
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Aurelio Cafaro | -- | 2525 | 2024-02-16 16:15:02 | | | |
| 2 | Sirius Huang | -1 word(s) | 2524 | 2024-02-17 02:23:30 | | |
Video Upload Options
Each time the virus starts a new round of expression/replication, even under effective antiretroviral therapy (ART), the transactivator of viral transcription Tat is one of the first HIV-1 protein to be produced, as it is strictly required for HIV replication and spreading. At this stage, most of the Tat protein exits infected cells, accumulates in the extracellular matrix and exerts profound effects on both the virus and neighbor cells, mostly of the innate and adaptive immune systems. Through these effects, extracellular Tat contributes to the acquisition of infection, spreading and progression to AIDS in untreated patients, or to non-AIDS co-morbidities in ART-treated individuals, who experience inflammation and immune activation despite virus suppression.
1. Introduction
2. The HIV-1 Tat Protein
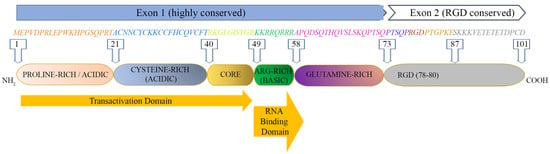
3. Mechanism and Kinetics of the Extracellular Release of Tat (eTat)
4. Role of eTat in HIV Acquisition, Dissemination and Reservoir Formation
4.1. HIV Acquisition
4.2. HIV Dissemination
4.3. Establishment and Maintenance of Latent Virus Reservoirs
References
- Trickey, A.; Sabin, C.A.; Burkholder, G.; Crane, H.; Monforte, A.D.; Egger, M.; Gill, M.J.; Grabar, S.; Guest, J.L.; Jarrin, I.; et al. Life expectancy after 2015 of adults with HIV on long-term antiretroviral therapy in Europe and North America: A collaborative analysis of cohort studies. Lancet HIV 2023, 10, e295–e307.
- Broyles, L.N.; Luo, R.; Boeras, D.; Vojnov, L. The risk of sexual transmission of HIV in individuals with low-level HIV viraemia: A systematic review. Lancet 2023, 402, 464–471.
- Aamer, H.A.; McClure, J.; Ko, D.; Maenza, J.; Collier, A.C.; Coombs, R.W.; Mullins, J.I.; Frenkel, L.M. Cells producing residual viremia during antiretroviral treatment appear to contribute to rebound viremia following interruption of treatment. PLoS Pathog. 2020, 16, e1008791.
- Veenhuis, R.T.; Abreu, C.M.; Costa, P.A.G.; Ferreira, E.A.; Ratliff, J.; Pohlenz, L.; Shirk, E.N.; Rubin, L.H.; Blankson, J.N.; Gama, L.; et al. Monocyte-derived macrophages contain persistent latent HIV reservoirs. Nat. Microbiol. 2023, 8, 833–844.
- Farhadian, S.F.; Lindenbaum, O.; Zhao, J.; Corley, M.J.; Im, Y.; Walsh, H.; Vecchio, A.; Garcia-Milian, R.; Chiarella, J.; Chintanaphol, M.; et al. HIV viral transcription and immune perturbations in the CNS of people with HIV despite ART. JCI Insight 2022, 7, e160267.
- Asowata, O.E.; Singh, A.; Ngoepe, A.; Herbert, N.; Fardoos, R.; Reddy, K.; Zungu, Y.; Nene, F.; Mthabela, N.; Ramjit, D.; et al. Irreversible depletion of intestinal CD4+ T cells is associated with T cell activation during chronic HIV infection. JCI Insight 2021, 6, e146162.
- Laspia, M.F.; Rice, A.P.; Mathews, M.B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 1989, 59, 283–292.
- Ensoli, B.; Barillari, G.; Salahuddin, S.Z.; Gallo, R.C.; Wong-Staal, F. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi’s sarcoma lesions of AIDS patients. Nature 1990, 345, 84–86.
- Ensoli, B.; Gendelman, R.; Markham, P.; Fiorelli, V.; Colombini, S.; Raffeld, M.; Cafaro, A.; Chang, H.K.; Brady, J.N.; Gallo, R.C. Synergy between basic fibroblast growth factor and HIV-1 Tat protein in induction of Kaposi’s sarcoma. Nature 1994, 371, 674–680.
