| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Paola Vittorioso | + 1564 word(s) | 1564 | 2020-04-10 15:55:28 | | | |
| 2 | Paola Vittorioso | + 1564 word(s) | 1564 | 2020-04-10 17:31:17 | | | | |
| 3 | chiara longo | + 1564 word(s) | 1564 | 2020-04-10 20:07:35 | | | | |
| 4 | Catherine Yang | Meta information modification | 1564 | 2020-04-13 05:53:34 | | | | |
| 5 | chiara longo | Meta information modification | 1564 | 2020-04-13 11:32:49 | | | | |
| 6 | chiara longo | Meta information modification | 1564 | 2020-04-13 12:09:50 | | | | |
| 7 | Nora Tang | -14 word(s) | 1550 | 2020-10-28 06:47:08 | | |
Video Upload Options
The DOF (DNA BINDING WITH ONE FINGER) family of plant-specific TF (transcription factors) was first identified in maize in 1995. Since then, DOF proteins have been shown to be present in the whole plant kingdom, including the unicellular alga Chlamydomonas reinhardtii. The DOF TF family is characterised by a highly conserved DNA binding domain (DOF domain), consisting of a CX2C-X21-CX2C motif, which is able to form a zinc finger structure. Early in the study of DOF proteins, their relevance for seed biology became clear. Indeed, the PBF (PROLAMIN BINDING FACTOR), one of the first DOF proteins characterised, controls the endosperm-specific expression of the zein genes in maize. Subsequently, several DOF proteins from both monocots and dicots have been shown to be primarily involved in seed development, dormancy and germination, as well as in seedling development and other light-mediated processes. In the last two decades, the molecular network underlying these processes have been outlined, and the main molecular players and their interactions have been identified. In this review, we will focus on the DOF TFs involved in these molecular networks, and on their interaction with other proteins.
1. Introduction
The first DOF (DNA BINDING WITH ONE FINGER) proteins were isolated because of their interaction with viral or bacterial sequences[1][2][3]. Indeed, the maize DOF1/MNB1a was originally identified as a protein binding to the cauliflower mosaic virus 35S (CaMV35S) promoter[4]; similarly, the Arabidopsis OBP1 (ocs Binding Factor/OBF BINDING PROTEIN 1) was shown to bind an element upstream of the ocs domain present in the CaMV35S promoter[2]. The tobacco NtBBF1 (Nicotiana tabacum rolB domainB Factor1) DOF protein was identified as the plant transcription factor binding to the promoter of the plant oncogene rolB[3], where it recognises a specific sequence in the region required for rolB expression in root meristematic cells and for induction by auxin[5][6]. All DOF proteins bind the highly conserved (T/A) AAAG consensus motif, identified through binding site-selection experiments using the maize DOF proteins DOF1, DOF2, DOF3 and PBF (PROLAMIN BINDING FACTOR)[7]. Although these first DOF proteins have been linked to viral and bacterial activity, subsequent studies highlighted their fundamental role in plant-specific processes (for a review, see Reference [8][9]).In Arabidopsis, seed development is divided in two phases: embryo/endosperm development and seed maturation[10]. Once the embryo is formed and cell division arrests, the seed enters the maturation phase characterised by an increase of the levels of ABA (abscisic acid) required for the establishment and maintenance of dormancy upon completion of maturation. Germination occurs when seeds are in optimal environmental conditions, mainly as far as water availability, light and temperature [11][12], and dormancy is released—under experimental conditions, dormancy can be released by a period of storage and a cold treatment at 4 ◦C for two days (stratification). The positive effect of light on seed germination is mediated mainly by the Red-light photoreceptor phytochrome B (phyB)[13], which controls the balance between ABA, which promotes dormancy, and GA (gibberellic acid), which stimulates germination by counteracting the effect of ABA[14][15][16]. Once germination is completed, seedling development undergoes photo- or skoto-morphogenesis, depending on the presence or absence of light, respectively[17].
