Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | SONG Hyoung-Kyu | -- | 1660 | 2024-02-02 04:00:30 | | | |
| 2 | Catherine Yang | Meta information modification | 1660 | 2024-02-02 07:04:43 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Danish, S.; Khan, A.; Dang, L.M.; Alonazi, M.; Alanazi, S.; Song, H.; Moon, H. Metaverse Applications in Bioinformatics. Encyclopedia. Available online: https://encyclopedia.pub/entry/54659 (accessed on 07 February 2026).
Danish S, Khan A, Dang LM, Alonazi M, Alanazi S, Song H, et al. Metaverse Applications in Bioinformatics. Encyclopedia. Available at: https://encyclopedia.pub/entry/54659. Accessed February 07, 2026.
Danish, Sufyan, Asfandyar Khan, L. Minh Dang, Mohammed Alonazi, Sultan Alanazi, Hyoung-Kyu Song, Hyeonjoon Moon. "Metaverse Applications in Bioinformatics" Encyclopedia, https://encyclopedia.pub/entry/54659 (accessed February 07, 2026).
Danish, S., Khan, A., Dang, L.M., Alonazi, M., Alanazi, S., Song, H., & Moon, H. (2024, February 02). Metaverse Applications in Bioinformatics. In Encyclopedia. https://encyclopedia.pub/entry/54659
Danish, Sufyan, et al. "Metaverse Applications in Bioinformatics." Encyclopedia. Web. 02 February, 2024.
Copy Citation
Bioinformatics and genomics are driving a healthcare revolution, particularly in the domain of drug discovery for anticancer peptides (ACPs). The integration of artificial intelligence (AI) has transformed healthcare, enabling personalized and immersive patient care experiences. These advanced technologies, coupled with the power of bioinformatics and genomic data, facilitate groundbreaking developments. The precise prediction of ACPs from complex biological sequences remains an ongoing challenge in the genomic area.
artificial intelligence
machine learning
bioinformatics
metaverse
1. Introduction
Healthcare is paramount for global well-being and crucial in fostering services and contributing to economic growth [1]. Technological advancements such as blockchain, augmented reality (AR), and virtual reality (VR) have transformed patient–physician interactions [2]. At the same time, the healthcare system in the metaverse integrates AI, telepresence, digital twinning, and blockchain to facilitate affordable treatments and improve patient outcomes [3]. Figure 1 illustrates the different applications of metaverse in healthcare. The metaverse also delivers personalized, immersive care by integrating real and virtual worlds [4]. Cancer remains a major global health threat, causing millions of deaths annually [5]. Although conventional treatments such as radiotherapy, target therapy, and chemotherapy can effectively target cancer cells, they also impose significant financial burdens and affect healthy cells [6]. To address these challenges, there is a growing need for an automated system to accurately identify ACPs from complex biological sequences, ensuring timely diagnosis and management before the condition worsens. Peptides have been recognized as effective cancer treatments due to their minimal impact on normal physiological activities and have been extensively studied in preclinical research for multiple purposes, including cardiovascular disease, diabetes, and other types of cancer [7]. In studies by Ijaz et al. [8], an early cervical cancer prediction model was developed using the CCPM. Saha et al. [9] studied different prognostic characters of GLS and GLS2 in cancer, showcasing their variance impact on clinical results across diverse cancer types. Figure 2 illustrates different cancer treatment options that involve targeting peptides.