- Ensoli, B.; Buonaguro, L.; Barillari, G.; Fiorelli, V.; Gendelman, R.; Morgan, R.A.; Wingfield, P.; Gallo, R.C. Release, uptake, and effects of extracellular HIV-1 Tat protein on cell growth and viral transactivation. J. Virol. 1993, 67, 277–287.
- Chang, H.C.; Samaniego, F.; Nair, B.C.; Buonaguro, L.; Ensoli, B. HIV-1 Tat protein exits from cells via a leaderless secretory pathway and binds to extracellular matrix-associated heparan sulfate proteoglycans through its basic region. AIDS 1997, 11, 1421–1431.
- Rayne, F.; Debaisieux, S.; Yezid, H.; Lin, Y.-L.; Mettling, C.; Konate, K.; Chazal, N.; Arold, S.T.; Pugnière, M.; Sanchez, F.; et al. Phosphatidylinositol-(4,5)-bisphosphate enables efficient secretion of HIV-1 Tat by infected T-cells. EMBO J. 2010, 29, 1348–1362.
- Deeks, S.G.; Tracy, R.; Douek, D.C. Systemic effects of inflammation on health during chronic HIV infection. Immunity 2013, 39, 633–645.
- Feinberg, M.B.; Baltimore, D.; Frankel, A.D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 1991, 88, 4045–4049.
- Mueller, N.; Pasternak, A.O.; Klaver, B.; Cornelissen, M.; Berkhout, B.; Das, A.T. The HIV-1 Tat Protein Enhances Splicing at the Major Splice Donor Site. J. Virol. 2018, 92, e01855-17.
- D’Orso, I.; Frankel, A.D. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010, 17, 815–821.
- López-Huertas, M.R.; Callejas, S.; Abia, D.; Mateos, E.; Dopazo, A.; Alcami, J.; Coiras, M. Modifications in host cell cytoskeleton structure and function mediated by intracellular HIV-1 Tat protein are greatly dependent on the second coding exon. Nucleic Acids Res. 2010, 38, 3287–3307.
- López-Huertas, M.R.; Mateos, E.; del Cojo, M.S.; Gómez-Esquer, F.; Díaz-Gil, G.; Rodríguez-Mora, S.; López, J.A.; Calvo, E.; López-Campos, G.; Alcamí, J.; et al. The presence of HIV-1 Tat protein second exon delays fas protein-mediated apoptosis in CD4+ T lymphocytes: A potential mechanism for persistent viral production. J. Biol. Chem. 2013, 288, 7626–7644.
- Kukkonen, S.; Martinez-Viedma, M.D.P.; Kim, N.; Manrique, M.; Aldovini, A. HIV-1 Tat second exon limits the extent of Tat-mediated modulation of interferon-stimulated genes in antigen presenting cells. Retrovirology 2014, 11, 30.
- Howcroft, T.K.; Strebel, K.; Martin, M.A.; Singer, D.S. Repression of MHC class I gene promoter activity by two-exon Tat of HIV. Science 1993, 260, 1320–1322.
- Kanazawa, S.; Okamoto, T.; Peterlin, B. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 2000, 12, 61–70.
- Mele, A.R.; Marino, J.; Dampier, W.; Wigdahl, B.; Nonnemacher, M.R. HIV-1 Tat Length: Comparative and Functional Considerations. Front. Microbiol. 2020, 11, 444.
- Epie, N.; Ammosova, T.; Sapir, T.; Voloshin, Y.; Lane, W.S.; Turner, W.; Reiner, O.; Nekhai, S. HIV-1 Tat interacts with LIS1 protein. Retrovirology 2005, 2, 6.
- Chen, D.; Wang, M.; Zhou, S.; Zhou, Q. HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J. 2002, 21, 6801–6810.
- Huo, L.; Li, D.; Sun, L.; Liu, M.; Shi, X.; Sun, X.; Li, J.; Dong, B.; Dong, X.; Zhou, J. Tat acetylation regulates its actions on microtubule dynamics and apoptosis in T lymphocytes. J. Pathol. 2010, 223, 28–36.
- Wei, P.; Garber, M.E.; Fang, S.-M.; Fischer, W.H.; Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998, 92, 451–462.
- Berkhout, B.; Silverman, R.H.; Jeang, K.-T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 1989, 59, 273–282.
- Gotora, P.T.; van der Sluis, R.; Williams, M.E. HIV-1 Tat amino acid residues that influence Tat-TAR binding affinity: A scoping review. BMC Infect. Dis. 2023, 23, 164.
