Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Stefano Raffaele Giannubilo | -- | 3231 | 2024-01-31 21:38:53 | | | |
| 2 | Lindsay Dong | Meta information modification | 3231 | 2024-02-01 01:31:12 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Giannubilo, S.R.; Cecati, M.; Marzioni, D.; Ciavattini, A. Circulating miRNAs and Preeclampsia. Encyclopedia. Available online: https://encyclopedia.pub/entry/54614 (accessed on 08 February 2026).
Giannubilo SR, Cecati M, Marzioni D, Ciavattini A. Circulating miRNAs and Preeclampsia. Encyclopedia. Available at: https://encyclopedia.pub/entry/54614. Accessed February 08, 2026.
Giannubilo, Stefano Raffaele, Monia Cecati, Daniela Marzioni, Andrea Ciavattini. "Circulating miRNAs and Preeclampsia" Encyclopedia, https://encyclopedia.pub/entry/54614 (accessed February 08, 2026).
Giannubilo, S.R., Cecati, M., Marzioni, D., & Ciavattini, A. (2024, January 31). Circulating miRNAs and Preeclampsia. In Encyclopedia. https://encyclopedia.pub/entry/54614
Giannubilo, Stefano Raffaele, et al. "Circulating miRNAs and Preeclampsia." Encyclopedia. Web. 31 January, 2024.
Copy Citation
miRNAs are single-stranded non-coding RNAs, 20–24 nt long, which control mRNA expression. Changes in miRNA expression can induce a variation in the relative mRNA level and influence cellular homeostasis, and the strong presence of miRNAs in all body fluids has made them useful biomarkers of several diseases. Preeclampsia is a multifactorial disease, but the etiopathogenesis remains unclear. The functions of trophoblasts, including differentiation, proliferation, migration, invasion and apoptosis, are essential for a successful pregnancy.
miRNA
preeclampsia
trophoblast
placenta
epigenetic
1. Introduction
The placenta is a transitory organ, necessary for development in utero in humans and mammals [1][2][3]. The important role of this organ is highlighted when normal placental development is compromised, leading to important pregnancy complications, such as preeclampsia (PE) [4][5], fetal growth restriction (FGR) [6][7][8], gestational diabetes mellitus (GDM) [9], preterm birth (PTB) [9][10] and gestational trophoblastic disease (GTD) [11][12].
Preeclampsia is a hypertensive disorder associated with pregnancy that affects 5–7% of pregnancies and is responsible for over 70,000 maternal deaths each year [13]. Parenteral or oral drug therapies are available and administered to preeclamptic women to stabilize the mother and fetus and reduce the risk of failure of several maternal organs, such as the liver, kidney and brain [14]. Nevertheless, to date, delivery at an optimal time is the only reliable treatment for preeclampsia [15].
Reduced arterial compliance and peripheral vasoconstriction characterize the preeclamptic condition from the early stages [16]; however, the exact pathophysiology of preeclampsia is still unknown. Probably, the initial defect in placentation and vascularization in the placenta bed due to poor trophoblast invasion of the spiral arteries [17][18][19] leads to an inappropriate activation of the immune system [20], as it occurs in recurrent miscarriage [21]. It is known that oxidative stress and inflammation are involved in endothelial dysfunction [22][23][24][25][26][27], a key process underlying several diseases including preeclampsia [28][29][30][31][32]. Hypoxia due to inadequate spiral artery remodeling compromises the placental endothelial function [33], which results in an oxidative stress condition [34][35], made worse by an increased production of nitric oxide [36][37]. Continued oxidative stress leads to chronic inflammation, which induces a premature senescence of preeclamptic placental tissue, as suggested by the short length of telomeric DNA [38].
The inability of preeclamptic placental tissue to regain a condition of equilibrium leads to an inevitable dysregulation of several metabolic pathways [39][40]. The understanding of this pathology is still elusive, although different studies tried to identify candidate genes involved in preeclampsia onset, measuring their expression in placental tissue [41][42].
Since preeclampsia can cause maternal and neonatal morbidity and mortality [43], several studies have been focused on finding efficient markers able to predict preeclampsia onset to improve the treatment of these pregnancies [5][44][45][46][47][48][49].
The presence of circulating RNAs released from many body tissues, the placenta included, offers a potential tool to indirectly observe any pathologies in real time, starting from the onset and during development. In this way, it possible to associate the physiological changes in the circulating RNA variation level in preeclamptic subjects with respect to healthy ones [50]. Circulating RNAs contain fragments that are transcribed but not translated called non-coding RNAs (ncRNAs). Bioinformatic analysis estimates that a quote of the human genome equal to 75–80% is transcribed into RNA molecules. Nevertheless, just 2% of transcribed RNA molecules are converted into proteins [51]. Usually, ncRNAs are divided into four different classes: PIWI-interacting RNAs (piRNAs), circular RNAs (circRNAs), long ncRNAs (lncRNAs) with a length over 200 nucleotides and small ncRNAs (known also as miRNAs) with a length under 200 nucleotides.
