Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Vijay kumar Srinivasalu | -- | 2878 | 2024-02-26 04:39:05 | | | |
| 2 | Lindsay Dong | + 1 word(s) | 2879 | 2024-02-26 09:36:39 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Khosla, D.; Misra, S.; Chu, P.L.; Guan, P.; Nada, R.; Gupta, R.; Kaewnarin, K.; Ko, T.K.; Heng, H.L.; Srinivasalu, V.K.; et al. Molecular Pathobiology of Cholangiocarcinoma. Encyclopedia. Available online: https://encyclopedia.pub/entry/55422 (accessed on 07 February 2026).
Khosla D, Misra S, Chu PL, Guan P, Nada R, Gupta R, et al. Molecular Pathobiology of Cholangiocarcinoma. Encyclopedia. Available at: https://encyclopedia.pub/entry/55422. Accessed February 07, 2026.
Khosla, Divya, Shagun Misra, Pek Lim Chu, Peiyong Guan, Ritambhra Nada, Rajesh Gupta, Khwanta Kaewnarin, Tun Kiat Ko, Hong Lee Heng, Vijay Kumar Srinivasalu, et al. "Molecular Pathobiology of Cholangiocarcinoma" Encyclopedia, https://encyclopedia.pub/entry/55422 (accessed February 07, 2026).
Khosla, D., Misra, S., Chu, P.L., Guan, P., Nada, R., Gupta, R., Kaewnarin, K., Ko, T.K., Heng, H.L., Srinivasalu, V.K., Kapoor, R., Singh, D., Klanrit, P., Sampattavanich, S., Tan, J., Kongpetch, S., Jusakul, A., Teh, B.T., Chan, J.Y., ...Hong, J.H.. (2024, February 26). Molecular Pathobiology of Cholangiocarcinoma. In Encyclopedia. https://encyclopedia.pub/entry/55422
Khosla, Divya, et al. "Molecular Pathobiology of Cholangiocarcinoma." Encyclopedia. Web. 26 February, 2024.
Copy Citation
Cholangiocarcinoma (CCA) manifests as a complex interplay of genetic and environmental factors, necessitating personalized approaches. CCA are driven by an intricate landscape of genetic mutations, epigenetic dysregulation, and post-transcriptional modification, which differs based on geography (e.g., for liver fluke versus non-liver fluke-driven CCA) and exposure to environmental carcinogens (e.g., exposure to aristolochic acid). Liquid biopsy, including circulating cell-free DNA, is a potential diagnostic tool for CCA, which warrants further investigations.
cholangiocarcinoma
pathobiology
liquid biopsy
1. Introduction
The molecular pathogenesis of cholangiocarcinoma (CCA) is complex and multifactorial. A multitude of molecular mechanisms have been implicated in the process of cholangio-carcinogenesis and may differ by anatomical location and etiology. The tumor cells undergo diverse genetic and epigenetic alterations resulting in enhanced proliferation signaling, dysregulation of apoptosis, angiogenesis, invasion, and stromal proliferation [1]. Prolonged biliary inflammation, cholestasis, and fibrosis incite aberrant activation of various receptors and deregulate intracellular signaling pathways. There is a need for a comprehensive understanding of the molecular mechanisms to stratify the patients using validated biomarkers for personalized treatment strategies.
The complex tumor microenvironment comprising a diverse array of tumor cells, highly invasive behavior, desmoplastic and hypovascular stroma, mutational landscape, development of therapeutic resistance, and intra and inter-tumoral heterogeneity has led to refractoriness to chemotherapy. Nearly half of the CCAs have targetable mutations [2][3][4]. Advances in pathology, molecular biology, and genetics have led to an improved understanding of the molecular mechanisms underlying CCA development. This enables the identification of specific molecular targets and prediction of treatment responses for more efficacious interventions. This knowledge is crucial for optimizing drug development and enhancing our overall comprehension of the biological processes underlying CCA’s development and progression.
2. Molecular Pathology
2.1. Pathological Features
CCA arises from biliary ducts. Since biliary ducts span from the canal of Hering to the common bile duct opening at the ampulla of Vater, the first step is to identify their anatomic location and growth patterns, followed by microscopic assessment for differentiation and subtype. This should be further supported by immunohistochemistry followed by background pathology to ascertain etiology and finally molecular subtyping.