- Tyagi, M.; Rusnati, M.; Presta, M.; Giacca, M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2001, 276, 3254–3261.
- Ruiz, A.P.; Ajasin, D.O.; Ramasamy, S.; DesMarais, V.; Eugenin, E.A.; Prasad, V.R. A Naturally Occurring Polymorphism in the HIV-1 Tat Basic Domain Inhibits Uptake by Bystander Cells and Leads to Reduced Neuroinflammation. Sci. Rep. 2019, 9, 3308.
- Gump, J.M.; June, R.K.; Dowdy, S.F. Revised role of glycosaminoglycans in TAT protein transduction domain-mediated cellular transduction. J. Biol. Chem. 2010, 285, 1500–1507.
- King, J.; Eugenin, E.; Buckner, C.; Berman, J. HIV tat and neurotoxicity. Microbes Infect. 2006, 8, 1347–1357.
- Brake, D.A.; Debouck, C.; Biesecker, G. Identification of an Arg-Gly-Asp (RGD) cell adhesion site in human immunodeficiency virus type 1 transactivation protein, tat. J. Cell Biol. 1990, 111, 1275–1281.
- Barillari, G.; Gendelman, R.; Gallo, R.C.; Ensoli, B. The Tat protein of human immunodeficiency virus type 1, a growth factor for AIDS Kaposi sarcoma and cytokine-activated vascular cells, induces adhesion of the same cell types by using integrin receptors recognizing the RGD amino acid sequence. Proc. Natl. Acad. Sci. USA 1993, 90, 7941–7945.
- Neuveut, C.; Jeang, K.T. Recombinant human immunodeficiency virus type 1 genomes with tat unconstrained by overlapping reading frames reveal residues in Tat important for replication in tissue culture. J. Virol. 1996, 70, 5572–5581.
- Neuveut, C.; Scoggins, R.M.; Camerini, D.; Markham, R.B.; Jeang, K.-T. Requirement for the second coding exon of Tat in the optimal replication of macrophage-tropic HIV-1. J. Biomed. Sci. 2003, 10, 651–660.
- Rodríguez-Mora, S.; Mateos, E.; Moran, M.; Martín, M.; López, J.A.; Calvo, E.; Terrón, M.C.; Luque, D.; Muriaux, D.; Alcamí, J.; et al. Intracellular expression of Tat alters mitochondrial functions in T cells: A potential mechanism to understand mitochondrial damage during HIV-1 replication. Retrovirology 2015, 12, 78.
- Wu, Y.; Marsh, J.W. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 2001, 293, 1503–1506.
- Chertova, E.; Chertov, O.; Coren, L.V.; Roser, J.D.; Trubey, C.M.; Bess, J.W.; Sowder, R.C.; Barsov, E.; Hood, B.L.; Fisher, R.J.; et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 2006, 80, 9039–9052.
- Schatz, M.; Marty, L.; Ounadjela, C.; Tong, P.B.V.; Cardace, I.; Mettling, C.; Milhiet, P.-E.; Costa, L.; Godefroy, C.; Pugnière, M.; et al. A Tripartite Complex HIV-1 Tat-Cyclophilin A-Capsid Protein Enables Tat Encapsidation That Is Required for HIV-1 Infectivity. J. Virol. 2023, 97, e0027823.
- Harrich, D.; Ulich, C.; García-Martínez, L.F.; Gaynor, R.B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997, 16, 1224–1235.
- Mele, A.R.; Marino, J.; Chen, K.; Pirrone, V.; Janetopoulos, C.; Wigdahl, B.; Klase, Z.; Nonnemacher, M.R. Defining the molecular mechanisms of HIV-1 Tat secretion: PtdIns(4,5)P2 at the epicenter. Traffic 2018, 19, 655–665.
- Ghanam, R.H.; Eastep, G.N.; Saad, J.S. Structural Insights into the Mechanism of HIV-1 Tat Secretion from the Plasma Membrane. J. Mol. Biol. 2023, 435, 167880.
- Jost, M.; Simpson, F.; Kavran, J.M.; Lemmon, M.A.; Schmid, S.L. Phosphatidylinositol-4,5-bisphosphate is required for endocytic coated vesicle formation. Curr. Biol. 1998, 8, 1399–1402.
- Botelho, R.J.; Teruel, M.; Dierckman, R.; Anderson, R.; Wells, A.; York, J.D.; Meyer, T.; Grinstein, S. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J. Cell Biol. 2000, 151, 1353–1368.
