Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Fang Weibo | -- | 2230 | 2024-02-21 12:03:14 | | | |
| 2 | Lindsay Dong | Meta information modification | 2230 | 2024-02-23 04:10:57 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Fang, W.; Liu, X.; Maiga, M.; Cao, W.; Mu, Y.; Yan, Q.; Zhu, Q. Digital PCR for Single-Cell Analysis. Encyclopedia. Available online: https://encyclopedia.pub/entry/55291 (accessed on 08 February 2026).
Fang W, Liu X, Maiga M, Cao W, Mu Y, Yan Q, et al. Digital PCR for Single-Cell Analysis. Encyclopedia. Available at: https://encyclopedia.pub/entry/55291. Accessed February 08, 2026.
Fang, Weibo, Xudong Liu, Mariam Maiga, Wenjian Cao, Ying Mu, Qiang Yan, Qiangyuan Zhu. "Digital PCR for Single-Cell Analysis" Encyclopedia, https://encyclopedia.pub/entry/55291 (accessed February 08, 2026).
Fang, W., Liu, X., Maiga, M., Cao, W., Mu, Y., Yan, Q., & Zhu, Q. (2024, February 21). Digital PCR for Single-Cell Analysis. In Encyclopedia. https://encyclopedia.pub/entry/55291
Fang, Weibo, et al. "Digital PCR for Single-Cell Analysis." Encyclopedia. Web. 21 February, 2024.
Copy Citation
Single-cell analysis provides an overwhelming strategy for revealing cellular heterogeneity and new perspectives for understanding the biological function and disease mechanism. Moreover, it promotes the basic and clinical research in many fields at a single-cell resolution. A digital polymerase chain reaction (dPCR) is an absolute quantitative analysis technology with high sensitivity and precision for DNA/RNA or protein. With the development of microfluidic technology, digital PCR has been used to achieve absolute quantification of single-cell gene expression and single-cell proteins. For single-cell specific-gene or -protein detection, digital PCR has shown great advantages.
digital PCR
microfluidic chip
single-cell analysis
1. Introduction
Cells are the basic units of life; however, the cells in the human body exhibit extensive heterogeneity, and many diseases come from single-cell mutation. A traditional analysis cannot clearly explain the behavior of individual cells. For example, in the case of cancer tumors, bulk analysis provides an overview of the average differences in gene expression, rather than the expression of genes in individual cells; this makes it difficult to find molecular differences which are only linked to specific cell types. Single-cell analysis can accurately provide information on intracellular substances and biochemical reactions within cells, reflecting specific relationships between cell functions and chemical components, as well as special roles for certain cells in living organisms [1].
The use of single-cell analysis is an interdisciplinary frontier field formed by the integration of analytical chemistry, biology, and medicine. It greatly promotes the understanding of life at the single-cell level and has vast applications in biomedical studies [2]. In the past decade, research on single-cell analysis has been highly favored by scholars; the number of papers published on PubMed on single-cell analysis in the past decade has shown a rapid growth trend yearly. Single-cell analysis enables scientists to study cells at the individual level, capturing the unique insights of each cell. It provides a more accurate and comprehensive picture of what happens to an organism at a specific point in time, which will help us better understand diseases. Single-cell analysis has involved in many fields, including basic research applications, such as cancer [3], immunology [4], neurology [5], stem cells [6], etc., and in clinical applications such as non-invasive prenatal diagnosis [7], in vitro fertilization [8], and circulating tumor cells (CTCs) [9].
The rapid development of single-cell analysis technology is driven by the rapid development of analytical tools such as cell separation, microfluidics, and single-cell sequencing. Microfluidic technology has been widely used for single-cell analyses [10]. The internal sizes of microfluidic devices are generally below 100 μm, and the reaction volume is typically at picoliter to nanoliter level, making them very suitable for single-cell analysis. With the development of the microfluidics, a digital polymerase chain reaction (dPCR) is a new option for single-cell analysis, and dPCR-based microfluidic devices have provided an efficient and accurate platform for single-cell analyses because of their accurate and automatic manipulation of the single-cell with high throughput and precision [11]. Digital PCR is a single-molecule amplification technique that is not applicable to the classical concept of concentration [12]. Single-molecule amplification is a stochastic issue. After sufficient amplification cycles and endpoint detection, the single molecule can be amplified efficiently, so the number of target molecules can be directly counted.