2.2. Cholangiocarcinoma Nomenclature according to Location in Biliary Tract Anatomy
Cholangiocarcinomas have three distinct anatomical categories depending on the site of the biliary tract from where it arises. The biliary tree has intrahepatic and extrahepatic components; hence, these tumors are also classified as intrahepatic and extrahepatic CCA. An extrahepatic biliary tree has two parts, termed perihilar (pCCA) and distal cholangiocarcinoma (dCCA). dCCA includes tumors between the origin of the cystic duct from the CBD and ampulla of Vater. Perihilar CCA includes segmental ducts (second-order bile ducts) and right and left hepatic ducts, their confluence upto the insertion of the cystic duct to form a common bile duct. Intrahepatic CCA includes tumors arising from the canal of Herring, bile ductules (20 μm), interlobular bile ducts (20–100 μm), septal (>100 μm and <300 μm) first-order branches. It is important to subcategorize, as each of these categories differs in their risk factors, epidemiological features, clinical presentations, and morphologic and molecular characteristics [5][6].
2.3. Growth Pattern
After deciding the location, the growth pattern of the tumor needs to be identified. The growth patterns of intrahepatic CCA can be mass forming lesions (60–80%), periductal infiltrating (15–35%), or intraductal growth types (8–29%) (Figure 1).
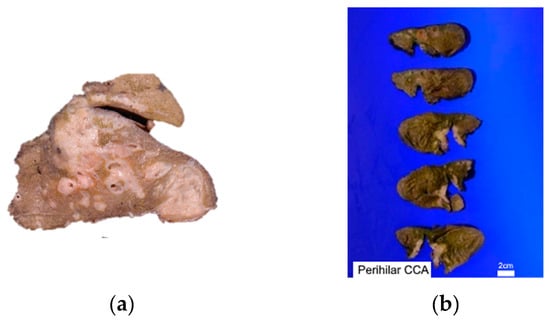
Figure 1. Gross photograph of (a) Intrahepatic CCA’s both mass-forming lesions and periductal infiltrative, (b) perihilar CCA-affected both right and left hepatic ducts.
Mass-forming lesions represent solid, non-encapsulated tumors found within the hepatic parenchyma. They exhibit a cut surface that is either white or greyish, often displaying central necrosis or scarring and well-defined borders. Evidence of intrahepatic metastases or the fusion of smaller lesions may be apparent, and the cut surface may manifest mucinous characteristics. These tumors are believed to originate from the small bile ducts within the liver. The periductal infiltrating type of intrahepatic CCA extends along the portal tracts, resulting in bile duct strictures with luminal narrowing. The intraductal growth variant of intrahepatic CCA presents as a papillary or polypoidal lesion within a dilated bile duct. Macroscopically, pCCA and dCCA share similar growth patterns. In about 80% of cases, they present as flat or ambiguously defined nodular sclerosing tumors, marked by thickening of the duct wall and widespread infiltration into surrounding structures. Moreover, they may appear as intraductal papillary tumors, signifying the malignant progression of intraductal papillary mucinous neoplasm (IPNB) [7].
2.4. Large and Small Duct Variants of Intrahepatic CCA
Intrahepatic CCA can be classified into a minimum of two primary categories based on their differentiation (see Figure 2): well/moderately-differentiated tubular/acinar adenocarcinoma, poorly-differentiated tubular/acinar adenocarcinoma, or less common morphological variations [8]. Another method of categorization involves differentiating between large and small duct types, which hold clinical, pathological, immunohistochemical, and molecular importance [7][9].