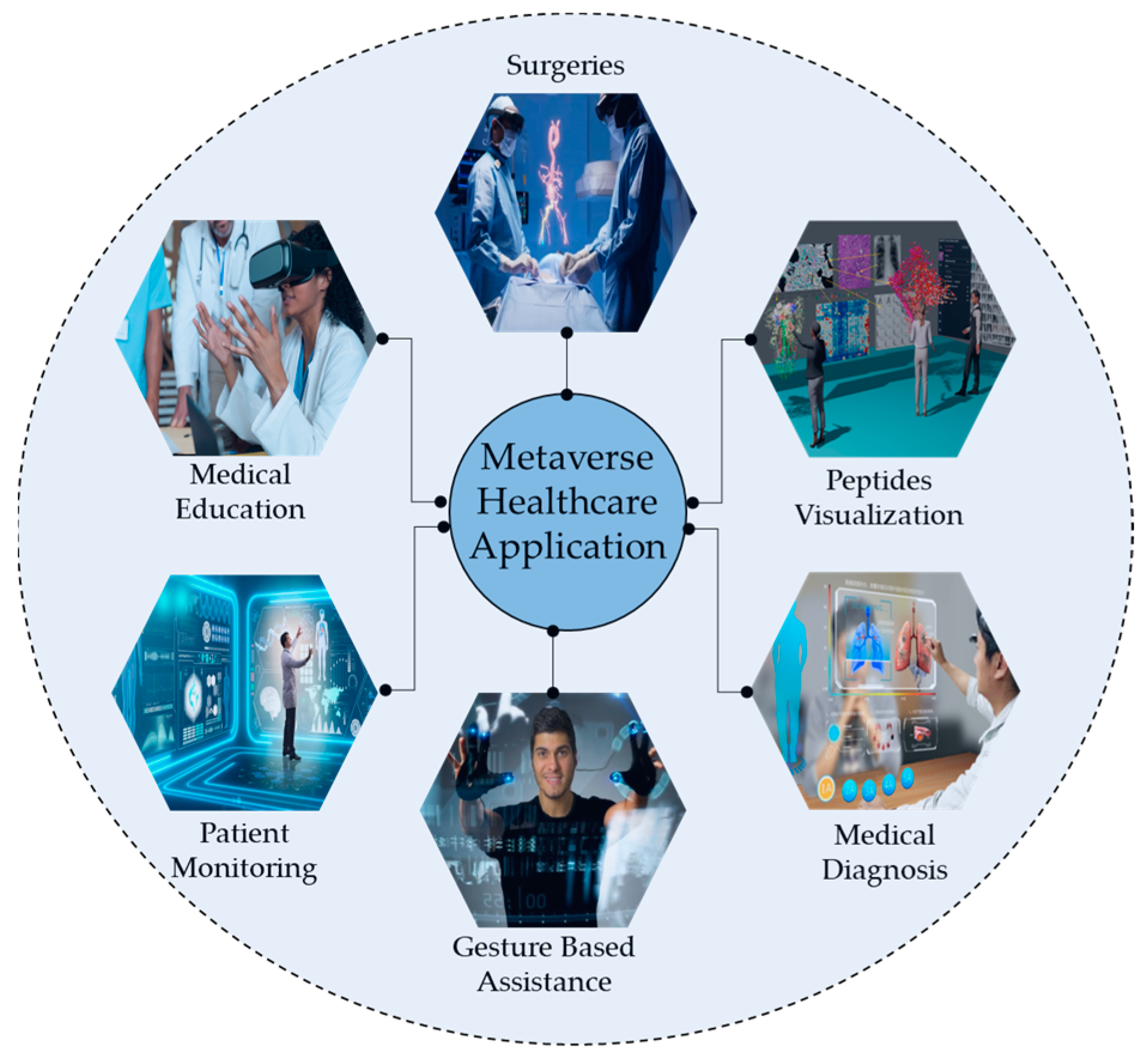
Figure 1. Several applications of the metaverse in healthcare and education for peptides visualization.
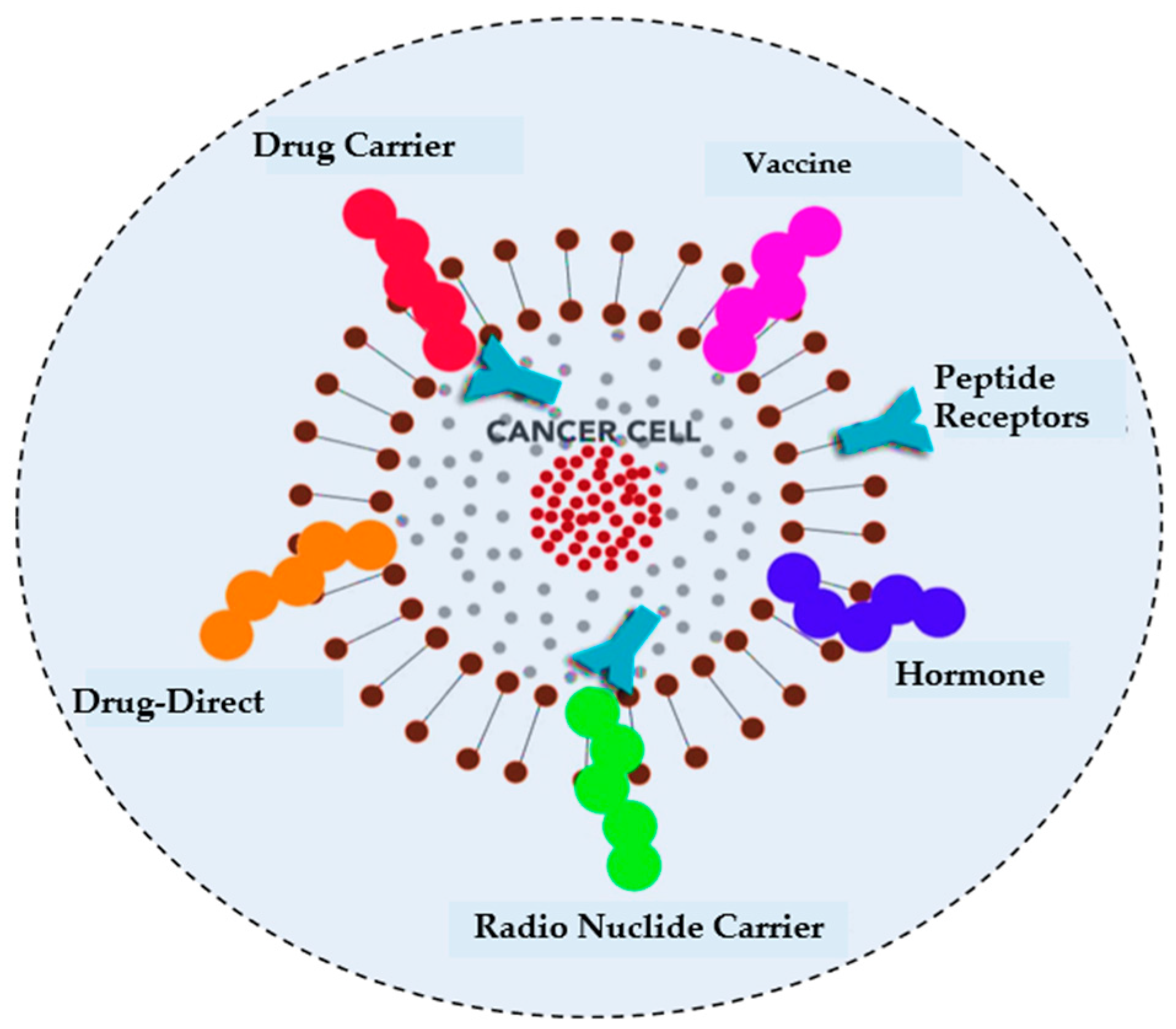
Figure 2. Peptide sequences offer a range of potential treatment options for cancer, which can be explored and evaluated for their effectiveness.
An ACP is a small sequence typically containing fewer than 50 amino acids [10]. A new direction for cancer treatment has been opened by determining ACPs because they have cationic properties that can easily target cancer-affected cells without interacting with normal cells [11]. However, ACPs have some potential drawbacks in the drug development process, i.e., high production costs and low stability [12]; despite this, they have some unique merits. ACPs are biological substances that naturally exist in a living organism; hence, they are more harmless than synthetic drugs and have better efficacy [13]. Over the last decade, peptide-based techniques have been used in numerous clinical trials for disease prevention and treatment [14], including heart disease, diabetes, and tumors. Experimental identification process of ACPs is very costly and time-consuming which is rarely utilized in clinical practice.
2. Metaverse Applications in Bioinformatics
Several intelligent models for identifying ACPs have been proposed by different researchers. For example, a silico model was proposed by [15], where AAC and binary profile-based information are used as a feature extraction method, obtaining 91.44% accuracy for binary classification. Similarly, Hajisharifi et al. [16] developed a novel approach, where PseAAC and local alignment kernel techniques are explored for feature extraction and achieved the outcomes of 89.7% and 83.82%, respectively. Next, this work is further boosted by Chen et al. [17], who introduced a sequence-based model that utilizes g-gap dipeptide composition and cross-validation techniques for superior performance; therefore, 94.77% accuracy was obtained for the discrimination of peptides. Akbar et al. [18] utilized an ensemble classifier for the discrimination of various peptides to obtain robust features: amphibolic PseAAC, reduced AAC, and g-gap DPC were considered, and a 96.45% classification score was obtained. Xu et al. [19] developed a model that considers ACP composition by incorporating binary encoding and physicochemical properties to obtain meaningful information but the performance is 91.86% which is a low discrimination score as compared to the other models. The sequence-based models have successfully identified novel ACP but due to technological advancement substantial work has been presented in the metaverse specifically for human healthcare.