2. Single-Cell Analysis
2.1. Single-Cell Isolation
Accurate and reliable single-cell isolation from the sample population is a prerequisite for all single-cell-based methods. Consequently, researchers have developed a series of single-cell separation and isolation methods with different advantages and limitations, including limited serial dilution [13], fluorescence-activated cell sorting (FACS) [14], manual micromanipulation [15], laser capture microdissection (LCM) [16], and microfluidics methods [17].
Limited serial dilution [13] is a commonly used monoclonal culture method, which can also be used to obtain individual cells. The cell suspension undergoes a series of dilutions based on the distribution and concentration of cells in the cell suspension. Ultimately, only one single-cell exists in a certain volume of suspension. This method is simple in operation, low in cost, but low in efficiency because achieving a single-cell in an aliquot is statistically based on Poisson distribution.
Fluorescence-activated cell sorting (FACS) is a high-throughput cell-sorting technology based on flow cytometry. It has been used for single-cell isolation [18][19]. In FACS systems, the cell suspension which has been fluorescent labeled is pressed into the flow cell and diluted with sheath fluid to an appropriate concentration. Then, through targeted vibration driving, this stream breaks into continuous droplets, some of which carry cells. Finally, droplets containing individual cells of interest can be collected using electrically charged plates.
Laser capture microdissection (LCM) is a technology that accurately separates single cells from tissue samples [20]. After staining tissue sections, they are examined under a microscope for target area selection, and the selected area is isolated using a laser. This method does not damage the tissue structure and directly obtains target cells from frozen or paraffin-embedded tissue sections. However, this method relies on a laser capture microdissection platform, which is very complex and expensive.
Manual micromanipulation is a method of manually selecting individual cells using a microscope. Through microscopic observation, well-formed cells can be selected from the prepared cell suspension, and individual cells can be sucked out using mouth pipette technology. Visualization operations allow highly active single cells with complete morphologies to be accurately obtained. However, this process requires skilled operators and is subject to human interference, resulting in low throughput.
Microfluidic devices have been widely applied in single-cell isolation in the past few years due to their ability to isolate and manipulate single cells with microscale and integrated flow channels [21]. Microwell array chip-based methods [22][23] and droplet-based methods [24][25] are mainstream microfluidic-based single-cell isolation methods. Microfluidic-based methods have the advantages of high separation throughput and low cost, and they are easy to operate using commercial instruments.
2.2. Single-Cell Lysis
After single-cell isolation, an appropriate single-cell lysis method is important for subsequent single-cell analyses. The aim of cell lysis is to obtain the single-cell DNA/RNA or protein, so adequate cell lysis is crucial for the accuracy of single-cell analysis. Major methods of single-cell lysis include chemical [26][27] and mechanical [28][29] methods. The former is relatively mild, while the latter is more intense, which is likely to cause DNA breakage in cells. Therefore, a suitable single-cell lysis method should be selected after considering several aspects, such as cell type, nucleic acid stability, and compatibility with downstream applications. Due to the difficulty of performing single-cell, and taking the downstream reaction into consideration, the chemical-lysis method is of top priority. After cell lysis, intra-cellular components of a single cell can be used for further analyses, such as proteomics, genomics, transcriptomics, and metabolomics.
3. Digital PCR
A digital polymerase chain reaction (dPCR) is an absolute quantitative analysis technology for nucleic acids, and it has high sensitivity and precision [30]. The reaction system is divided into a large number of independent micro reaction units for PCR amplification, with each unit containing 0 or 1 template. Then, the concentration of the initial sample can be calculated according to the positive fluorescence unit’s ratio and a Poisson statistical analysis. Compared to traditional PCR technology, digital PCR technology does not rely on the standard curve, has higher sensitivity and accuracy, and can realize the absolute quantitative analysis of a sample. Chamber-based digital PCR (cdPCR) [31][32][33] and droplet-based digital PCR (ddPCR) [34][35] are the two major methods of digital PCR. Among them, the method based on cdPCR is generally achieved through microfluidic chips with a large number of reaction chambers, and the method based on ddPCR utilizes droplets as the basic reaction unit.