2. DOF TFs Regulate Seed Storage Protein Accumulation and Mobilisation
Expression of prolamin genes is tightly controlled. The main cis-element present in their promoters is an endosperm-specific box[18][19], which consists of two motifs: a GLM (GCN4-like motif) (5′ G(A)TGA(G) GTCAT 3′) that shares homology with yeast GCN4[20], and a 7 bp P-box (Prolamin box) (5′TGTAAAG3′)[21][22][23]. The endosperm nuclear factors binding the P-box on the promoters of barley prolamin genes were among the first DNA binding factors identified in plants[24][25][26][27]. The corresponding PBF (PROLAMIN BINDING FACTOR) gene was first isolated from maize and shown to encode a DOF protein that interacts with O2 (Opaque2)[28]. The barley, wheat and rice homologues of maize PBF—BPBF, WPBF and RPBF respectively—were also shown to control the expression of prolamin genes[29][30]. BPBF has also been shown to interact with GAMYB, a barley transcription factor belonging to the R2R3 MYB family[31], and to cooperatively induce expression of the Hor2 (Hordein2) prolamin gene[31]. Interestingly, besides BPBF, the barley DOF protein, SAD (SCUTELLUM and ALEURONE-expressed DOF), bind the Hor2 promoter and interact in vivo with GAMYB[32]. At the onset of seed germination, stored compounds, mainly proteins and starch, are hydrolysed by proteases and hydrolytic enzymes secreted from aleurone cells[33][34]. GAs induce the expression of these proteases- and hydrolases-encoding genes; the promoters of these genes are characterised by a DOF binding site[35][36]. GAs also induce GAMYB transcription; GAMYB induces expression of hydrolase genes. Interestingly, SAD interact with GAMYB and cooperates in the positive control of hydrolase genes expression. Also BPBF is involved in the transcriptional control of these genes, but with an antagonistic function with respect to SAD[37]. Indeed, in regulating the expression of hydrolase genes in aleurone cells, BPBF functions as a repressor, whereas SAD functions as a transcriptional activator, although both interact with GAMYB[37][38].
3. Interaction DOF-DELLA Represses Seed Germination
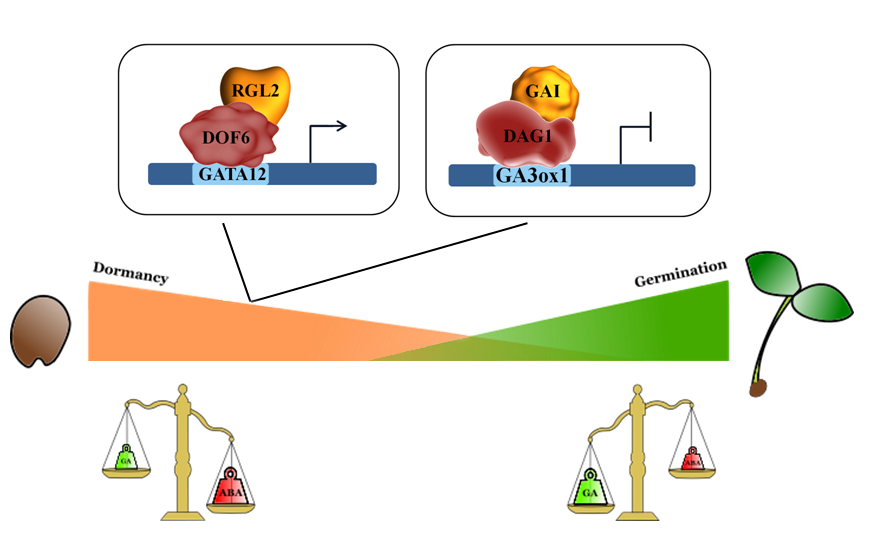
DELLA proteins are repressors of GA signaling and of GA-mediated processes, such as seed germination. As for the seed germination process, it was shown that the DELLA protein primarily involved in this process is RGL2 (RGA-LIKE2)[39][40][41]. Studies on the RGL2 target gene GATA12, encoding a GATA zinc finger transcription factor repressor of GA-mediated seed germination, led to the identification of the RGL2-DOF6 (At3g45610; DOF3.2/DOF6) complex responsible for GATA12 induction in freshly harvested seeds. ChIP (Chromatin Immunoprecipitation) assays proved that both RGL2 and DOF6 are required to induce GATA12 expression[42] (Figure 1). Another DOF protein, which represses seed germination through direct interaction with a DELLA protein, is DAG1 (DOF AFFECTING GERMINATION 1), which was convincingly demonstrated to be involved in seed germination[43]. DAG1 cooperates with GAI (GIBBERELLIC ACID INTENSITIVE) to negatively regulate GA3ox1. Indeed, GAI is necessary for the binding of DAG1 to the DOF sites in the GA3ox1 promoter, and it directly interacts with DAG1[44](Figure 1).