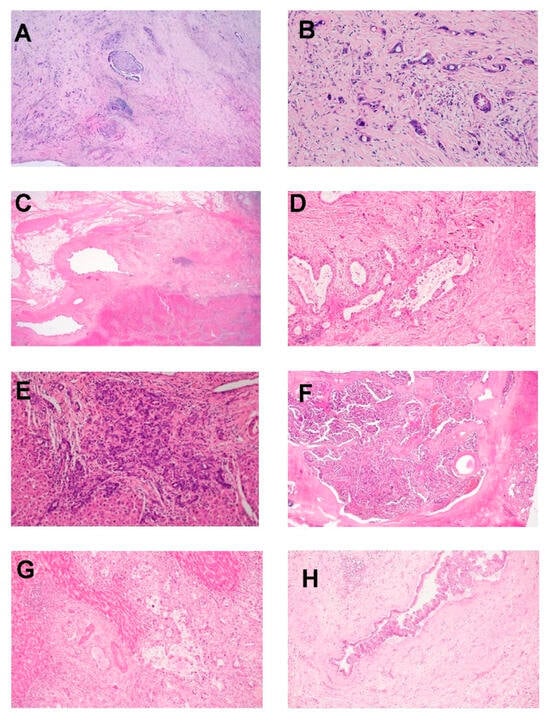
Figure 2. Photomicrographs show (A) iCCA-LD composed of glands with abundant desmoplastic stroma, perineural invasion, and portal vein thrombosis with tumor infiltration. (B) iCCA-LD showing malignant glands lined by cuboidal dysplastic cells having intracytoplasmic mucin; (C) perihilar CCA with tumor arising from the segmental intrahepatic duct, extrahepatic bile duct is normal; (D) tumor was composed of large glands lined by cuboidal–columnar cells with abundant intraluminal mucin; (E) iCCA-SD composed of small cuboidal cells without any mucin with hyperchromatic nuclei forming anastomosing cords; (F) perihilar CCA with an intraductal papillary configuration; (G) iCCA infiltration into adjacent hepatocytes; (H) bile duct showing evidence of biliary epithelial neoplasia-low grade with a papillary pattern that is a precursor lesion for extrahepatic CCA (H&E-A-H, 4×-A,F, ×20-B,D, ×1.25-C, ×10 F,G,H original magnification).
The large duct variant of intrahepatic CCA (LD-iCCA) originates from the intrahepatic bile ducts or their associated peribiliary glands. These tumors exhibit similarities to the biliary epithelium, featuring cuboidal to tall columnar cells with cytoplasmic mucin, forming extensive acini with open luminal spaces. Furthermore, they display a significant presence of desmoplastic stroma [10].
The small bile duct type of intrahepatic CCA (SD-iCCA) resembles cholangiolar cells and arises from progenitor cells or mature hepatocytes by trans-differentiation. These tumors are composed of small monotonous or anastomosing glands lined with cuboidal cells. Tumor cells have uniform nuclei with vesicular nuclei, scant to moderate eosinophilic or amphophilic cytoplasm, and no mucin production. There are no recognizable precursor lesions for SD-iCCA, whereas biliary intra-epithelial neoplasia (BIN-low and high grade) and IDPN (intraductal papillary neoplasm) are precursor lesions of LD-iCCA. Predisposing risk factors are also different for both these sub-categories; SD-iCCA are associated with chronic liver diseases/cirrhosis (especially viral hepatitis) and non-biliary cirrhosis (hemochromatosis, alcoholic liver disease, metabolic syndrome, obesity, and diabetes mellitis). LD-iCCA is associated with chronic biliary disease, precursor lesions, and hepatolithiasis. Caroli’s disease, congenital fibrosis, and bile duct cysts are predisposing factors for LD-iCCA with corresponding morphologies. SD-iCCA nearly always has a mass-forming macroscopic growth pattern. LD-iCCA has variable macroscopic growth patterns with mucin production, poorer differentiation, perineural/lymphatic invasion, and lymph node metastases [7].
Both large and small duct iCCA are stained positive with EMA (MUC1), hepatocyte nuclear factor-1β (HNF-1β), CK7, and CK 19. CK20 immunostain is typically negative or focally positive. Several studies have documented distinct immunohistochemical characteristics of LD-iCCA and SD-iCCA [11][12]. SD-iCCA are positive with NCAM (CD56), C-reactive protein, N-cadherin, and IDH1/2. LD-iCCA are positive with MUC-5AC, MUC 6, S-100, TFF1, MMP, and KRAS.
It is imperative to distinguish intrahepatic CCA from metastases originating from colorectal carcinoma, upper gastrointestinal tract malignancy, or tumors with a pancreatobiliary origin. Generally, intrahepatic CCA displays a lack of reactivity for CDX2 and SAT-B2, although there are instances where mild focal positivity for these markers may be observed. The immunostains CDX2 and SAT-B2 are instrumental in negating a diagnosis of metastatic colorectal adenocarcinoma, given its typical strong positivity for CK20, CDX2, and SAT-B2 but negativity for CK7 and CK19.