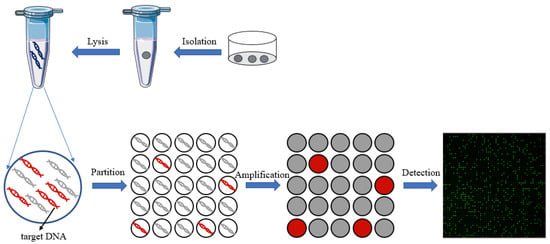
In recent years, with the development of microfluidic technology, digital PCR has been widely used in gene mutation detection [36], copy number variation detection [37], virus microbial detection [38], genetically modified organisms’ detection [39][40], and other fields [41]. Compared to qPCR, digital PCR can achieve absolute quantification without standard curves, and it is an end-point-detection method which can increase detection sensitivity compared to qPCR reaction inhibition variations. On the other hand, the reaction units of dPCR are nanoliter chambers and droplets, which will help increase primer relative concentration to capture the low-abundance targets. Thus, dPCR technology can reduce the expression bias of different abundance targets in qPCR. All of these advantages make dPCR suitable for single-cell analysis. The principle of digital PCR for single-cell analysis is shown in Figure 1.

Figure 1. The principle of digital PCR for single-cell analysis.
4. Digital PCR for Single-Cell Analysis
4.1. Chamber-Based Digital PCR (cdPCR) for Single-Cell Analysis
Chamber-based digital PCR (cdPCR) is one of the main methods commonly used for digital PCR [42]. Compared to droplet digital PCR (ddPCR), cdPCR has a more stable physical partition and is often simpler in operation. Therefore, it has promoted the application of digital PCR, and many cdPCR devices were developed for different purposes [43][44].
There are various types of RNA, from different biogenesis, and they possess distinct molecular characteristics. Traditional RNA analysis methods based on bulk cells cannot reflect the state of all cells or a group of cells in the sample. Studying RNA at the single-cell level can provide cell heterogeneity in tissues [45]. White et al. presented a high-throughput dPCR microfluidic device for the analysis of mRNA, micro-RNAs, and RNA editing events in single cells [46]. The cDNA from each single cell is distributed into a dedicated dPCR array consisting of 1020 independent 25 pL chambers, using surface-tension-based sample partitioning. They demonstrated its application in the absolute quantification of cDNA derived from mRNA and miRNA across over 1200 single cells. Then, they applied the chip-based single-cell dPCR to perform the measurements of single-nucleotide RNA editing of EEF2K in single K562 cells.
Cancer poses a serious threat to the health and safety of all humanity [47]; according to the latest estimated data from the International Agency for Research on Cancer (IARC) of the World Health Organization, there will be 19.29 million new cancer cases worldwide in 2021. Single-cell analysis has become a widely used tool in cancer research. It is used to characterize the cellular and molecular composition of tumors.
4.2. Droplet-Based Digital PCR (ddPCR) for Single-Cell Analysis
Droplet-based digital PCR (ddPCR) is another method commonly used for digital PCR, and ddPCR has been commercialized, such as Bio-Rad QX series and Naica crystal. Compared to cdPCR, it is simple to operate, has lower costs, and can achieve high-throughput detection [48].
Mitochondria are the energy-producing organelle in cells and the main site for aerobic respiration. They are involved in various cellular functions, including energy conversion, tricarboxylic acid cycle, storage of calcium ions, regulation of membrane potential and control of programmed cell death, cell proliferation and metabolism, etc. Mitochondrial disease is a genetic disease that interferes with the production of energy in the body and is currently incurable. Single-cell mitochondrial analysis helps to understand heterogeneity dynamics and disease treatment. Maeda et al. developed a ddPCR method to determine mitochondrial heteroplasmy in a single cell [49]. Five strains of cells derived from patients diagnosed with mitochondrial disease were examined in this study to evaluate the heteroplasmy of mtDNA in a single cell. This method could reveal the existence of mtDNA variances at as low as 1% frequency in a single cell without cloning steps.
Messenger RNA (mRNA) are a large class of RNA molecules that transmits genetic information from DNA to the ribosome, where it serves as a template for protein synthesis. Absolute quantification of mRNA at the single-cell level helps to understand cellular heterogeneity and thus better understand the biological mechanism.
Thereafter, Sun et al. proposed a robust one-step ddPCR-based method to quantify mRNA mutation in single cells [50]. The researchers use a rationally designed peptide nucleic acid (PNA) to capture wild-type RNA without affecting the RT-PCR of mutant mRNA (mutRNA). Applying this strategy, they quantified mutRNA in three types of single cells including human melanoma cell line (SKMEL-28), cervical cancer cells (HeLa), and thyroid cancer cells (ARO). This method enables precise mRNA mutation detection in single cells with as low as 0.01% mutated mRNA in a high background of wild-type mRNA. MicroRNAs (miRNAs) are a class of endogenous small RNAs with a length of approximately 20–24 nucleotides, which play various important regulatory roles in cells. Therefore, the single-cell analysis of miRNAs at the single-cell level is crucial as it helps to better understand the relationship between miRNAs and cellular function.