Figure 1. DOF proteins interact with DELLA factors to negatively regulate seed germination.
4. DOF Proteins in Seedling Development and Other Light-Mediated Processes
Seedling development depends on environmental conditions. Indeed, once germination is completed, seedlings undergo two possible developmental programs, photomorphogenesis or skotomorphogenesis, depending on the presence or absence of light, respectively[17]. Photomorphogenesis is characterised by inhibition of hypocotyl elongation, open and expanded cotyledons and chloroplast development, whereas skotomorphogenesis is characterised by long hypocotyls and small unfolded cotyledons. Light, mainly through the photoreceptors phyA (and phyB phytochrome A and B), mediates these developmental programs via downstream signalling molecules, and through the control of hormonal levels.
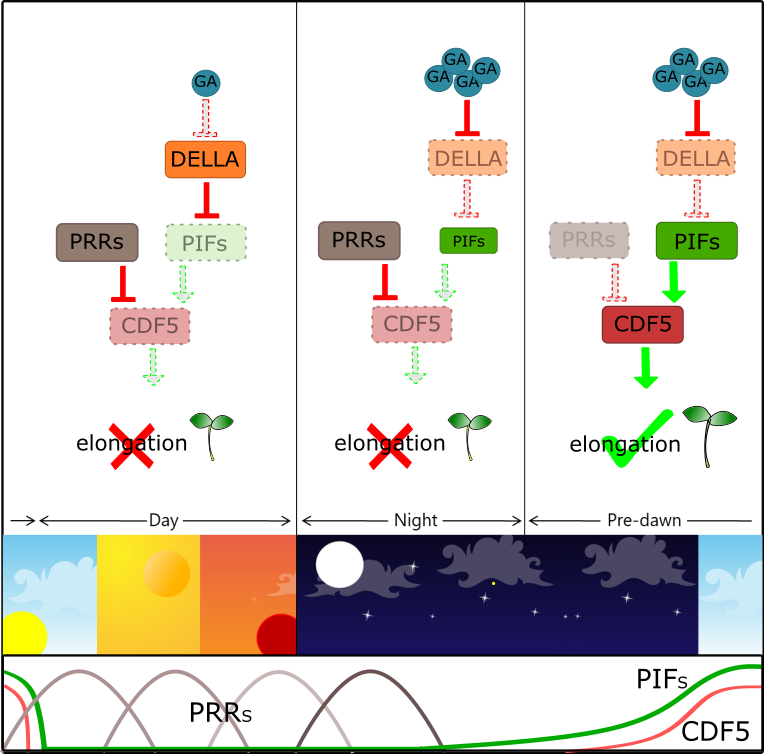
COG1 (COGWHEEL 1) is the first DOF protein which was shown to be involved in both phyA- and phyB-mediated seedling development. It has been shown that COG1 promotes hypocotyl elongation, through PIF4 and PIF5 (PHYTOCHROME INTERACTING FACTOR4 and 5 ), which in turn induce BRs (Brassinosteroids) biosynthesis. CDF5 (CYCLING DOF FACTOR 5) belongs to a sub-family of DOF proteins homologous to CDF1, which negatively controls expression of the floral activator-encoding gene CO (CONSTANS)[45]. Interestingly, CDF and COG1 belong to the same DOF phylogenetic clade, previously referred to as group II[46]. It was recently shown that CDF5 promotes cell expansion and hypocotyl elongation in a light- and clock-dependent manner[47]. CDF5 acts downstream of both the PIFs and the PRR9/7/5 (PSEUDO-RESPONSE REGULATORS 9/7/5) antagonistic pathway, which promote and, respectively, inhibit hypocotyl growth. Both PIFs and PRRs target CDF5: PIFs induce pre-dawn expression of CDF5 to promote hypocotyl growth, whereas PRRs directly repress CDF5 from morning to dusk to prevent overgrowth of the hypocotyl[47](Figure 2). Similarly, DAG1 has been recently shown to promote cell expansion and hypocotyl elongation[48]: light-grown dag1 mutant seedlings have hypocotyls significantly shorter than the wild-type, suggesting that DAG1 is a negative regulator in the light-mediated inhibition of hypocotyl elongation[49]. Indeed, DAG1 promotes hypocotyl elongation through the control of ABA, ethylene and auxin signaling; Gene Ontology analysis of the 257 DE genes in dag1 hypocotyls compared to the wild-type, revealed that “response to abscisic acid”, “ethylene biosynthetic process” as well as “response to ethylene” and “response to auxin” were among the most significantly enriched categories[48].