Recent research has explored the metaverse’s potential in healthcare, covering various applications and challenges. Bansal et al. [20] outlined its uses in immersive training experiences, telemedicine, and clinical care, emphasizing extended reality (XR) technologies and hardware. Another study [21] focused on ophthalmology, utilizing VR and digital twins (DTs) for personalized care. Next, Ali et al. [22] proposed a secure metaverse architecture with blockchain and eXplainable AI (XAI) for transparent disease prediction. Moreover, Razdan and Sharma [23] proposed a comprehensive metaverse architecture, emphasizing big data processing and security measures, while a reference architecture with distinct layers is also presented for metaverse applications, ensuring robust governance and quality of service.
Moreover, some researchers have used alternative methods such as the generalized chaos game representation [24] short-term memory models with binary profile features and k-mer sparse matrices using two novel benchmark datasets (ACP740 and ACP240) [25]. Chen et al. introduced an ACP-DA model, composed of DL and augmentation approaches, resulting 82.03% and 88.33% on ACP740 and ACP240 datasets, respectively [26]. Next, Ye et al. [27] proposed an ensemble learning model using numerous datasets [16][28] with which 95.4% and 92.4% accuracies were obtained after comprehensive experiments. Moreover, in [29] ETree classfier and AAC feature extractor are used for the discrimination of different biological sequences. Akbar et al. also explored ensemble classification for ACPs and attained 96.45% accuracy using an evolutionary genetic algorithm (iACP-GAEnsC) [18]. Shahid Akbar et al. developed cACP-DeepGram by utilizing three statistical feature representation schemes and achieved satisfactory outcomes of 96.94% accuracy [30]. Shahid Akbar et al. also introduced cACP, a discriminatory computational technique employing diverse statistical feature representation schemes, and feature selection PCA, and achieved a tremendous classification score of 96.91% [31]. Shahid Akbar et al. made a notable contribution with their proposed cACP-2LFS, a novel sequential discriminative model designed for ACP classification. In addition, their model utilizes the K-space amino acid pair (KSAAP) for extracting correlated descriptors and incorporates a two-level feature selection (2LFS) method, resulting in impressive accuracy rates of 94.11% and 93.72% using independent and LEE datasets, respectively. These achievements highlight its potential applications in the fields of medicine, proteomics, and research academia [32]. Despite these attempts, some recent studies have explored DL models for the efficient classification of ACPs from complex biological sequences. Ahmed et al. presented a novel multi-head deep CNN by extracting discriminative physicochemical properties from diverse peptides, followed by evolutionary-based features, leading 83.0%, 86.0%, and 91.0% accuracies levels on ACP-240, ACP-740, and a combination both datasets, respectively [33]. Hulam et al. proposed DL-based method that uses dipeptide deviation from the expected mean (DDE) as a feature extractor for precise predictions of ACPs, outperforming the 85.88% and 84.8% accuracies score using ACP240 and ACP740 datasets [34]. Advanced DL methods such as MLACP 2.0 [35] incorporate seven distinctive classifiers and multiple feature encoding methods. In addition, other mainstream approaches applied neural networks and multitask learning, where they incorporate hybrid sequencing information [36][37].
Recently, ensemble classifiers have gained popularity in ACP classification, where a decision has been made based on the majority vote of multiple machine learning algorithms, such as an SVM, an RF, and AdaBoost. Ensemble classifiers leverage individual algorithms for their strengths and mitigate their weaknesses for improved performance. The results obtained with this approach are superior to those from a single algorithm [38]. Multi-algorithm ensemble clustering aggregates the results of different clustering algorithms to produce more accurate and robust consensus clustering results. Ensemble clustering has many applications, such as image segmentation and gene expression analysis, and can make the clustering process more robust and accurate by embracing ensemble methods [39]. Various integrated methods have been used to evaluate regression [40][41] and classification [42][43] problems in the literature, including weighted averaging [44], weighted voting [45], simple averaging [46], and majority voting [47]. Other techniques can also be used for protein analysis, such as combining individual sequence-based models to generate a final prediction. To predict the biological functions and properties of peptides and proteins, sequence-based statistical predictors require machine learning algorithms and feature extraction techniques. Predictors of protein–protein interactions, protein subcellular localization, and ACPs have shown promising results in several applications, and developing new therapeutics requires the development of such predictors [48].