The challenge lies in differentiating between CCA and metastatic pancreatic ductal adenocarcinoma, as well as upper gastrointestinal tract carcinomas, through immunostains, as both types of tumors typically exhibit positivity for CK7 and CK19. Fernández Moro et al. [13] proposed a comprehensive immunohistochemical panel including CK19, CK20, MUC2, MUC5AC, CA19–9, mCEA, CA125, and SMAD4 to aid in the differentiation of metastatic and pancreatobiliary adenocarcinomas.
2.5. Molecular Genetics
CCA encompasses a highly heterogeneous genomic mutational landscape that is associated with poor outcomes in patients [3][4][14][15][16]. In a previous study on 489 CCAs spanning 10 countries, comprehensive integrative clustering revealed 4 clusters that are defined by distinct etiologies with separate genetic, epigenetic, and clinical features [14]. In brief, clusters 1 and 2 were mostly fluke-positive CCA patients with enriched TP53 mutation, ERBB2 amplification, and elevated ERBB2 expression. A wide array of genetic alterations exists, including mutations in certain tumor suppressors such as ARID1A, SMAD4, and PTEN, implying a diverse selective pressure that drives the pathogenesis of CCA. On the other hand, cluster 3 and 4, which were primarily comprised of fluke-negative CCA patients, were enriched in BAP1 and IDH1/2 mutations and FGFR alterations. Other than geography, the mutational landscape of CCA also differs by anatomical location. Despite the fact that oncogenic mutations in CCA, such as TP53, KRAS, IDH1, ARID1A, and CDKN2A/B are commonly found in many other cancer types, it is apparent that the alteration frequencies differ substantially between intrahepatic CCA and extrahepatic CCA (eCCA). Furthermore, FGFR fusions were detected only exclusively in intrahepatic CCA, and the fusions were found to be mutually exclusive with FGFR/BRAF/ERBB2/KRAS mutations [17]. CCA pathogenesis could occur due to post-transcriptional modification (PTM). In a recent integrative analysis of 348 fluke-negative CCA samples (including 87 from pCCA and 261 from intrahepatic CCA), pathway analysis of driver mutations revealed enrichment in RTK-RAS, Wnt, PI3K, cell cycle, TP53, TGF-beta, and HIPPO pathways. Further analysis indicated recurring mutations in genes participating in PTM (METTL14 and RBM10) in pCCA [18]. Functional studies demonstrated that METTL14R298H mutation-mediated m6A modification disrupted the repression of the MACF1/β-catenin pathway, thus, indicating the involvement of PTM in driving the occurrence and pathogenesis of CCA.
Next, epigenetic dysregulation also orchestrates CCA pathogenesis. Driver mutations in genes related to chromatin modification and DNA methylation, such as SMARCA4, PBRM1, BAP1, ARID1A, WHSC1, DNMT3A, and EZH2 were frequently identified [14][18], implying that the mutation in these epigenetic modifiers could be the primary event preceding epigenetic dysregulation, thus regulating the transcriptome driving the CCA. Distinct patterns of DNA hypermethylation targeting either promoter CpG islands or promoter CpG shores were also observed in fluke-positive CCA cases and fluke-negative CCA cases, respectively [14], which highlights the prognostic value of DNA methylation in defining the molecular subtypes of CCA. More recently, genome-wide changes in DNA methylation and enhancer activities have proven valuable in deciphering different molecular subtypes to guide the therapeutic interventions in many other cancers [19][20][21]. In an epigenetic study by Tang and co-workers [22] on intrahepatic CCA, the collagen type XII alpha 1 chain (COL12A1) was identified as a specific biomarker for enrichment in the epithelial–mesenchymal transition pathway and advanced tumor stage. Aberrant expression of COL12A1 was attributed to promoter hypermethylation-induced downregulation of miR-424-5p. The in vivo tumor growth and COL12A1 expression were alleviated by the treatment with the miR-424-5p agonist. This highlighted the potential of epigenetic profiling studies in exploring promising druggable targets for epigenetic therapy of CCA. Taken together, crosstalk between genetic, epigenetic, and PTM is evident in driving the pathogenesis of CCA.