The name piRNAs was derived from their ability to bind to PIWI family proteins. They are between 24 and 30 nucleotides in length, and they are known to participate in the regulation of cell proliferation, apoptosis and the cell cycle [52]. LncRNAs and circRNAs are more than 200 nucleotides in length but have different shapes. As suggested by their name, lncRNAs are linear unlike circRNAs, which are ringlike. Both lncRNAs and circRNAs are transcribed from regions located at exons, introns, intergenic regions or 5′/3′-untranslational regions and interact with DNA, RNA and proteins by folding themselves into complicated second structures [53]. LncRNAs and circRNAs can modulate gene expression through several strategies. They can prevent mRNA degradation, avoiding the miRNA, by bind to the targeted mRNA. They can regulate gene expression by modulating transcription factors to tie to promoters [54]. In addition, they can exert a regulative action on several signaling pathways, acting as a scaffold to regulate the interactions between proteins [55]. miRNAs are small ncRNAs, which control mRNA expression. Changes in miRNA expression can induce a variation in the relative mRNA level and influence cellular homeostasis [56][57].
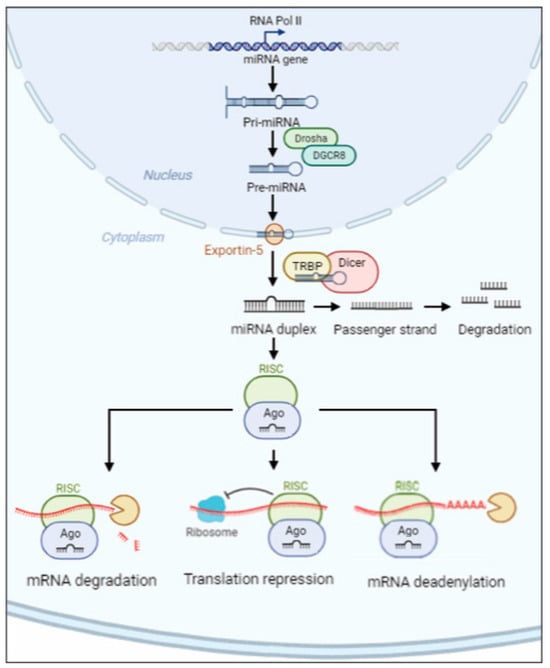
2. miRNA Biogenesis, Mechanisms of Export and Annotation Criteria
miRNAs are single-stranded non-coding RNAs, 20–24 nt long, which originate from the primary miRNA transcript (pri-miRNA) [58]. In the nucleus, an RNase III endonuclease (named Drosha) combined to DGCR8 (DiGeorge critical region 8) protein (named Pasha) begins the maturation of the pri-miRNA, liberating a ~60–70-nt stem loop intermediate (pre-miRNA). The complex Exportin-5–Ran-GTP moves the pre-miRNA precursor hairpin from the nucleus to the cytoplasm. The RNase Dicer associated with the transactivation response element RNA-binding protein (TRBP) digests the pre-miRNA to find its mature length. Finally, the passenger strand of the mature miRNA is digested, while the functional strand is loaded with Argonaute (Ago2) proteins into the RNA-induced silencing complex (RISC). RISC silences the mRNA target through mRNA deadenylation, translational repression or cleavage driven by the functional strand of the mature miRNA, which recognizes 3′-untranslated region (3′-UTR) [59][60]. Although the 3′UTR sequence is the most prevalent site for the recognition of mRNA target, miRNAs can also interact with 5′-untranslated region (5′-UTR) [61] (Figure 1).

Figure 1. Schematic overview of miRNA biosynthesis. The miRNA pathway produces pri-miRNA transcripts from miRNA genes encoded in exonic, intronic or intergenic regions. In the nucleus, Drosha and DGCR8 digest the pri-miRNA into pre-miRNA. The pre-miRNA is driven into the cytoplasm by the complex Exportin-5–Ran-GTP. Once the mature length is obtained, the functional strand is loaded with Ago2 protein into RISC complex which silences the mRNA’s target through mRNA deadenylation, translational repression or degradation.
In recent years, circulating miRNAs have gained the attention of activity research because they function in the same manner as classical signaling, made of growth factors, hormones and cytokines [62]. Circulating miRNAs are surprisingly stable and able to carry out their suppressive function against mRNA target into the recipient cells.