Figure 2. CDF5 promotes hypocotyl elongation in a clock-dependent manner.
5. DOF in Early Steps of Arabidopsis Development
Arabidopsis DOF genes are mainly expressed in the vascular system, in the xylem, in the phloem or both (for a review, see Reference [50]). Recently, two Arabidopsis DOF proteins (DOF2.4 and DOF5.1) have been identified as mobile factors involved in root procambial development and were named PEAR1 and 2 (PHLOEM EARLY DOF1 and 2). The corresponding PEAR1 and 2 genes are highly expressed in PSE (protophloem sieve elements)[51]. PEAR1 and 2 were shown to trigger periclinal cell division by controlling genes that promote radial growth, such as SMXL3 (SUPPRESSOR OF MAX2 1-LIKE3), encoding a key regulator of phloem formation. In addition, four homologues (DOF3.2, DOF5.3, DOF1.1 and DOF5.6, have been identified as PSE-specifically or PSE-abundantly expressed DOF genes with a broader protein localisation. These DOF factors redundantly function in the promotion of periclinal cell divisions in PSE cells[51]. The action of PEAR factors is counteracted by the HD-ZIP III transcription factors PHB, CNA and REV (PHABULOSA, CORONA and REVOLUTA). In addition, this molecular network involves a double-negative feedback loop where PEAR1 induces transcription of the PHB, CNA, and REV genes and the corresponding PHB, CNA and REV proteins negatively control PEAR1 transcription and protein movement[51].
References
- Shuichi Yanagisawa; A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Research 1995, 23, 3403-3410, 10.1093/nar/23.17.3403.
- Bei Zhang; Wei Chen; Rhonda C Foley; Michael Büttner; Karam B. Singh; Interactions between Distinct Types of DNA Binding Proteins Enhance Binding to ocs Element Promoter Sequences. The Plant Cell 1995, 7, 2241, 10.2307/3870165.
- Angelo Paolis; Sabrina Sabatini; Luca Pascalis; Paolo Costantino; Imerio Capone; A rolB regulatory factor belongs to a new class of single zinc finger plant proteins. The Plant Journal 1996, 10, 215-223, 10.1046/j.1365-313x.1996.10020215.x.
- S Yanagisawa; K Izui; Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif.. Journal of Biological Chemistry 1993, 268, , null.
- I. Capone; Maura Cardarelli; D. Mariotti; M. Pomponi; A. De Paolis; Paolo Costantino; Different promoter regions control level and tissue specificity of expression of Agrobacterium rhizogenes rolB gene in plants. Plant Molecular Biology 1991, 16, 427-436, 10.1007/bf00023993.
- Imerio Capone; Giovanna Frugis; Paolo Costantino; Maura Cardarelli; Expression in different populations of cells of the root meristem is controlled by different domains of the rolB promoter. Plant Molecular Biology 1994, 25, 681-691, 10.1007/bf00029606.
- Shuichi Yanagisawa; Robert J. Schmidt; Diversity and similarity among recognition sequences of Dof transcription factors.. The Plant Journal 1999, 17, 209-214, 10.1046/j.1365-313x.1999.00363.x.
- Chen Dong; Huigang Hu; Jianghui Xie; Genome-wide analysis of the DNA-binding with one zinc finger (Dof) transcription factor family in bananas. Genome 2016, 59, 1085-1100, 10.1139/gen-2016-0081.
- S. Gupta; N. Malviya; H. Kushwaha; J. Nasim; N. C. Bisht; V. K. Singh; D. Yadav; Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549-562, 10.1007/s00425-014-2239-3.
- Marilyn A. L. West; John J. Harada; Embryogenesis in Higher Plants: An Overview. The Plant Cell 1993, 5, 1361, 10.2307/3869788.