Several computational methods have been developed to classify ACPs and non-ACPs, but there is still room for improvement. To overcome the research gap mentioned later in the paper, this study proposed a new collection of computational approaches to improve the classification performance for new biological sequences. The suggested system could lead to a better understanding of ACPs and aid in the development of new therapeutics.
-
The imbalanced nature of datasets in anticancer peptide classification poses a significant hurdle for many existing machine learning methods. Biased models can emerge, favoring classes with a higher number of instances, thereby compromising the model’s ability to accurately identify and classify less-represented classes. This imbalance issue is particularly critical in the context of anticancer peptides, where a thorough understanding of diverse instances is crucial for effective classification.
-
Prevailing methods often lean towards simplicity, employing single-feature extractions and classifiers. While this simplicity aids in model interpretability and computational efficiency, it may fall short of capturing the intricate and nuanced patterns inherent in anticancer peptides. The complex nature of these peptides demands more sophisticated approaches that can discern subtle variations and relationships within the data, enhancing the model’s discriminatory power.
-
Current strategies aimed at enhancing classification accuracy often resort to fusion techniques. While these techniques offer potential improvements, they may inadvertently introduce homogeneity in the utilized information, leading to limiting the model’s ability to discern diverse and subtle characteristics crucial for accurate anticancer peptide classification. Striking a balance between fusion for improved accuracy and preserving the diversity of information remains a key challenge in developing robust models.
-
Some machine learning-based methods in anticancer peptide classification may exhibit a tendency to overlook the expansive landscape of feature extraction models and selection techniques. A more comprehensive exploration of this landscape is imperative to ensure that potentially more effective approaches are not neglected. The diversity among anticancer peptides demands a thorough examination of various feature extraction methods and selection techniques to uncover the most suitable combination for accurate classification.
References
- Arora, T.; Grey, I. Health behaviour changes during COVID-19 and the potential consequences: A mini-review. J. Health Psychol. 2020, 25, 1155–1163.
- Maki, O.; Alshaikhli, M.; Gunduz, M.; Naji, K.K.; Abdulwahed, M. Development of digitalization road map for healthcare facility management. IEEE Access 2022, 10, 14450–14462.
- Kapoor, A.; Guha, S.; Das, M.K.; Goswami, K.C.; Yadav, R. Digital Healthcare: The Only Solution for Better Healthcare during COVID-19 Pandemic? Elsevier: Amsterdam, The Netherlands, 2020; Volume 72, pp. 61–64.
- Alshamrani, M. IoT and artificial intelligence implementations for remote healthcare monitoring systems: A survey. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 4687–4701.
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48.
- Harris, F.; Dennison, S.R.; Singh, J.; Phoenix, D.A. On the selectivity and efficacy of defense peptides with respect to cancer cells. Med. Res. Rev. 2013, 33, 190–234.
- Karbalaeemohammad, S.; Naderi-Manesh, H. Two novel anticancer peptides from Aurein1. 2. Int. J. Pept. Res. Ther. 2011, 17, 159–164.
- Ijaz, M.F.; Attique, M.; Son, Y. Data-driven cervical cancer prediction model with outlier detection and over-sampling methods. Sensors 2020, 20, 2809.
- Saha, S.K.; Islam, S.R.; Abdullah-Al-Wadud, M.; Islam, S.; Ali, F.; Park, K.S. Multiomics analysis reveals that GLS and GLS2 differentially modulate the clinical outcomes of cancer. J. Clin. Med. 2019, 8, 355.
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2008, 1778, 357–375.
- Mader, J.S.; Hoskin, D.W. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert Opin. Investig. Drugs 2006, 15, 933–946.
- Gaspar, D.; Veiga, A.S.; Castanho, M.A. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294.
- Huang, Y.; Feng, Q.; Yan, Q.; Hao, X.; Chen, Y. Alpha-helical cationic anticancer peptides: A promising candidate for novel anticancer drugs. Mini Rev. Med. Chem. 2015, 15, 73–81.
- Thundimadathil, J. Cancer treatment using peptides: Current therapies and future prospects. J. Amino Acids 2012, 2012, 967347.