2.6. Diagnosis and Evaluation
The clinical manifestations of CCA depend on the location of the tumor within the biliary tract. Extrahepatic tumors present earlier with features of biliary obstruction (jaundice, dark-colored urine, clay-colored stools, and pruritus) in contrast to intrahepatic CCA, where biliary obstruction is less likely. Patients with intrahepatic CCA present late with non-specific symptoms of dull aching abdominal pain, weight loss, or abdominal mass due to a mass effect or invasion into the hepatic parenchyma. Patients presenting with jaundice or upper abdominal pain should undergo liver function tests, which include bilirubin levels (total/conjugated/unconjugated), serum aminotransferases, and alkaline phosphatase (ALP). Patients with extrahepatic CCA will have elevated conjugated bilirubin and alkaline phosphatase. Transaminase levels may also increase in the later course of the disease due to chronic biliary obstruction. Intrahepatic CCAs usually have normal levels of bilirubin but abnormal levels of ALP. The tumor markers include carbohydrate antigen (CA) 19-9 and carcinoembryonic antigen (CEA). However, the sensitivity of these markers is limited for detecting early-stage CCA as these could be elevated in other benign and malignant conditions as well. Alpha-fetoprotein (AFP) can be helpful in differentiating intrahepatic CCA from hepatocellular carcinoma (HCC) as it has high specificity for diagnosing HCC.
Cross-sectional imaging is important to identify the location, extent of involvement, and also to assess the feasibility of surgery. Most patients will undergo transabdominal ultrasonography as obstructive jaundice is a common presentation. Ultrasonography (USG) will help in confirming biliary duct dilation, identifying the site of obstruction and also ruling out gallstones. Multidetector computed tomography (MDCT) is commonly used for diagnosis and staging purposes due to wider availability. Magnetic resonance imaging (MRI) with magnetic resonance cholangiopancreatography (MRCP) provides a non-invasive assessment of the hepatopancreaticobiliary tract. CCA appears as T1 hypointense and T2 hyperintense on an MRI, with proximal ductal dilatation with intense delayed contrast enhancement. MRCP allows for the three-dimensional visualization of bile ducts and vascular structures. It should be performed prior to biliary drainage in order to accurately pick up the pathology. PET or PET/CT does not provide any additional information with respect to the staging of tumors but leads to a change in surgical management due to upstaging in 20–25% of patients [23]. Ductal dilatation of greater than 6 mm with an intact gallbladder in the absence of gallstones points toward a biliary obstructive lesion. Dilatation of the intrahepatic ducts is seen with proximal extrahepatic CCAs, and dilatation of both intrahepatic and extrahepatic ducts may be seen with distal CCAs. An abrupt ductal diameter change suggests tumors at the site of the narrowing.
Tissue diagnosis can pose challenges, particularly in perihilar lesions. Tissue samples can be obtained by brush cytology, fine needle aspiration, or percutaneous approach. Endoscopic ultrasound (EUS) or endoscopic retrograde cholangiopancreatography (ERCP) is preferred in patients with distal extrahepatic obstruction as it enables the localization and assessment of the extent of the tumor and facilitates brush cytology or biopsy. ERCP has the added advantage of therapeutic intervention, such as stent placement.
2.7. Liquid Biopsy in Diagnostics
Liquid biopsies are blood tests for identifying circulating tumor cells, cell-free nucleic acids, and secreted proteins that are found in body fluids such as blood, urine, and bile. Unlike tissue biopsy, liquid biopsy is generally less intrusive, less costly, and safer. There is an increased interest in novel biomarkers from liquid biopsies to guide the diagnosis and treatment of CCA. There have been recent developments in assessing cell-free DNA (cfDNA) as novel CCA biomarkers.
cfDNA is composed of mainly short (50–250 bp) double-stranded DNA fragments that are found in low abundance in body fluids. In healthy individuals, most cfDNA is derived from circulating leukocytes [24]. In cancer patients, a proportion of cfDNA is derived from cancer cells, termed circulating tumor DNA (ctDNA). Depending on the tumor burden, the proportion of cfDNA from a cancer patient’s liquid biopsy can range from less than 1% to as high as 90% [24]. Both real-time quantitative PCR (qPCR) and next-generation sequencing (NGS) platforms can be used to detect mutations in CCA-associated genes (e.g., ARID1A, PBRM1, MTOR, FGFR2, and TP53) in plasma cfDNA from CCA patients [25]. However, the concordance between mutations detected in tumors and those found in cfDNA is highly variable.
cfDNAs from serial liquid biopsies are useful to monitor changes to the tumor mutational profile before, during, and after the treatment period, which can guide designing treatment options. Ettrich et al. [26] analyzed paired plasma cfDNA and tumor samples from CCA patients before and after treatments. They showed that there was a significant concordance between mutations found in tumors and those in the corresponding plasma cfDNA. Furthermore, this study showed that about 63% of the treatment-naive patients had a shift in their mutational profile during treatment, which was also detected in the corresponding ctDNA.