- Koornneef, M and Karssen, C.M. Seed Dormancy and Germination in Arabidopsis. Cold Spring Harb. Cold Spring Harb. Lab. Press 1994, 6, 313–334.
- Mishra M And Yadav As Singh D; Standardizing the Methods for Breaking Seed Dormancy to Enhance Germination of Gloriosa superba Seeds. Expert Opinion on Environmental Biology 2016, 4, , 10.4172/2325-9655.1000123.
- T. Shinomura; A. Nagatani; J. Chory; M. Furuya; The Induction of Seed Germination in Arabidopsis thaliana Is Regulated Principally by Phytochrome B and Secondarily by Phytochrome A. Plant Physiology 1994, 104, 363-371, 10.1104/pp.104.2.363.
- T. Toyomasu; Phytochrome Regulates Gibberellin Biosynthesis during Germination of Photoblastic Lettuce Seeds. Plant Physiology 1998, 118, 1517-1523, 10.1104/pp.118.4.1517.
- Shinjiro Yamaguchi; Maria W. Smith; Robert G. S. Brown; Yuji Kamiya; Tai-Ping Sun; Phytochrome Regulation and Differential Expression of Gibberellin 3b-Hydroxylase Genes in Germinating Arabidopsis Seeds. The Plant Cell 1998, 10, 2115, 10.2307/3870788.
- Mitsunori Seo; Atsushi Hanada; Ayuko Kuwahara; Akira Endo; Masanori Okamoto; Yukika Yamauchi; Helen North; Annie Marion‐Poll; Tai-Ping Sun; Tomokazu Koshiba; Yuji Kamiya; Shinjiro Yamaguchi; Eiji Nambara; Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. The Plant Journal 2006, 48, 354-366, 10.1111/j.1365-313x.2006.02881.x.
- Ari Sadanandom; Éva Ádám; Beatriz Orosa; András Viczián; Cornelia Klose; Cunjin Zhang; Eve-Marie Josse; László Kozma-Bognár; Ferenc Nagy; SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana.. Proceedings of the National Academy of Sciences 2015, 112, 11108-13, 10.1073/pnas.1415260112.
- Brian Forde; A. Heyworth; J. Pywell; M. Kreis; Nucleotide sequence of a B1 hordein gene and the identification of possible upstream regulatory elements in endosperm storage protein genes from barley, wheat and maize. Nucleic Acids Research 1985, 13, 7327-7339, 10.1093/nar/13.20.7327.
- M Kreis; B G Forde; S Rahman; B J Miflin; P R Shewry; Molecular evolution of the seed storage proteins of barley, rye and wheat.. Journal of Molecular Biology 1985, 183, , null.
- Hill, D.E.; Hope, I.A.; Macke, J.P.; Struhl, K. Saturation mutagenesis of the yeast bis3 regulatory site: Requirements for transcriptional induction and for binding by GCN4 activator protein. Science (80-. ). 1986, 234, 451–457.
- A. Boronat; M.C. Martínez; Manuel Reina; P. Puigdomenech; J. Palau; Isolation and sequencing of a 28 kD glutelin-2 gene from maize. common elements in the 5′ flanking regions among zein and glutelin genes. Plant Science 1986, 47, 95-102, 10.1016/0168-9452(86)90055-5.
- Gary A. Thompson; Brian A. Larkins; Structural elements regulating zein gene expression. BioEssays 1989, 10, 108-113, 10.1002/bies.950100404.
- Laura M. M. Ottoboni; Adilson Leite; Jose A. Yunes; Maria Luiza P. N. Targon; Gonçalo A. De Souza Filho; Paulo Arruda; Sequence analysis of 22 kDa-like α-coixin genes and their comparison with homologous zein and kafirin genes reveals highly conserved protein structure and regulatory elements. Plant Molecular Biology 1993, 21, 765-778, 10.1007/bf00027110.
- U.-G. Maier; J.W.S. Brown; C. Toloczyki; G. Feix; Binding of a nuclear factor to a consensus sequence in the 5' flanking region of zein genes from maize. The EMBO Journal 1987, 6, 17-22, null.