5. Conclusions
Single-cell analysis helps to understand cellular heterogeneity, and single-cell data can provide a deeper understanding of biological processes. Many single-cell analysis methods have been developed in the past few years. Chamber-based digital PCR and droplet-based digital PCR are the two main implementation methods with different advantages. Chamber-based digital PCR devices can achieve high-throughput single-cell analysis through a simple chip design. These chips have various structures with flexible operations and can achieve direct sample loading without external power sources, such as pumps. Physical partitioning in space is also a major advantage for multiple single-cell’s analyses. On the other hand, droplet-based digital PCR devices can generate a large number of droplets in a short period of time to achieve ultra-high-throughput single-cell analysis. Furthermore, these methods have relatively standardized processes due to the mature commercial digital PCR systems.
However, digital PCR for single-cell analysis also faces some urgent issues that need to be addressed. There is no standardized protocol of digital PCR for single-cell analysis. Many detection variations exist in terms of different methods, which will hinder the digital PCR to achieve large-scale applications for single-cell analysis. On the other hand, the whole process of single-cell analysis normally consists of a couple of steps, including single-cell isolation, single-cell lysis, and DNA/RNA amplification and analysis. So, the system complexity will significantly increase when more functional modules are integrated into a dPCR device for a higher level of automatic analysis.
References
- Hodzic, E. Single-cell analysis: Advances and future perspectives. Bosn. J. Basic Med. Sci. 2016, 16, 313–314.
- Pan, X. Single Cell Analysis: From Technology to Biology and Medicine. Single Cell Biol. 2015, 3, 106.
- Ren, X.; Zhang, L.; Zhang, Y.; Li, Z.; Siemers, N.; Zhang, Z. Insights Gained from Single-Cell Analysis of Immune Cells in the Tumor Microenvironment. Annu. Rev. Immunol. 2021, 39, 583–609.
- Giladi, A.; Amit, I. Single-Cell Genomics: A Stepping Stone for Future Immunology Discoveries. Cell 2018, 172, 14–21.
- Dulken, B.W.; Buckley, M.T.; Navarro Negredo, P.; Saligrama, N.; Cayrol, R.; Leeman, D.S.; George, B.M.; Boutet, S.C.; Hebestreit, K.; Pluvinage, J.V.; et al. Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 2019, 571, 205–210.
- Dong, F.; Hao, S.; Zhang, S.; Zhu, C.; Cheng, H.; Yang, Z.; Hamey, F.K.; Wang, X.; Gao, A.; Wang, F.; et al. Differentiation of transplanted haematopoietic stem cells tracked by single-cell transcriptomic analysis. Nat. Cell Biol. 2020, 22, 630–639.
- Cayrefourcq, L.; Vincent, M.; Pierredon, S.; Moutou, C.; Imbert-Bouteille, M.; Haquet, E.; Puechberty, J.; Willems, M.; Liautard-Haag, C.; Molinari, N.; et al. Single Circulating Fetal Trophoblastic Cells Eligible for Non Invasive Prenatal Diagnosis: The Exception Rather than the Rule. Sci. Rep. 2020, 10, 9861.
- Gao, Y.; Zhang, J.; Liu, Z.; Qi, S.; Guo, X.; Wang, H.; Cheng, Y.; Tian, S.; Ma, M.; Peng, H.; et al. Single-cell Sequencing Reveals Clearance of Blastula Chromosomal Mosaicism in In Vitro Fertilization Babies. Genom. Proteom. Bioinform. 2022, 20, 1224–1231.
- Sun, Y.; Wu, L.; Liu, S.; Jiang, M.; Hu, B.; Zhou, K.; Guo, W.; Xu, Y.; Zhong, Y.; Zhou, X.; et al. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat. Commun. 2021, 12, 4091.
- Liu, L.; Dong, X.; Tu, Y.; Miao, G.; Zhang, Z.; Zhang, L.; Wei, Z.; Yu, D.; Qiu, X. Methods and platforms for analysis of nucleic acids from single-cell based on microfluidics. Microfluid. Nanofluidics 2021, 25, 87.
- Zhang, J.; Xue, J.; Luo, N.; Chen, F.; Chen, B.; Zhao, Y. Microwell array chip-based single-cell analysis. Lab. Chip. 2023, 23, 1066–1079.
- Maioli, M.; Varadi, G.; Kurdi, R.; Caglioti, L.; Palyi, G. Limits of the Classical Concept of Concentration. J. Phys. Chem. B 2016, 120, 7438–7445.