Besides plasma, several studies have analyzed bile-derived cfDNA. Driescher et al. [27] showed that bile cfDNA was more sensitive than plasma cfDNA in detecting mutations reported in the matched tumors (96.2% vs. 31.6%). Shen et al. [28] and Arechederra et al. [29] reported significant concordance between mutations detected in tumors and those found in the corresponding bile cfDNA. Apart from detecting CCA-specific mutations in cfDNA, there are currently a few other studies on detecting CCA-specific DNA methylation in cfDNA. Wasenang et al. [30] used a qPCR-based methylation-sensitive high-resolution melting (MS-HRM) approach to assess for the presence of CCA-specific methylated sites on OPCML, HOXA9, and HOXD9 genes in serum-derived cfDNA. Additionally, the Circulating Cell-free Genome Atlas (CCGA) consortium published two studies that used NGS-based targeted methylation panels to study bisulfite-converted cfDNA in a variety of cancers, including CCA [31][32]. They showed that cancer type-specific methylomes were detected in cfDNA albeit with different sensitivity and accuracy for each cancer type. Apart from cfDNA, cell-free RNA, especially microRNA (miRNA) found in exosomes, can be novel CCA biomarker candidates. Recent studies have shown that body fluids, such as plasma and bile, contain a significant amount of miRNAs and some of them could be used to differentiate between CCA patients and healthy individuals [33][34].
In summary, cfDNA and ctDNA from liquid biopsy are attractive diagnostic and monitoring tools for cancer therapy. Despite many positive developments in the use of liquid biopsy for CCA, more research is needed to improve the sensitivity and specificity before its routine clinical usage.
References
- Fava, G. Molecular mechanisms of cholangiocarcinoma. World J. Gastrointest. Pathophysiol. 2010, 1, 12–22.
- Ong, C.K.; Subimerb, C.; Pairojkul, C.; Wongkham, S.; Cutcutache, I.; Yu, W.; McPherson, J.R.; E Allen, G.; Ng, C.C.Y.; Wong, B.H.; et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat. Genet. 2012, 44, 690–693.
- Montal, R.; Sia, D.; Montironi, C.; Leow, W.Q.; Esteban-Fabró, R.; Pinyol, R.; Torres-Martin, M.; Bassaganyas, L.; Moeini, A.; Peix, J.; et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J. Hepatol. 2020, 73, 315–327.
- Deng, M.; Ran, P.; Chen, L.; Wang, Y.; Yu, Z.; Cai, K.; Feng, J.; Qin, Z.; Yin, Y.; Tan, S.; et al. Proteogenomic characterization of cholangiocarcinoma. Hepatology 2023, 77, 411–429.
- Marcano-Bonilla, L.; Mohamed, E.A.; Mounajjed, T.; Roberts, L.R. Biliary tract cancers: Epidemiology, molecular pathogenesis and genetic risk associations. Chin. Clin. Oncol. 2016, 5, 61.
- Sempoux, C.; Jibara, G.; Ward, S.C.; Fan, C.; Qin, L.; Roayaie, S.; Fiel, M.I.; Schwartz, M.; Thung, S.N. Intrahepatic cholangiocarcinoma: New insights in pathology. Semin. Liver Dis. 2011, 31, 49–60.
- Nakanuma, Y.; Kakuda, Y. Pathologic classification of cholangiocarcinoma: New concepts. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 277–293.
- Nakanuma, Y.; Sato, Y.; Harada, K.; Sasaki, M.; Xu, J.; Ikeda, H. Pathological classification of intrahepatic cholangiocarcinoma based on a new concept. World J. Hepatol. 2010, 2, 419–427.