- M C Hammond-Kosack; M J Holdsworth; M W Bevan; In vivo footprinting of a low molecular weight glutenin gene (LMWG-1D1) in wheat endosperm.. The EMBO Journal 1993, 12, 545-554, null.
- Ueda, T.; Wang, Z.; Pham, N.; Messing, J. Identification of a transcriptional activator-binding element in the 27-kilodalton zein promoter, the -300 element. Mol. Cell. Biol. 1994, 14, 4350–4359.
- Z Wang; T Ueda; J Messing; Characterization of the maize prolamin box-binding factor-1 (PBF-1) and its role in the developmental regulation of the zein multigene family.. Gene 1998, 223, , null.
- Jesús Vicente-Carbajosa; Stephen P. Moose; Ronald L. Parsons; Robert J. Schmidt; A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proceedings of the National Academy of Sciences 1997, 94, 7685-7690, 10.1073/pnas.94.14.7685.
- Montana Mena; Jesús Vicente-Carbajosa; Robert J. Schmidt; Pilar Carbonero; An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm.. The Plant Journal 1998, 16, 53-62, 10.1046/j.1365-313x.1998.00275.x.
- Masayuki P. Yamamoto; Yasuyuki Onodera; Satoru M. Touno; Fumio Takaiwa; Synergism between RPBF Dof and RISBZ1 bZIP Activators in the Regulation of Rice Seed Expression Genes1[W]. Plant Physiology 2006, 141, 1694-1707, 10.1104/pp.106.082826.
- Diaz, I.; Vicente-Carbajosa, J.; Abraham, Z.; Martínez, M.; Moneda, I.I. La; Carbonero, P. The GAMYP protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 2002, 29, 453–464.
- Isabel Diaz; Manuel Martinez; Ines Isabel-LaMoneda; Ignacio Rubio Somoza; Pilar Carbonero; The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. The Plant Journal 2005, 42, 652-662, 10.1111/j.1365-313x.2005.02402.x.
- G B Fincher; Molecular and Cellular Biology Associated with Endosperm Mobilization in Germinating Cereal Grains. Annual Review of Plant Biology 1989, 40, 305-346, 10.1146/annurev.pp.40.060189.001513.
- R.W Skadsen; Physiological and molecular genetic mechanisms regulating hydrolytic enzyme gene expression in cereal grains. Physiologia Plantarum 1998, 104, 486-502, 10.1034/j.1399-3054.1998.1040326.x.
- Gubler, F.; Jacobsen, J. V. Gibberellin-responsive elements in the promoter of a barley high-pI α-amylase gene. Plant Cell 1992, 4, 1435–1441.
- Chung-An Lu; Eng-Kiat Lim; Su-May Yu; Sugar Response Sequence in the Promoter of a Rice α-Amylase Gene Serves as a Transcriptional Enhancer. Journal of Biological Chemistry 1998, 273, 10120-10131, 10.1074/jbc.273.17.10120.
- Inés Isabel-LaMoneda; Isabel Diaz; Manuel Martinez; Montaña Mena; Pilar Carbonero; SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. The Plant Journal 2003, 33, 329-340, 10.1046/j.1365-313x.2003.01628.x.
- Montaña Mena; Francisco Javier Cejudo; Inés Isabel-LaMoneda; Pilar Carbonero; A Role for the DOF Transcription Factor BPBF in the Regulation of Gibberellin-Responsive Genes in Barley Aleurone1. Plant Physiology 2002, 130, 111-119, 10.1104/pp.005561.
- Sorcheng Lee; Hui Cheng; Kathryn E. King; Weefuen Wang; Yawen He; Alamgir Hussain; Jane Lo; Nicholas P. Harberd; Jinrong Peng; Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genome Research 2002, 16, 646-658, 10.1101/gad.969002.
- Petra Stamm; Pratibha Ravindran; Bijayalaxmi Mohanty; Ee Ling Tan; Hao Yu; Prakash P. Kumar; Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biology 2012, 12, 179-179, 10.1186/1471-2229-12-179.
- Urszula Piskurewicz; Yusuke Jikumaru; Natsuko Kinoshita; Eiji Nambara; Yuji Kamiya; Luis Lopez-Molina; The Gibberellic Acid Signaling Repressor RGL2 Inhibits Arabidopsis Seed Germination by Stimulating Abscisic Acid Synthesis and ABI5 Activity[W]. The Plant Cell 2008, 20, 2729-2745, 10.1105/tpc.108.061515.