- Gross, A.; Schoendube, J.; Zimmermann, S.; Steeb, M.; Zengerle, R.; Koltay, P. Technologies for Single-Cell Isolation. Int. J. Mol. Sci. 2015, 16, 16897–16919.
- Jaitin, D.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A.; et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 2014, 343, 776–779.
- Hohnadel, M.; Maumy, M.; Chollet, R. Development of a micromanipulation method for single cell isolation of prokaryotes and its application in food safety. PLoS ONE 2018, 13, e198208.
- Espina, V.; Heiby, M.; Pierobon, M.; Liotta, L.A. Laser capture microdissection technology. Expert Rev. Mol. Diagn. 2007, 7, 647–657.
- Huang, Q.; Mao, S.; Khan, M.; Lin, J. Single-cell assay on microfluidic devices. Analyst 2019, 144, 808–823.
- Pensold, D.; Zimmer-Bensch, G. Methods for Single-Cell Isolation and Preparation. Adv. Exp. Med. Biol. 2020, 1255, 7–27.
- Brunner, A.D.; Thielert, M.; Vasilopoulou, C.; Ammar, C.; Coscia, F.; Mund, A.; Hoerning, O.B.; Bache, N.; Apalategui, A.; Lubeck, M.; et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Mol. Syst. Biol. 2022, 18, e10798.
- Gautam, V.; Chatterjee, S.; Sarkar, A.K. Single Cell Type Specific RNA Isolation and Gene Expression Analysis in Rice Using Laser Capture Microdissection (LCM)-Based Method. Methods Mol. Biol. 2021, 2238, 275–283.
- Xu, X.; Wang, J.; Wu, L.; Guo, J.; Song, Y.; Tian, T.; Wang, W.; Zhu, Z.; Yang, C. Microfluidic Single-Cell Omics Analysis. Small 2019, 16, 1903905.
- Xu, C.; Wang, K.; Huang, P.; Liu, D.; Guan, Y. Single-Cell Isolation Microfluidic Chip Based on Thermal Bubble Micropump Technology. Sensors 2023, 23, 3623.
- Torres, A.J.; Hill, A.S.; Love, J.C. Nanowell-Based Immunoassays for Measuring Single-Cell Secretion: Characterization of Transport and Surface Binding. Anal. Chem. 2014, 86, 11562–11569.
- Wang, Y.; Wang, X.; Pan, T.; Li, B.; Chu, J. Label-free single-cell isolation enabled by microfluidic impact printing and real-time cellular recognition. Lab. Chip. 2021, 21, 3695–3706.
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214.
- Fan, H.C.; Wang, J.; Potanina, A.; Quake, S.R. Whole-genome molecular haplotyping of single cells. Nat. Biotechnol. 2011, 29, 51–57.
- Wood, D.K.; Weingeist, D.M.; Bhatia, S.N.; Engelward, B.P. Single cell trapping and DNA damage analysis using microwell arrays. Proc. Natl. Acad. Sci. USA 2010, 107, 10008–10013.
- Li, Z.G.; Liu, A.Q.; Klaseboer, E.; Zhang, J.B.; Ohl, C.D. Single cell membrane poration by bubble-induced microjets in a microfluidic chip. Lab. Chip. 2013, 13, 1144–1150.
- Kim, J.; Hong, J.W.; Kim, D.P.; Shin, J.H.; Park, I. Nanowire-integrated microfluidic devices for facile and reagent-free mechanical cell lysis. Lab. Chip. 2012, 12, 2914–2921.
- Vogelstein, B.; Kinzler, K. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241.
- Yin, J.; Zou, Z.; Hu, Z.; Zhang, S.; Zhang, F.; Wang, B.; Lv, S.; Mu, Y. A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens. Lab. Chip. 2020, 20, 979–986.
- Zhu, Q.; Qiu, L.; Yu, B.; Xu, Y.; Gao, Y.; Pan, T.; Tian, Q.; Song, Q.; Jin, W.; Jin, Q.; et al. Digital PCR on an integrated self-priming compartmentalization chip. Lab. Chip. 2014, 14, 1176–1185.
- Zhu, Q.; Gao, Y.; Yu, B.; Ren, H.; Qiu, L.; Han, S.; Jin, W.; Jin, Q.; Mu, Y. Self-priming compartmentalization digital LAMP for point-of-care. Lab. Chip. 2012, 12, 4755–4763.