Experimental data demonstrated that miRNAs can be released outside the cell under four different forms: (a) miRNAs combined to Argonaute2 protein (Ago2) [63][64], (b) miRNAs tied to RNA-binding protein nucleophosmin (NPM1) [65], (c) miRNAs tied to high-density lipoprotein (HDL) [66], and (d) miRNAs closed within extracellular vesicles (EVs) such as exosomes and microvesicles.
The circulating miRNAs detected in plasma, and not included in EV, are mostly bound to proteins. The Ago2 protein not only takes part in miRNA maturation via RISC complex but also exercises a shielding effect on extracellular miRNAs against RNases. Particularly, Arroyo et al. suggested that the entire Ago2–miRNA complex may be able to modulate gene expression in recipient cells [64].
Like Ago2, NPM1 also has a protective role for circulating miRNAs. Wang et al. hypothesized that NPM1 is involved not only in the packaging and release of miRNAs outside the cell but that it remains bound to them in the peripheral circulation [65]. Nevertheless, the biological function of circulating miRNAs associated with both Ago2 and NPM1 proteins in pregnancy and pregnancy-related diseases is still unknown.
HDL is one of the five different types of lipoproteins, including chylomicrons, very-low-density lipoprotein (VLDL), intermediate-density lipoprotein (IDL) and low-density lipoprotein (LDL). HDLs have a micellular constitution and organization, like most polar lipids, and a mass composed of 50–60% proteins, mostly represented by apolipoproteins A-I (APOA1) [65]. It is widely known that HDLs also vehicle miRNAs, not only lipids and proteins [66]. The load of miRNAs in HDFs is different in composition and concentration between normal and pathological subjects and equally attributable to a specific disease [67][68][69]. It has been demonstrated that the miRNAs-HDL complex is internalized only if the recipient cell expresses the scavenger receptor BI (SR-BI) [66].
EVs are small lipid particles released from most human cell types, both malignant and healthy [70]. Interestingly, EVs have also been suggested as a promising class of therapeutic agents [71]. EVs mediate intercellular communication by moving heterogeneous molecules (i.e., DNA, RNAs, miRNAs, proteins and lipids). The recognition of miRNAs sorted for cargo into EVs relies on heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) or synaptotagmin-binding cytoplasmic RNA interaction protein (SYNCRIP).
A passive release of miRNAs can occur in a state of chronic inflammation, apoptosis or necrosis, leading to cell lysis. In this case, the miRNAs are released into biological fluids associated with Ago proteins [63]. A quantitative analysis of the passive release of miRNAs has not yet been carried out.
miRNAs are identified by a code made of the “miR” prefix and a unique identifying number. miRNAs belonging to certain species are indicated with an additional three-letter prefix; for example, hsa-miR-124 is a miRNA belonging to the human species (the prefix hsa means Homo Sapiens). The identifying numbers are given sequentially, meaning that miR-124 was discovered and named before miR-456. miRNAs with nearly identical sequences, except for one or two nucleotides, can be given with an additional lowercase letter [72]. Let-7 is a fundamental miRNA tumor suppressor.
3. miRNAs, Implantation and Preeclampsia
The possibility that miRNAs may play a crucial role in the pathogenesis of preeclampsia stems from the consideration that these molecules actively participate in the implantation processes of pregnancy and, on the other hand, that preeclampsia finds its pathophysiological basis in the very early stages of pregnancy. Nucleic acids have been detected in uterine fluid and, more specifically, in EVs containing miRNAs, suggesting a role of miRNAs in embryo–endometrial communication [73]. Using biopsy material and modern transcriptomics, it has also been shown that miRNAs are dysregulated in the endometrium of women with recurrent implantation failure [74]. Trophoblast migration and invasion, and cellular adaptations in the physiological changes underlying gestation, involve EVs as key modulators. Endometrial luminal epithelial cells [75] and proper communication between these cells determine the success or failure of pregnancy [76]. Exosome nanovesicles can transfer information (e.g., hsa-miR-30b, hsa-miR-200c, hsamiR-17 and hsa-miR-106a, miRNAs involved in endometrial receptivity and implantation) from the endometrium to the blastocyst, thereby promoting implantation [77][78]. In addition, extracellular vesicles secreted by the endometrium are internalized by the embryo to enhance adhesion and invasion [79], mediating embryo–endometrial communication [80]. During the early invasion phase (8 to 10 days after conception), the cytotrophoblast differentiates into an extravillous interstitial trophoblast and invades the decidua. At this point, feto–maternal communication occurs between the extravillous trophoblast and the decidualized endometrial stroma [81]. One of the most probable hypotheses to describe the etiology of preeclampsia is based on a failure of extravillous trophoblasts to invade the uterine spiral arteries in the placental bed. Insufficient placental vascular remodeling induces placental hypoperfusion, which is critical for the pathogenesis of preeclampsia. Placental insufficiency has also been associated with abnormal levels of extracellular fetal DNA, mRNA transcripts and circulating C19MC microRNAs (miR-516b-5p, miR-517-5p, miR-520a-5p, miR-525-5p and miR-526a) [82]. Exosomes secreted by cytotrophoblasts, which express placenta-specific miRNAs, including syncytin-2, have been implicated in embryo implantation through the promotion of Treg differentiation and suppression of the nuclear factor B signaling pathway and, thus, the immune and inflammatory response [83]. The specific loading of miRNAs in maternal plasma exosomes obtained in the first trimester of pregnancy in women who developed preeclampsia [84] suggests a potential role of miRNAs in the pathogenesis of preeclampsia as early as the first trimester.