- Lendvai, G.; Szekerczés, T.; Illyés, I.; Dóra, R.; Kontsek, E.; Gógl, A.; Kiss, A.; Werling, K.; Kovalszky, I.; Schaff, Z.; et al. Cholangiocarcinoma: Classification, Histopathology and Molecular Carcinogenesis. Pathol. Oncol. Res. 2020, 26, 3–15.
- Sigel, C.S.; Drill, E.; Zhou, Y.; Basturk, O.; Askan, G.; Pak, L.M.; Vakiani, E.; Wang, T.; Boerner, T.; Do, R.K.; et al. Intrahepatic Cholangiocarcinomas Have Histologically and Immunophenotypically Distinct Small and Large Duct Patterns. Am. J. Surg. Pathol. 2018, 42, 1334–1345.
- Patil, P.A.; Taddei, T.; Jain, D.; Zhang, X. HNF-1β is a More Sensitive and Specific Marker Than C-Reactive Protein for Identifying Biliary Differentiation in Primary Hepatic Carcinomas. Arch. Pathol. Lab. Med. 2022, 146, 220–226.
- Hayashi, A.; Misumi, K.; Shibahara, J.; Arita, J.; Sakamoto, Y.; Hasegawa, K.; Kokudo, N.; Fukayama, M. Distinct Clinicopathologic and Genetic Features of 2 Histologic Subtypes of Intrahepatic Cholangiocarcinoma. Am. J. Surg. Pathol. 2016, 40, 1021–1030.
- Fernández Moro, C.; Fernandez-Woodbridge, A.; Alistair D’souza, M.; Zhang, Q.; Bozoky, B.; Vasan, S.K.; Catalano, P.; Heuchel, R.; Shtembari, S.; Del Chiaro, M.; et al. Correction: Immunohistochemical Typing of Adenocarcinomas of the Pancreatobiliary System Improves Diagnosis and Prognostic Stratification. PLoS ONE 2017, 12, e0171283.
- Jusakul, A.; Cutcutache, I.; Yong, C.H.; Lim, J.Q.; Ni Huang, M.; Padmanabhan, N.; Nellore, V.; Kongpetch, S.; Ng, A.W.T.; Ng, L.M.; et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017, 7, 1116–1135.
- Normanno, N.; Martinelli, E.; Melisi, D.; Pinto, C.; Rimassa, L.; Santini, D.; Scarpa, A. Role of molecular genetics in the clinical management of cholangiocarcinoma. ESMO Open 2022, 7, 100505.
- Chen, G.; Cai, Z.; Dong, X.; Zhao, J.; Lin, S.; Hu, X.; Liu, F.-E.; Liu, X.; Zhang, H. Genomic and Transcriptomic Landscape of Tumor Clonal Evolution in Cholangiocarcinoma. Front. Genet. 2020, 11, 195.
- Kongpetch, S.; Jusakul, A.; Lim, J.Q.; Ng, C.C.Y.; Chan, J.Y.; Rajasegaran, V.; Lim, T.H.; Lim, K.H.; Choo, S.P.; Dima, S.; et al. Lack of Targetable FGFR2 Fusions in Endemic Fluke-Associated Cholangiocarcinoma. JCO Glob. Oncol. 2020, 6, 628–638.
- Zhang, Y.; Ma, Z.; Li, C.; Wang, C.; Jiang, W.; Chang, J.; Han, S.; Lu, Z.; Shao, Z.; Wang, Y.; et al. The genomic landscape of cholangiocarcinoma reveals the disruption of post-transcriptional modifiers. Nat. Commun. 2022, 13, 3061.
- Li, J.; Liang, Y.; Fan, J.; Xu, C.; Guan, B.; Zhang, J.; Guo, B.; Shi, Y.; Wang, P.; Tan, Y.; et al. DNA methylation subtypes guiding prognostic assessment and linking to responses the DNA methyltransferase inhibitor SGI-110 in urothelial carcinoma. BMC Med. 2022, 20, 222.
- Southekal, S.; Shakyawar, S.K.; Bajpai, P.; Elkholy, A.; Manne, U.; Mishra, N.K.; Guda, C. Molecular Subtyping and Survival Analysis of Osteosarcoma Reveals Prognostic Biomarkers and Key Canonical Pathways. Cancers 2023, 15, 2134.