- Tyagi, A.; Kapoor, P.; Kumar, R.; Chaudhary, K.; Gautam, A.; Raghava, G. In silico models for designing and discovering novel anticancer peptides. Sci. Rep. 2013, 3, 2984.
- Hajisharifi, Z.; Piryaiee, M.; Beigi, M.M.; Behbahani, M.; Mohabatkar, H. Predicting anticancer peptides with Chou′ s pseudo amino acid composition and investigating their mutagenicity via Ames test. J. Theor. Biol. 2014, 341, 34–40.
- Chen, W.; Ding, H.; Feng, P.; Lin, H.; Chou, K.-C. iACP: A sequence-based tool for identifying anticancer peptides. Oncotarget 2016, 7, 16895.
- Akbar, S.; Hayat, M.; Iqbal, M.; Jan, M.A. iACP-GAEnsC: Evolutionary genetic algorithm based ensemble classification of anticancer peptides by utilizing hybrid feature space. Artif. Intell. Med. 2017, 79, 62–70.
- Xu, L.; Liang, G.; Wang, L.; Liao, C. A novel hybrid sequence-based model for identifying anticancer peptides. Genes 2018, 9, 158.
- Bansal, G.; Rajgopal, K.; Chamola, V.; Xiong, Z.; Niyato, D. Healthcare in metaverse: A survey on current metaverse applications in healthcare. IEEE Access 2022, 10, 119914–119946.
- Tan, T.F.; Li, Y.; Lim, J.S.; Gunasekeran, D.V.; Teo, Z.L.; Ng, W.Y.; Ting, D.S. Metaverse and virtual health care in ophthalmology: Opportunities and challenges. Asia-Pac. J. Ophthalmol. 2022, 11, 237–246.
- Ali, S.; Abdullah; Armand, T.P.T.; Athar, A.; Hussain, A.; Ali, M.; Yaseen, M.; Joo, M.-I.; Kim, H.-C. Metaverse in healthcare integrated with explainable ai and blockchain: Enabling immersiveness, ensuring trust, and providing patient data security. Sensors 2023, 23, 565.
- Razdan, S.; Sharma, S. Internet of medical things (IoMT): Overview, emerging technologies, and case studies. IETE Tech. Rev. 2022, 39, 775–788.
- Ge, L.; Liu, J.; Zhang, Y.; Dehmer, M. Identifying anticancer peptides by using a generalized chaos game representation. J. Math. Biol. 2019, 78, 441–463.
- Yi, H.-C.; You, Z.-H.; Zhou, X.; Cheng, L.; Li, X.; Jiang, T.-H.; Chen, Z.-H. ACP-DL: A deep learning long short-term memory model to predict anticancer peptides using high-efficiency feature representation. Mol. Ther. Nucleic Acids 2019, 17, 1–9.
- Chen, X.-g.; Zhang, W.; Yang, X.; Li, C.; Chen, H. Acp-da: Improving the prediction of anticancer peptides using data augmentation. Front. Genet. 2021, 12, 698477.
- Ge, R.; Feng, G.; Jing, X.; Zhang, R.; Wang, P.; Wu, Q. Enacp: An ensemble learning model for identification of anticancer peptides. Front. Genet. 2020, 11, 760.
- Boopathi, V.; Subramaniyam, S.; Malik, A.; Lee, G.; Manavalan, B.; Yang, D.-C. mACPpred: A support vector machine-based meta-predictor for identification of anticancer peptides. Int. J. Mol. Sci. 2019, 20, 1964.
- Agrawal, P.; Bhagat, D.; Mahalwal, M.; Sharma, N.; Raghava, G.P. AntiCP 2.0: An updated model for predicting anticancer peptides. Brief. Bioinform. 2021, 22, bbaa153.
- Akbar, S.; Hayat, M.; Tahir, M.; Khan, S.; Alarfaj, F.K. cACP-DeepGram: Classification of anticancer peptides via deep neural network and skip-gram-based word embedding model. Artif. Intell. Med. 2022, 131, 102349.
- Akbar, S.; Rahman, A.U.; Hayat, M.; Sohail, M. cACP: Classifying anticancer peptides using discriminative intelligent model via Chou’s 5-step rules and general pseudo components. Chemom. Intell. Lab. Syst. 2020, 196, 103912.