- Galimberti, S.; Balducci, S.; Guerrini, F.; Del Re, M.; Cacciola, R. Digital Droplet PCR in Hematologic Malignancies: A New Useful Molecular Tool. Diagnostics 2022, 12, 1305.
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005.
- Sho, S.; Court, C.M.; Kim, S.; Braxton, D.R.; Hou, S.; Muthusamy, V.R.; Watson, R.R.; Sedarat, A.; Tseng, H.; Tomlinson, J.S. Digital PCR Improves Mutation Analysis in Pancreas Fine Needle Aspiration Biopsy Specimens. PLoS ONE 2017, 12, e170897.
- Brik, A.; Weber, D.G.; Casjens, S.; Rozynek, P.; Meier, S.; Behrens, T.; Stamatis, G.; Darwiche, K.; Theegarten, D.; Brüning, T.; et al. Digital PCR for the Analysis of MYC Copy Number Variation in Lung Cancer. Dis. Markers 2020, 2020, 4176376.
- Tiwari, A.; Ahmed, W.; Oikarinen, S.; Sherchan, S.P.; Heikinheimo, A.; Jiang, G.; Simpson, S.L.; Greaves, J.; Bivins, A. Application of digital PCR for public health-related water quality monitoring. Sci. Total Environ. 2022, 837, 155663.
- Demeke, T.; Dobnik, D. Critical assessment of digital PCR for the detection and quantification of genetically modified organisms. Anal. Bioanal. Chem. 2018, 410, 4039–4050.
- Niu, C.; Xu, Y.; Zhang, C.; Zhu, P.; Huang, K.; Luo, Y.; Xu, W. Ultrasensitive Single Fluorescence-Labeled Probe-Mediated Single Universal Primer–Multiplex–Droplet Digital Polymerase Chain Reaction for High-Throughput Genetically Modified Organism Screening. Anal. Chem. 2018, 90, 5586–5593.
- Denis, J.A.; Guillerm, E.; Coulet, F.; Larsen, A.K.; Lacorte, J. The Role of BEAMing and Digital PCR for Multiplexed Analysis in Molecular Oncology in the Era of Next-Generation Sequencing. Mol. Diagn. Ther. 2017, 21, 587–600.
- Quan, P.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271.
- Yin, J.; Zou, Z.; Yin, F.; Liang, H.; Hu, Z.; Fang, W.; Lv, S.; Zhang, T.; Wang, B.; Mu, Y. A Self-Priming Digital Polymerase Chain Reaction Chip for Multiplex Genetic Analysis. Acs Nano 2020, 14, 10385–10393.
- Wang, W.; Feng, M.; He, F.; Song, J.; Song, Q.; Xia, D.; Liu, R.; Yao, H.; Han, J. The Viral Load of Human Cytomegalovirus Infection in Children following Hematopoietic Stem Cell Transplant by Chip Digital PCR. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 1–6.
- Ziegenhain, C.; Vieth, B.; Parekh, S.; Reinius, B.; Guillaumet-Adkins, A.; Smets, M.; Leonhardt, H.; Heyn, H.; Hellmann, I.; Enard, W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell 2017, 65, 631–643.
- White, A.K.; Heyries, K.A.; Doolin, C.; VanInsberghe, M.; Hansen, C.L. High-Throughput Microfluidic Single-Cell Digital Polymerase Chain Reaction. Anal. Chem. 2013, 85, 7182–7190.
- Zaimy, M.A.; Saffarzadeh, N.; Mohammadi, A.; Pourghadamyari, H.; Izadi, P.; Sarli, A.; Moghaddam, L.K.; Paschepari, S.R.; Azizi, H.; Torkamandi, S.; et al. New methods in the diagnosis of cancer and gene therapy of cancer based on nanoparticles. Cancer Gene Ther. 2017, 24, 233–243.
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J. Med. Virol. 2021, 93, 4182–4197.
- Maeda, R.; Kami, D.; Maeda, H.; Shikuma, A.; Gojo, S. High throughput single cell analysis of mitochondrial heteroplasmy in mitochondrial diseases. Sci. Rep. 2020, 10, 10821.
- Sun, Y.; Tian, H.; Liu, C.; Yang, D.; Li, Z. A Clamp-Based One-Step Droplet Digital Reverse Transcription PCR (ddRT-PCR) for Precise Quantitation of Messenger RNA Mutation in Single Cells. ACS Sens. 2018, 3, 1795–1801.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
765
Revisions:
2 times
(View History)
Update Date:
23 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