- Debaisieux, S.; Lachambre, S.; Gross, A.; Mettling, C.; Besteiro, S.; Yezid, H.; Henaff, D.; Chopard, C.; Mesnard, J.-M.; Beaumelle, B. HIV-1 Tat inhibits phagocytosis by preventing the recruitment of Cdc42 to the phagocytic cup. Nat. Commun. 2015, 6, 6211.
- Tryoen-Tóth, P.; Chasserot-Golaz, S.; Tu, A.; Gherib, P.; Bader, M.-F.; Beaumelle, B.; Vitale, N. HIV-1 Tat protein inhibits neurosecretion by binding to phosphatidylinositol 4,5-bisphosphate. J. Cell Sci. 2013, 126, 454–463.
- Harwig, A.; Jongejan, A.; van Kampen, A.H.C.; Berkhout, B.; Das, A.T. Tat-dependent production of an HIV-1 TAR-encoded miRNA-like small RNA. Nucleic Acids Res. 2016, 44, 4340–4353.
- Sampey, G.C.; Saifuddin, M.; Schwab, A.; Barclay, R.; Punya, S.; Chung, M.-C.; Hakami, R.M.; Zadeh, M.A.; Lepene, B.; Klase, Z.A.; et al. Exosomes from HIV-1-infected Cells Stimulate Production of Pro-inflammatory Cytokines through Trans-activating Response (TAR) RNA. J. Biol. Chem. 2016, 291, 1251–1266.
- Chen, L.; Feng, Z.; Yue, H.; Bazdar, D.; Mbonye, U.; Zender, C.; Harding, C.V.; Bruggeman, L.; Karn, J.; Sieg, S.F.; et al. Exosomes derived from HIV-1-infected cells promote growth and progression of cancer via HIV TAR RNA. Nat. Commun. 2018, 9, 4585.
- Chettimada, S.; Lorenz, D.R.; Misra, V.; Dillon, S.T.; Reeves, R.K.; Manickam, C.; Morgello, S.; Kirk, G.D.; Mehta, S.H.; Gabuzda, D. Exosome markers associated with immune activation and oxidative stress in HIV patients on antiretroviral therapy. Sci. Rep. 2018, 8, 7227.
- Johnson, T.P.; Patel, K.; Johnson, K.R.; Maric, D.; Calabresi, P.A.; Hasbun, R.; Nath, A. Induction of IL-17 and nonclassical T-cell activation by HIV-Tat protein. Proc. Natl. Acad. Sci. USA 2013, 110, 13588–13593.
- Dahl, V.; Lee, E.; Peterson, J.; Spudich, S.S.; Leppla, I.; Sinclair, E.; Fuchs, D.; Palmer, S.; Price, R.W. Raltegravir treatment intensification does not alter cerebrospinal fluid HIV-1 infection or immunoactivation in subjects on suppressive therapy. J. Infect. Dis. 2011, 204, 1936–1945.
- Xiao, H.; Neuveut, C.; Tiffany, H.L.; Benkirane, M.; Rich, E.A.; Murphy, P.M.; Jeang, K.-T. Selective CXCR4 antagonism by Tat: Implications for in vivo expansion of coreceptor use by HIV-1. Proc. Natl. Acad. Sci. USA 2000, 97, 11466–11471.
- Poggi, A.; Zocchi, M.R. HIV-1 Tat triggers TGF-beta production and NK cell apoptosis that is prevented by pertussis toxin B. J. Immunol. Res. 2006, 13, 369–372.
- Westendorp, M.O.; Frank, R.; Ochsenbauer, C.; Stricker, K.; Dhein, J.; Walczak, H.; Debating, K.-M.; Krammer, P.H. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature 1995, 375, 497–500.
- Marchiò, S.; Alfano, M.; Primo, L.; Gramaglia, D.; Butini, L.; Gennero, L.; De Vivo, E.; Arap, W.; Giacca, M.; Pasqualini, R.; et al. Cell surface-associated Tat modulates HIV-1 infection and spreading through a specific interaction with gp120 viral envelope protein. Blood 2005, 105, 2802–2811.
- Weinberger, L.S.; Burnett, J.C.; Toettcher, J.E.; Arkin, A.P.; Schaffer, D.V. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 2005, 122, 169–182.