 Combined tests, such as the measurement of mean arterial pressure (MAP), the ratio of soluble Fms-like tyrosine kinase-1 to placental growth factor (sFlt-1/PlGF) and the uterine artery pulsatility index (UTPI), are already widely validated [101]. The strategy of combining biochemical and biophysical data stems from the consideration that it is unlikely that preeclampsia can be detected early by a single predictive parameter with sufficient accuracy to be clinically useful. The association between miRNAs and biophysical parameters was evaluated by demonstrating a negative correlation between miR-942 levels and maternal blood pressure and between miR-143 levels and the uterine artery Doppler pulsatility index [93].
Combined tests, such as the measurement of mean arterial pressure (MAP), the ratio of soluble Fms-like tyrosine kinase-1 to placental growth factor (sFlt-1/PlGF) and the uterine artery pulsatility index (UTPI), are already widely validated [101]. The strategy of combining biochemical and biophysical data stems from the consideration that it is unlikely that preeclampsia can be detected early by a single predictive parameter with sufficient accuracy to be clinically useful. The association between miRNAs and biophysical parameters was evaluated by demonstrating a negative correlation between miR-942 levels and maternal blood pressure and between miR-143 levels and the uterine artery Doppler pulsatility index [93].
4. miRNAs and Clinical Preeclampsia
4.1. miRNAs and Diagnosed Preeclampsia
One of the first demonstrations of the possibility of using circulating miRNAs as a marker for preeclamptic pregnancy was by Gunel et al. They measured the expression level of miR-210 and miR-152 from the maternal plasma of both healthy and preeclamptic pregnant women. The results demonstrated an upregulation for miR-210 and a downregulation for miR-152 in women with preeclamptic pregnancy [85]. In the study of Campos et al., a lower expression of circulating miR-196b in maternal plasma was correlated to a preeclamptic condition in pregnant women [86]. Both miR-195-5p [87] and miR-885-5p [88] overexpression was correlated to a preeclamptic condition in women. Investigating the plasma of women with diagnosed pregnancy complicated by preeclampsia, Sheng et al. observed an upregulation for miR-206 [89]. The study realized by Akgor et al. analyzed the circulating level of miRNAs in women with diagnosed pregnancy complicated by preeclampsia. The authors observed a panel of miRNAs, many of which are already known as potential biomarkers for the non-invasive diagnosis of preeclampsia. However, they observed, for the first time, an upregulation of two novel miRNAs (miR-191-5p and miR-197) associated with the preeclamptic condition [90]. Ayoub et al. found an upregulation for both miR-186 and miR-181a in the serum of women with pregnancy complicated by preeclampsia [91]. The dysregulated expression of circulating miRNAs depends on an epigenetic mechanism, as observed by Sekar et al. They found an increased level of miR-510 in the plasma of women with preeclamptic pregnancy correlated to a decreased methylation status of its promoter [92].
Conversely, Luque et al. investigated miRNA circulating in maternal plasma. Evidence demonstrated that the investigated miRNAs (miR-192, miR-125b, miR-143, miR-126, miR-221, miR-942 and miR-127) were not a useful tool to predict preeclampsia, considering that their serum levels demonstrated no significant differences between the preeclampsia and control groups [93]. In the same way, Let7a-5p is not associated with preeclampsia because its expression in maternal plasma is not significantly different between uncomplicated and preeclamptic pregnancy [94].
4.2. miRNAs and Onset of Preeclampsia
miRNAs have been investigated to distinguish between the early (before 34 weeks of pregnancy) and late (after 34 weeks of pregnancy) onset of preeclampsia.