- Akbar, S.; Hayat, M.; Tahir, M.; Chong, K.T. cACP-2LFS: Classification of anticancer peptides using sequential discriminative model of KSAAP and two-level feature selection approach. IEEE Access 2020, 8, 131939–131948.
- Ahmed, S.; Muhammod, R.; Khan, Z.H.; Adilina, S.; Sharma, A.; Shatabda, S.; Dehzangi, A. ACP-MHCNN: An accurate multi-headed deep-convolutional neural network to predict anticancer peptides. Sci. Rep. 2021, 11, 23676.
- Ghulam, A.; Ali, F.; Sikander, R.; Ahmad, A.; Ahmed, A.; Patil, S. ACP-2DCNN: Deep learning-based model for improving prediction of anticancer peptides using two-dimensional convolutional neural network. Chemom. Intell. Lab. Syst. 2022, 226, 104589.
- Park, H.W.; Pitti, T.; Madhavan, T.; Jeon, Y.-J.; Manavalan, B. MLACP 2.0: An updated machine learning tool for anticancer peptide prediction. Comput. Struct. Biotechnol. J. 2022, 20, 4473–4480.
- Chen, J.; Cheong, H.H.; Siu, S.W. xDeep-AcPEP: Deep learning method for anticancer peptide activity prediction based on convolutional neural network and multitask learning. J. Chem. Inf. Model. 2021, 61, 3789–3803.
- Sun, M.; Yang, S.; Hu, X.; Zhou, Y. ACPNet: A deep learning network to identify anticancer peptides by hybrid sequence information. Molecules 2022, 27, 1544.
- Onan, A.; Korukoğlu, S.; Bulut, H. A multiobjective weighted voting ensemble classifier based on differential evolution algorithm for text sentiment classification. Expert Syst. Appl. 2016, 62, 1–16.
- Dimitriadou, E.; Weingessel, A.; Hornik, K. A cluster ensembles framework. In Design and Application of Hybrid Intelligent Systems; IOS Press: Amsterdam, The Netherlands, 2003.
- Khan, S.U.; Haq, I.U.; Khan, Z.A.; Khan, N.; Lee, M.Y.; Baik, S.W. Atrous Convolutions and Residual GRU Based Architecture for Matching Power Demand with Supply. Sensors 2021, 21, 7191.
- Khan, S.U.; Khan, N.; Ullah, F.U.M.; Kim, M.J.; Lee, M.Y.; Baik, S.W. Towards intelligent building energy management: AI-based framework for power consumption and generation forecasting. Energy Build. 2023, 279, 112705.
- Hussain, A.; Khan, S.U.; Rida, I.; Khan, N.; Baik, S.W. Human Centric Attention with Deep Multiscale Feature Fusion Framework for Activity Recognition in Internet of Medical Things. Inf. Fusion 2023, 102211.
- Hussain, A.; Khan, S.U.; Khan, N.; Shabaz, M.; Baik, S.W. AI-driven behavior biometrics framework for robust human activity recognition in surveillance systems. Eng. Appl. Artif. Intell. 2024, 127, 107218.
- Hussain, A.; Amin, S.U.; Lee, H.; Khan, A.; Khan, N.F.; Seo, S. An Automated Chest X-Ray Image Analysis for Covid-19 and Pneumonia Diagnosis using Deep Ensemble Strategy. IEEE Access 2023, 11, 97207–97220.
- Ekbal, A.; Saha, S. Weighted vote based classifier ensemble selection using genetic algorithm for named entity recognition. In Proceedings of the International Conference on Application of Natural Language to Information Systems, Cardiff, UK, 23–25 June 2010; pp. 256–267.
- Opitz, D.; Maclin, R. Popular ensemble methods: An empirical study. J. Artif. Intell. Res. 1999, 11, 169–198.
- Hayat, M.; Khan, A.; Yeasin, M. Prediction of membrane proteins using split amino acid and ensemble classification. Amino Acids 2012, 42, 2447–2460.
- Chen, W.; Ding, H.; Zhou, X.; Lin, H.; Chou, K.-C. iRNA (m6A)-PseDNC: Identifying N6-methyladenosine sites using pseudo dinucleotide composition. Anal. Biochem. 2018, 561, 59–65.
More
Information
Subjects:
Biodiversity Conservation
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
709
Revisions:
2 times
(View History)
Update Date:
02 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