- Hong, J.H.; Yong, C.H.; Heng, H.L.; Chan, J.Y.; Lau, M.C.; Chen, J.; Lee, J.Y.; Lim, A.H.; Li, Z.; Guan, P.; et al. Integrative multiomics enhancer activity profiling identifies therapeutic vulnerabilities in cholangiocarcinoma of different etiologies. Gut 2023, gutjnl-2023-330483.
- Tang, Z.; Yang, Y.; Zhang, Q.; Liang, T. Epigenetic dysregulation-mediated COL12A1 upregulation predicts worse outcome in intrahepatic cholangiocarcinoma patients. Clin. Epigenetics. 2023, 15, 13.
- Corvera, C.U.; Blumgart, L.H.; Akhurst, T.; DeMatteo, R.P.; D’angelica, M.; Fong, Y.; Jarnagin, W.R. 18F-fluorodeoxyglucose positron emission tomography influences management decisions in patients with biliary cancer. J. Am. Coll. Surg. 2008, 206, 57–65.
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765.
- Wintachai, P.; Lim, J.Q.; Techasen, A.; Lert-Itthiporn, W.; Kongpetch, S.; Loilome, W.; Chindaprasirt, J.; Titapun, A.; Namwat, N.; Khuntikeo, N.; et al. Diagnostic and Prognostic Value of Circulating Cell-Free DNA for Cholangiocarcinoma. Diagnostics 2021, 11, 999.
- Ettrich, T.J.; Schwerdel, D.; Dolnik, A.; Beuter, F.; Blätte, T.J.; Schmidt, S.A.; Stanescu-Siegmund, N.; Steinacker, J.; Marienfeld, R.; Kleger, A.; et al. Genotyping of circulating tumor DNA in cholangiocarcinoma reveals diagnostic and prognostic information. Sci. Rep. 2019, 9, 13261.
- Driescher, C.; Fuchs, K.; Haeberle, L.; Goering, W.; Frohn, L.; Opitz, F.V.; Haeussinger, D.; Knoefel, W.T.; Keitel, V.; Esposito, I. Bile-Based Cell-Free DNA Analysis Is a Reliable Diagnostic Tool in Pancreatobiliary Cancer. Cancers 2020, 13, 39.
- Shen, N.; Zhang, D.; Yin, L.; Qiu, Y.; Liu, J.; Yu, W.; Fu, X.; Zhu, B.; Xu, X.; Duan, A.; et al. Bile cell-free DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol. Rep. 2019, 42, 549–560.
- Arechederra, M.; Rullán, M.; Amat, I.; Oyon, D.; Zabalza, L.; Elizalde, M.; Latasa, M.U.; Mercado, M.R.; Ruiz-Clavijo, D.; Saldaña, C.; et al. Next-generation sequencing of bile cell-free DNA for the early detection of patients with malignant biliary strictures. Gut 2022, 71, 1141–1151.
- Wasenang, W.; Chaiyarit, P.; Proungvitaya, S.; Limpaiboon, T. Serum cell-free DNA methylation of OPCML and HOXD9 as a biomarker that may aid in differential diagnosis between cholangiocarcinoma and other biliary diseases. Clin. Epigenetics 2019, 11, 39.
- Liu, L.; Toung, J.; Jassowicz, A.; Vijayaraghavan, R.; Kang, H.; Zhang, R.; Kruglyak, K.; Huang, H.; Hinoue, T.; Shen, H.; et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann. Oncol. 2018, 29, 1445–1453.
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759.
- Li, L.; Masica, D.; Ishida, M.; Tomuleasa, C.; Umegaki, S.; Kalloo, A.N.; Georgiades, C.; Singh, V.K.; Khashab, M.; Amateau, S.; et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology 2014, 60, 896–907.
- Correa-Gallego, C.; Maddalo, D.; Doussot, A.; Kemeny, N.; Kingham, T.P.; Allen, P.J.; D’Angelica, M.I.; DeMatteo, R.P.; Betel, D.; Klimstra, D.; et al. Circulating Plasma Levels of MicroRNA-21 and MicroRNA-221 Are Potential Diagnostic Markers for Primary Intrahepatic Cholangiocarcinoma. PLoS ONE 2016, 11, e0163699.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
438
Revisions:
2 times
(View History)
Update Date:
26 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