Miura et al. studied the expression level of all ten miRNAs belonging to C19MC, isolated from maternal blood samples at 27–34 weeks of gestation. The authors demonstrated that, except for miR-518b and miR-519d, the remaining miRNAs were upregulated in women with an early onset of preeclampsia compared with women with a late onset of preeclampsia [95]. Dong et al. confirmed the usefulness of miRNAs as a biomarker in the detection of preeclampsia onset. Particularly, they measured the expression level of both miR-21 and miR-31 in the maternal plasma of preeclamptic women. The results demonstrated that an increase in circulating miR-31 was associated with early onset preeclampsia; meanwhile, an increase in circulating miR-21 was related to late-onset preeclampsia [96]. Decreased levels of circulating miR-126 were associated with early onset preeclampsia compared to gestation-matched controls [97].
Research activity has led to the identification of miRNAs being added to the list of potential predictors for the severity of preeclampsia. Pan et al. analyzed the expression level of miRNAs in the plasma of women with normal pregnancy and pregnancy complicated by mild preeclampsia. Plasma was collected before and after parturition. They demonstrated that the parturition influenced the expression of miRNAs in the plasma of the same women and that the expression level of miR-141 and miR-221 was different between normal and preeclamptic plasma, both before and after parturition [98]. Li et al. demonstrated that the differential expression of circulating miRNAs investigated was related to the severity of the preeclamptic condition. In fact, both miR-141 and miR-29a are significantly overexpressed in the plasma of women affected by mild preeclampsia. On the contrary, miR-144 was significantly downregulated in the plasma of women affected by mild preeclampsia and severe preeclampsia with respect to uncomplicated pregnancy [99].
4.3. miRNAs and Prediction of Preeclampsia
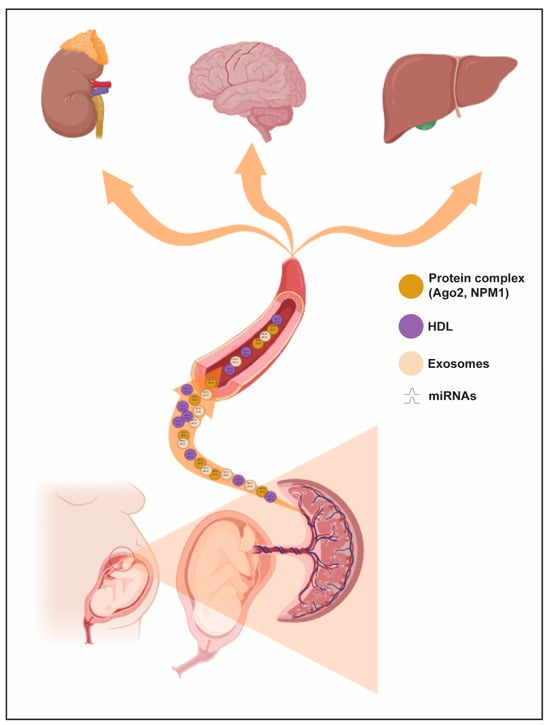
The ability to predict preeclampsia is a major challenge in contemporary obstetrics, and resources are now focused on the first trimester of pregnancy, where prophylactic strategies can help reduce the incidence of this disorder [100] (Figure 2).

Figure 2. Trafficking routes of circulating miRNAs during pregnancy. Circulating miRNAs may be delivered to maternal circulation and affect various cell events in maternal targeting organs (e.g., kidney, brain and liver).
5. miRNAs, Preeclampsia and Epigenetics
Preeclampsia, in addition to being one of the most frequent causes of maternal and fetal morbidity and mortality in pregnancy, has long-term negative implications for both mother and offspring [102]. Epidemiological studies indicate that women who experience preeclampsia during pregnancy have an increased vascular and metabolic risk later, as do the children of preeclamptic mothers [103]. These epidemiological considerations underlie epigenetic studies of preeclamptic disease. Although preeclampsia is a very complex disease, a great deal of evidence confirms that endothelial dysfunction is a central feature of pathogenesis and a factor that epigenetically may lead to an increased cardiovascular risk in later life. Epigenetics, or how the environment influences gene expression without altering the DNA sequence, is one of the mechanisms by which gestational hypoxia enables adaptive responses to change in the placental environment in preeclampsia. Epigenetic modifications are one of the potential mechanisms, including aberrant miRNA expression, through which the exposure to an altered environment in utero results in the development of chronic disease. The actions of miRNAs, DNA methylation and histone modification are the three most studied epigenetic processes [104]. In vitro studies have shown that miRNA expression is modulated by hypoxia, cell signaling pathways and epigenetic modifications through promoter methylation [105]. The downregulation of miRNAs also results from the hypermethylation of promoter regions [106][107]. Hypomethylation of the miR-141-3p promoter has been reported to increase the expression of miR-141-3p, which, in turn, induces inflammasome formation, a decreased expression of MMP2/9 and the inhibition of trophoblast proliferation and invasion [108]. From this point of view, the degree of miRNA methylation might have an epigenetic effect. From the perspective of endothelial dysfunction with possible epigenetic spillover, maternal and cord-derived endothelial progenitor cells (EPCs) from preeclamptic pregnancies show an aberrant miRNAs profile compared with healthy pregnancies [109]. EPCs are essential for maintaining a healthy endothelium throughout an individual’s lifetime. Decreased cell numbers and colony-forming units of maternal EPCs are described as a sign of impaired endothelial repair capacity in preeclampsia [110]. The importance of studying miRNA changes in the epigenetic domain stems from the possible therapeutic developments in preeclampsia and women’s future cardiovascular risk. The relevance of miRNAs in vascular neovascularization has been demonstrated by several knockdown approaches of enzymes involved in miRNA biogenesis [111].