- Buonaguro, L.; Buonaguro, F.M.; Giraldo, G.; Ensoli, B. The human immunodeficiency virus type 1 Tat protein transactivates tumor necrosis factor beta gene expression through a TAR-like structure. J. Virol. 1994, 68, 2677–2682.
- Nappi, F.; Chiozzini, C.; Bordignon, V.; Borsetti, A.; Bellino, S.; Cippitelli, M.; Barillari, G.; Caputo, A.; Tyagi, M.; Giacca, M.; et al. Immobilized HIV-1 Tat protein promotes gene transfer via a transactivation-independent mechanism which requires binding of Tat to viral particles. J. Gene Med. 2009, 11, 955–965.
- Zauli, G.; Gibellini, D.; Celeghini, C.; Mischiati, C.; Bassini, A.; La Placa, M.; Capitani, S. Pleiotropic effects of immobilized versus soluble recombinant HIV-1 Tat protein on CD3-mediated activation, induction of apoptosis, and HIV-1 long terminal repeat transactivation in purified CD4+ T lymphocytes. J. Immunol. 1996, 157, 2216–2224.
- Ott, M.; Emiliani, S.; Van Lint, C.; Herbein, G.; Lovett, J.; Chirmule, N.; McCloskey, T.; Pahwa, S.; Verdin, E. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science 1997, 275, 1481–1485.
- Li, C.J.; Ueda, Y.; Shi, B.; Borodyansky, L.; Huang, L.; Li, Y.-Z.; Pardee, A.B. Tat protein induces self-perpetuating permissivity for productive HIV-1 infection. Proc. Natl. Acad. Sci. USA 1997, 94, 8116–8120.
- Gavioli, R.; Gallerani, E.; Fortini, C.; Fabris, M.; Bottoni, A.; Canella, A.; Bonaccorsi, A.; Marastoni, M.; Micheletti, F.; Cafaro, A.; et al. HIV-1 tat protein modulates the generation of cytotoxic T cell epitopes by modifying proteasome composition and enzymatic activity. J. Immunol. 2004, 173, 3838–3843.
- Campbell, G.R.; Loret, E.P. What does the structure-function relationship of the HIV-1 Tat protein teach us about developing an AIDS vaccine? Retrovirology 2009, 6, 50.
- Fanales-Belasio, E.; Moretti, S.; Nappi, F.; Barillari, G.; Micheletti, F.; Cafaro, A.; Ensoli, B. Native HIV-1 Tat protein targets monocyte-derived dendritic cells and enhances their maturation, function, and antigen-specific T cell responses. J. Immunol. 2002, 168, 197–206.
- Fanales-Belasio, E.; Moretti, S.; Fiorelli, V.; Tripiciano, A.; Cossut, M.R.P.; Scoglio, A.; Collacchi, B.; Nappi, F.; Macchia, I.; Bellino, S.; et al. HIV-1 Tat Addresses Dendritic Cells to Induce a Predominant Th1-Type Adaptive Immune Response That Appears Prevalent in the Asymptomatic Stage of Infection. J. Immunol. 2009, 182, 2888–2897.
- Li, J.C.-B.; Yim, H.C.-H.; Lau, A.S. Role of HIV-1 Tat in AIDS pathogenesis: Its effects on cytokine dysregulation and contributions to the pathogenesis of opportunistic infection. AIDS 2010, 24, 1609–1623.
- Chopard, C.; Tong, P.B.V.; Tóth, P.; Schatz, M.; Yezid, H.; Debaisieux, S.; Mettling, C.; Gross, A.; Pugnière, M.; Tu, A.; et al. Cyclophilin A enables specific HIV-1 Tat palmitoylation and accumulation in uninfected cells. Nat. Commun. 2018, 9, 2251.
- Baggaley, R.F.; White, R.G.; Boily, M.-C. HIV transmission risk through anal intercourse: Systematic review, meta-analysis and implications for HIV prevention. Leuk. Res. 2010, 39, 1048–1063.
- Vittinghoff, E.; Scheer, S.; O’Malley, P.; Colfax, G.; Holmberg, S.D.; Buchbinder, S.P. Combination antiretroviral therapy and recent declines in AIDS incidence and mortality. J. Infect. Dis. 1999, 179, 717–720.
- Jin, F.; Jansson, J.; Law, M.; Prestage, G.P.; Zablotska, I.; Imrie, J.C.; Kippax, S.C.; Kaldor, J.M.; Grulich, A.E.; Wilson, D.P. Per-contact probability of HIV transmission in homosexual men in Sydney in the era of HAART. AIDS 2010, 24, 907–913.
