9, 1505.
- Klinhom-On, N.; Seubwai, W.; Sawanyawisuth, K.; Obchoei, S.; Mahalapbutr, P.; Wongkham, S. FOXM1 inhibitor, Siomycin A, synergizes and restores 5-FU cytotoxicity in human cholangiocarcinoma cell lines via targeting thymidylate synthase. Life Sci. 2021, 286, 120072.
- Xie, T.; Geng, J.; Wang, Y.; Wang, L.; Huang, M.; Chen, J.; Zhang, K.; Xue, L.; Liu, X.; Mao, X.; et al. FOXM1 evokes 5-fluorouracil resistance in colorectal cancer depending on ABCC10. Oncotarget 2017, 8, 8574–8589.
- Khan, I.; Halasi, M.; Patel, A.; Schultz, R.; Kalakota, N.; Chen, Y.H.; Aardsma, N.; Liu, L.; Crispino, J.D.; Mahmud, N.; et al. FOXM1 contributes to treatment failure in acute myeloid leukemia. JCI Insight 2018, 3, e121583.