6. Conclusions
Several non-coding RNAs and, thus, miRNAs are differentially expressed in the pathophysiology of women’s health in general [112] and in pregnancy and the placenta in particular. In recent years, the number of identified miRNAs has significantly increased; however, the exact mechanisms remain to be elucidated, mainly because of the cell-specific functions exhibited by many miRNAs. A deeper understanding of miRNAs and their relationships with gene modifications will help in determining the mechanism by which these molecules contribute to placental development. The identification of miRNAs that may act as potential non-invasive biomarkers for the prediction of pregnancy outcomes in the first trimester, especially among high-risk women, may have implications for research, identifying signaling pathways for further investigation and clinical implications, facilitating early diagnosis and timely interventions. This challenge is not easy to address since preeclampsia is probably not a single disease but may present in several different forms, and there are many difficulties in finding, measuring and reproducing miRNA results.
References
- Tossetta, G.; Avellini, C.; Licini, C.; Giannubilo, S.R.; Castellucci, M.; Marzioni, D. High temperature requirement A1 and fibronectin: Two possible players in placental tissue remodelling. Eur. J. Histochem. 2016, 60, 2724.
- Huppertz, B. The anatomy of the normal placenta. J. Clin. Pathol. 2008, 61, 1296–1302.
- Tossetta, G.; Fantone, S.; Busilacchi, E.M.; Di Simone, N.; Giannubilo, S.R.; Scambia, G.; Giordano, A.; Marzioni, D. Modulation of matrix metalloproteases by ciliary neurotrophic factor in human placental development. Cell Tissue Res. 2022, 390, 113–129.
- Tossetta, G.; Fantone, S.; Giannubilo, S.R.; Marinelli Busilacchi, E.; Ciavattini, A.; Castellucci, M.; Di Simone, N.; Mattioli-Belmonte, M.; Marzioni, D. Pre-eclampsia onset and SPARC: A possible involvement in placenta development. J. Cell Physiol. 2019, 234, 6091–6098.
- Gesuita, R.; Licini, C.; Picchiassi, E.; Tarquini, F.; Coata, G.; Fantone, S.; Tossetta, G.; Ciavattini, A.; Castellucci, M.; Di Renzo, G.C.; et al. Association between first trimester plasma htra1 level and subsequent preeclampsia: A possible early marker? Pregnancy Hypertens. 2019, 18, 58–62.
- Cardaropoli, S.; Paulesu, L.; Romagnoli, R.; Ietta, F.; Marzioni, D.; Castellucci, M.; Rolfo, A.; Vasario, E.; Piccoli, E.; Todros, T. Macrophage migration inhibitory factor in fetoplacental tissues from preeclamptic pregnancies with or without fetal growth restriction. Clin. Dev. Immunol. 2012, 2012, 639342.
- Marzioni, D.; Todros, T.; Cardaropoli, S.; Rolfo, A.; Lorenzi, T.; Ciarmela, P.; Romagnoli, R.; Paulesu, L.; Castellucci, M. Activating protein-1 family of transcription factors in the human placenta complicated by preeclampsia with and without fetal growth restriction. Placenta 2010, 31, 919–927.
- Todros, T.; Marzioni, D.; Lorenzi, T.; Piccoli, E.; Capparuccia, L.; Perugini, V.; Cardaropoli, S.; Romagnoli, R.; Gesuita, R.; Rolfo, A.; et al. Evidence for a role of TGF-beta1 in the expression and regulation of alpha-SMA in fetal growth restricted placentae. Placenta 2007, 28, 1123–1132.