- Boily, M.-C.; Baggaley, R.F.; Wang, L.; Masse, B.; White, R.G.; Hayes, R.J.; Alary, M. Heterosexual risk of HIV-1 infection per sexual act: Systematic review and meta-analysis of observational studies. Lancet Infect. Dis. 2009, 9, 118–129.
- Ward, H.; Rönn, M. Contribution of sexually transmitted infections to the sexual transmission of HIV. Curr. Opin. HIV AIDS 2010, 5, 305–310.
- Quinn, T.C.; Wawer, M.J.; Sewankambo, N.; Serwadda, D.; Li, C.; Wabwire-Mangen, F.; Meehan, M.O.; Lutalo, T.; Gray, R.H. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 2000, 342, 921–929.
- Baeten, J.M.; Kahle, E.; Lingappa, J.R.; Coombs, R.W.; Delany-Moretlwe, S.; Nakku-Joloba, E.; Mugo, N.R.; Wald, A.; Corey, L.; Donnell, D.; et al. Genital HIV-1 RNA predicts risk of heterosexual HIV-1 transmission. Sci. Transl. Med. 2011, 3, 77ra29.
- Wawer, M.J.; Gray, R.H.; Sewankambo, N.K.; Serwadda, D.; Li, X.; Laeyendecker, O.; Kiwanuka, N.; Kigozi, G.; Kiddugavu, M.; Lutalo, T.; et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J. Infect. Dis. 2005, 191, 1403–1409.
- Hollingsworth, T.D.; Anderson, R.M.; Fraser, C. HIV-1 transmission, by stage of infection. J. Infect. Dis. 2008, 198, 687–693.
- Caputo, V.; Libera, M.; Sisti, S.; Giuliani, B.; Diotti, R.A.; Criscuolo, E. The initial interplay between HIV and mucosal innate immunity. Front. Immunol. 2023, 14, 1104423.
- Kariuki, S.M.; Selhorst, P.; Ariën, K.K.; Dorfman, J.R. The HIV-1 transmission bottleneck. Retrovirology 2017, 14, 22.
- Oberle, C.S.; The Swiss HIV Cohort Study (SHCS); Joos, B.; Rusert, P.; Campbell, N.K.; Beauparlant, D.; Kuster, H.; Weber, J.; Schenkel, C.D.; Scherrer, A.U.; et al. Tracing HIV-1 transmission: Envelope traits of HIV-1 transmitter and recipient pairs. Retrovirology 2016, 13, 62.
- Monini, P.; Cafaro, A.; Srivastava, I.K.; Moretti, S.; Sharma, V.A.; Andreini, C.; Chiozzini, C.; Ferrantelli, F.; Cossut, M.R.P.; Tripiciano, A.; et al. HIV-1 Tat Promotes Integrin-Mediated HIV Transmission to Dendritic Cells by Binding Env Spikes and Competes Neutralization by Anti-HIV Abs. PLoS ONE 2012, 7, e48781.
- Ward, A.B.; Wilson, I.A. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem. Sci. 2015, 40, 101–107.
- Huet, T.; Dazza, M.-C.; Brun-Vézinet, F.; Roelants, G.E.; Wain-Hobson, S. A highly defective HIV-1 strain isolated from a healthy Gabonese individual presenting an atypical Western blot. AIDS 1989, 3, 707–716.
- Anderson, E.M.; Simonetti, F.R.; Gorelick, R.J.; Hill, S.; Gouzoulis, M.A.; Bell, J.; Rehm, C.; Pérez, L.; Boritz, E.; Wu, X.; et al. Dynamic Shifts in the HIV Proviral Landscape During Long Term Combination Antiretroviral Therapy: Implications for Persistence and Control of HIV Infections. Viruses 2020, 12, 136.
- Botha, J.C.; Demirov, D.; Gordijn, C.; Katusiime, M.G.; Bale, M.J.; Wu, X.; Wells, D.; Hughes, S.H.; Cotton, M.F.; Mellors, J.W.; et al. The largest HIV-1-infected T cell clones in children on long-term combination antiretroviral therapy contain solo LTRs. mBio 2023, 14, e01116-23.