- Pratibha Ravindran; Vivek Verma; Petra Stamm; Prakash P. Kumar; A Novel RGL2–DOF6 Complex Contributes to Primary Seed Dormancy in Arabidopsis thaliana by Regulating a GATA Transcription Factor. Molecular Plant 2017, 10, 1307-1320, 10.1016/j.molp.2017.09.004.
- Maura Papi; Sabrina Sabatini; David Bouchez; Christine Camilleri; Paolo Costantino; Paola Vittorioso; Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genome Research 2000, 14, 28-33, null.
- Alessandra Boccaccini; Silvia Santopolo; Davide Capauto; Riccardo Lorrai; Emanuele Minutello; Giovanna Serino; Paolo Costantino; Paola Vittorioso; The DOF Protein DAG1 and the DELLA Protein GAI Cooperate in Negatively Regulating the AtGA3ox1 Gene. Molecular Plant 2014, 7, 1486-1489, 10.1093/mp/ssu046.
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. Plant science: FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science (80-. ). 2005, 309, 293–297.
- Miguel Angel Moreno-Risueno; Manuel Martinez; Jesús Vicente-Carbajosa; Pilar Carbonero; The family of DOF transcription factors: from green unicellular algae to vascular plants. Molecular Genetics and Genomics 2006, 277, 379-390, 10.1007/s00438-006-0186-9.
- Guiomar Martín; Arnau Rovira; Nil Veciana; Judit Soy; Gabriela Toledo-Ortiz; Charlotte M. M. Gommers; Marc Boix; Rossana Henriques; Eugenio G. Minguet; David Alabadí; Karen Halliday; Pablo Leivar; Elena Monte; Circadian Waves of Transcriptional Repression Shape PIF-Regulated Photoperiod-Responsive Growth in Arabidopsis. Current Biology 2018, 28, 311-318.e5, 10.1016/j.cub.2017.12.021.
- Riccardo Lorrai; Francesco Gandolfi; Alessandra Boccaccini; Veronica Ruta; Marco Possenti; Anna Tramontano; Paolo Costantino; Rosalba Lepore; Paola Vittorioso; Genome-wide RNA-seq analysis indicates that the DAG1 transcription factor promotes hypocotyl elongation acting on ABA, ethylene and auxin signaling. Scientific Reports 2018, 8, 15895, 10.1038/s41598-018-34256-3.
- Stefano Gabriele; Annalisa Rizza; Julie Martone; Patrizia Circelli; Paolo Costantino; Paola Vittorioso; The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. The Plant Journal 2009, 61, 312-323, 10.1111/j.1365-313x.2009.04055.x.
- Rozenn Le Hir; Catherine Bellini; The Plant-Specific Dof Transcription Factors Family: New Players Involved in Vascular System Development and Functioning in Arabidopsis. Frontiers in Plant Science 2013, 4, , 10.3389/fpls.2013.00164.
- Shunsuke Miyashima; Pawel Roszak; Iris Sevilem; Koichi Toyokura; Bernhard Blob; Jung-Ok Heo; Nathan Mellor; Hanna Elina Help; Sofia Otero; Wouter Smet; Mark Boekschoten; Guido J. Hooiveld; Kayo Hashimoto; Ondřej Smetana; Riccardo Siligato; Eva-Sophie Wallner; Ari Pekka Mähönen; Yuki Kondo; Charles W. Melnyk; Thomas Greb; Keiji Nakajima; Rosangela Sozzani; Anthony Bishopp; Bert De Rybel; Yrjo Helariutta; Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 2019, 565, 490-494, 10.1038/s41586-018-0839-y.
- Eva-Sophie Wallner; Vadir López Salmerón; Ilya Belevich; Gernot Poschet; Ilona Jung; Karin Grünwald; Iris Sevilem; Eija Jokitalo; Ruediger Hell; Yrjo Helariutta; Javier Agustí; Ivan Lebovka; Thomas Greb; Strigolactone- and Karrikin-Independent SMXL Proteins Are Central Regulators of Phloem Formation.. Current Biology 2017, 27, 1241-1247, 10.1016/j.cub.2017.03.014.