- Tossetta, G.; Fantone, S.; Gesuita, R.; Di Renzo, G.C.; Meyyazhagan, A.; Tersigni, C.; Scambia, G.; Di Simone, N.; Marzioni, D. HtrA1 in Gestational Diabetes Mellitus: A Possible Biomarker? Diagnostics 2022, 12, 2705.
- Giannubilo, S.R.; Licini, C.; Picchiassi, E.; Tarquini, F.; Coata, G.; Fantone, S.; Tossetta, G.; Ciavattini, A.; Castellucci, M.; Giardina, I.; et al. First trimester HtrA1 maternal plasma level and spontaneous preterm birth. J. Matern. Fetal Neonatal Med. 2022, 35, 780–784.
- Capparuccia, L.; Marzioni, D.; Giordano, A.; Fazioli, F.; De Nictolis, M.; Busso, N.; Todros, T.; Castellucci, M. PPARgamma expression in normal human placenta, hydatidiform mole and choriocarcinoma. Mol. Hum. Reprod. 2002, 8, 574–579.
- Crescimanno, C.; Marzioni, D.; Paradinas, F.J.; Schrurs, B.; Muhlhauser, J.; Todros, T.; Newlands, E.; David, G.; Castellucci, M. Expression pattern alterations of syndecans and glypican-1 in normal and pathological trophoblast. J. Pathol. 1999, 189, 600–608.
- Dimitriadis, E.; Rolnik, D.L.; Zhou, W.; Estrada-Gutierrez, G.; Koga, K.; Francisco, R.P.V.; Whitehead, C.; Hyett, J.; da Silva Costa, F.; Nicolaides, K.; et al. Pre-eclampsia. Nat. Rev. Dis. Primers 2023, 9, 8.
- Odigboegwu, O.; Pan, L.J.; Chatterjee, P. Use of Antihypertensive Drugs During Preeclampsia. Front. Cardiovasc. Med. 2018, 5, 50.
- Mol, B.W.J.; Roberts, C.T.; Thangaratinam, S.; Magee, L.A.; de Groot, C.J.M.; Hofmeyr, G.J. Pre-eclampsia. Lancet 2016, 387, 999–1011.
- Thadhani, R.; Ecker, J.L.; Kettyle, E.; Sandler, L.; Frigoletto, F.D. Pulse pressure and risk of preeclampsia: A prospective study. Obstet. Gynecol. 2001, 97, 515–520.
- Fantone, S.; Mazzucchelli, R.; Giannubilo, S.R.; Ciavattini, A.; Marzioni, D.; Tossetta, G. AT-rich interactive domain 1A protein expression in normal and pathological pregnancies complicated by preeclampsia. Histochem. Cell Biol. 2020, 154, 339–346.
- Tossetta, G.; Fantone, S.; Giannubilo, S.R.; Ciavattini, A.; Senzacqua, M.; Frontini, A.; Marzioni, D. HTRA1 in Placental Cell Models: A Possible Role in Preeclampsia. Curr. Issues Mol. Biol. 2023, 45, 3815–3828.
- Fantone, S.; Giannubilo, S.R.; Marzioni, D.; Tossetta, G. HTRA family proteins in pregnancy outcome. Tissue Cell 2021, 72, 101549.
- Ali, S.M.; Khalil, R.A. Genetic, immune and vasoactive factors in the vascular dysfunction associated with hypertension in pregnancy. Expert. Opin. Ther. Targets 2015, 19, 1495–1515.
- Cecati, M.; Emanuelli, M.; Giannubilo, S.R.; Quarona, V.; Senetta, R.; Malavasi, F.; Tranquilli, A.L.; Saccucci, F. Contribution of adenosine-producing ectoenzymes to the mechanisms underlying the mitigation of maternal-fetal conflicts. J. Biol. Regul. Homeost. Agents 2013, 27, 519–529.
- Ferreira, L.B.; Williams, K.A.; Best, G.; Haydinger, C.D.; Smith, J.R. Inflammatory cytokines as mediators of retinal endothelial barrier dysfunction in non-infectious uveitis. Clin. Transl. Immunol. 2023, 12, e1479.
- Mateuszuk, L.; Campagna, R.; Kutryb-Zajac, B.; Kus, K.; Slominska, E.M.; Smolenski, R.T.; Chlopicki, S. Reversal of endothelial dysfunction by nicotinamide mononucleotide via extracellular conversion to nicotinamide riboside. Biochem. Pharmacol. 2020, 178, 114019.
- Tossetta, G.; Piani, F.; Borghi, C.; Marzioni, D. Role of CD93 in Health and Disease. Cells 2023, 12, 1778.