- Lorenzo-Redondo, R.; Fryer, H.R.; Bedford, T.; Kim, E.Y.; Archer, J.; Kosakovsky Pond, S.L.K.; Chung, Y.S.; Penugonda, S.; Chipman, J.G.; Fletcher, C.V.; et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 2016, 530, 51–56.
- Fletcher, C.V.; Staskus, K.; Wietgrefe, S.W.; Rothenberger, M.; Reilly, C.; Chipman, J.G.; Beilman, G.J.; Khoruts, A.; Thorkelson, A.; Schmidt, T.E.; et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. USA 2014, 111, 2307–2312.
- Sigal, A.; Kim, J.T.; Balazs, A.B.; Dekel, E.; Mayo, A.; Milo, R.; Baltimore, D. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 2011, 477, 95–98.
- Martinez-Picado, J.; Zurakowski, R.; Buzón, M.J.; Stevenson, M. Episomal HIV-1 DNA and its relationship to other markers of HIV-1 persistence. Retrovirology 2018, 15, 15.
- Lassen, K.G.; Ramyar, K.X.; Bailey, J.R.; Zhou, Y.; Siliciano, R.F. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006, 2, e68.
- Wietgrefe, S.W.; Anderson, J.; Duan, L.; Southern, P.J.; Zuck, P.; Wu, G.; Howell, B.J.; Reilly, C.; Kroon, E.; Chottanapund, S.; et al. Initial productive and latent HIV infections originate in vivo by infection of resting T cells. J. Clin. Investig. 2023, 133, e171501.
- Weinberger, L.S.; Dar, R.D.; Simpson, M.L. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat. Genet. 2008, 40, 466–470.
- Razooky, B.S.; Pai, A.; Aull, K.; Rouzine, I.M.; Weinberger, L.S. A Hardwired HIV Latency Program. Cell 2015, 160, 990–1001.
- Zerbato, J.M.; Purves, H.V.; Lewin, S.R.; Rasmussen, T.A. Between a shock and a hard place: Challenges and developments in HIV latency reversal. Curr. Opin. Virol. 2019, 38, 1–9.
- Moranguinho, I.; Valente, S.T. Block-And-Lock: New Horizons for a Cure for HIV-1. Viruses 2020, 12, 1443.
- Ensoli, B.; Bellino, S.; Tripiciano, A.; Longo, O.; Francavilla, V.; Marcotullio, S.; Cafaro, A.; Picconi, O.; Paniccia, G.; Scoglio, A.; et al. Therapeutic immunization with HIV-1 Tat reduces immune activation and loss of regulatory T-cells and improves immune function in subjects on HAART. PLoS ONE 2010, 5, e13540.
- Mediouni, S.; Darque, A.; Baillat, G.; Ravaux, I.; Dhiver, C.; Tissot-Dupont, H.; Mokhtari, M.; Moreau, H.; Tamalet, C.; Brunet, C.; et al. Antiretroviral therapy does not block the secretion of the human immunodeficiency virus tat protein. Infect. Disord. Drug Targets 2012, 12, 81–86.
- Nicoli, F.; Gallerani, E.; Sforza, F.; Finessi, V.; Chachage, M.; Geldmacher, C.; Cafaro, A.; Ensoli, B.; Caputo, A.; Gavioli, R. The HIV-1 Tat protein affects human CD4+ T-cell programing and activation, and favors the differentiation of naïve CD4+ T cells. AIDS 2018, 32, 575–581.
- Shan, L.; Deng, K.; Gao, H.; Xing, S.; Capoferri, A.A.; Durand, C.M.; Rabi, S.A.; Laird, G.M.; Kim, M.; Hosmane, N.N.; et al. Transcriptional reprogramming during effector-to-memory transition renders CD4+ T cells permissive for latent HIV-1 infection. Immunity 2017, 47, 766–775.
- Zauli, G.; Gibellini, D.; Caputo, A.; Bassini, A.; Negrini, M.; Monne, M.; Mazzoni, M.; Capitani, S. The human immunodeficiency virus type-1 Tat protein upregulates Bcl-2 gene expression in Jurkat T-cell lines and primary peripheral blood mononuclear cells. Blood 1995, 86, 3823–3834.
- Ensoli, B.; Moretti, S.; Borsetti, A.; Maggiorella, M.T.; Buttò, S.; Picconi, O.; Tripiciano, A.; Sgadari, C.; Monini, P.; Cafaro, A. New insights into pathogenesis point to HIV-1 Tat as a key vaccine target. Arch. Virol. 2021, 166, 2955–2974.