- Campagna, R.; Vignini, A. NAD(+) Homeostasis and NAD(+)-Consuming Enzymes: Implications for Vascular Health. Antioxidants 2023, 12, 376.
- Campagna, R.; Mateuszuk, L.; Wojnar-Lason, K.; Kaczara, P.; Tworzydlo, A.; Kij, A.; Bujok, R.; Mlynarski, J.; Wang, Y.; Sartini, D.; et al. Nicotinamide N-methyltransferase in endothelium protects against oxidant stress-induced endothelial injury. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119082.
- Szczesny-Malysiak, E.; Stojak, M.; Campagna, R.; Grosicki, M.; Jamrozik, M.; Kaczara, P.; Chlopicki, S. Bardoxolone Methyl Displays Detrimental Effects on Endothelial Bioenergetics, Suppresses Endothelial ET-1 Release, and Increases Endothelial Permeability in Human Microvascular Endothelium. Oxid. Med. Cell Longev. 2020, 2020, 4678252.
- Piani, F.; Tossetta, G.; Cara-Fuentes, G.; Agnoletti, D.; Marzioni, D.; Borghi, C. Diagnostic and Prognostic Role of CD93 in Cardiovascular Disease: A Systematic Review. Biomolecules 2023, 13, 910.
- Ristovska, E.C.; Genadieva-Dimitrova, M.; Todorovska, B.; Milivojevic, V.; Rankovic, I.; Samardziski, I.; Bojadzioska, M. The Role of Endothelial Dysfunction in the Pathogenesis of Pregnancy-Related Pathological Conditions: A Review. Pril (Makedon. Akad. Nauk. Umet. Odd. Med. Nauki) 2023, 44, 113–137.
- Shen, J.; San, W.; Zheng, Y.; Zhang, S.; Cao, D.; Chen, Y.; Meng, G. Different types of cell death in diabetic endothelial dysfunction. Biomed. Pharmacother. 2023, 168, 115802.
- Tossetta, G.; Fantone, S.; Delli Muti, N.; Balercia, G.; Ciavattini, A.; Giannubilo, S.R.; Marzioni, D. Preeclampsia and severe acute respiratory syndrome coronavirus 2 infection: A systematic review. J. Hypertens. 2022, 40, 1629–1638.
- Fantone, S.; Tossetta, G.; Di Simone, N.; Tersigni, C.; Scambia, G.; Marcheggiani, F.; Giannubilo, S.R.; Marzioni, D. CD93 a potential player in cytotrophoblast and endothelial cell migration. Cell Tissue Res. 2022, 387, 123–130.
- Cevher Akdulum, M.F.; Demirdag, E.; Arik, S.I.; Safarova, S.; Erdem, M.; Bozkurt, N.; Erdem, A. Is the First-Trimester Systemic Immune-Inflammation Index Associated With Preeclampsia? Cureus 2023, 15, e44063.
- Tossetta, G.; Fantone, S.; Piani, F.; Crescimanno, C.; Ciavattini, A.; Giannubilo, S.R.; Marzioni, D. Modulation of NRF2/KEAP1 Signaling in Preeclampsia. Cells 2023, 12, 1545.
- Mazzanti, L.; Cecati, M.; Vignini, A.; D’Eusanio, S.; Emanuelli, M.; Giannubilo, S.R.; Saccucci, F.; Tranquilli, A.L. Placental expression of endothelial and inducible nitric oxide synthase and nitric oxide levels in patients with HELLP syndrome. Am. J. Obstet. Gynecol. 2011, 205, 236.e1-7.
- Fantone, S.; Ermini, L.; Piani, F.; Di Simone, N.; Barbaro, G.; Giannubilo, S.R.; Gesuita, R.; Tossetta, G.; Marzioni, D. Downregulation of argininosuccinate synthase 1 (ASS1) is associated with hypoxia in placental development. Hum. Cell 2023, 36, 1190–1198.
- Cecati, M.; Giannubilo, S.R.; Saccucci, F.; Sartini, D.; Ciavattini, A.; Emanuelli, M.; Tranquilli, A.L. Potential Role of Placental Klotho in the Pathogenesis of Preeclampsia. Cell Biochem. Biophys. 2016, 74, 49–57.
- Maynard, S.E.; Karumanchi, S.A. Angiogenic factors and preeclampsia. Semin. Nephrol. 2011, 31, 33–46.
- Van Niekerk, G.; Christowitz, C.; Engelbrecht, A.M. Insulin-mediated immune dysfunction in the development of preeclampsia. J. Mol. Med. 2021, 99, 889–897.
- Leavey, K.; Benton, S.J.; Grynspan, D.; Kingdom, J.C.; Bainbridge, S.A.; Cox, B.J. Unsupervised Placental Gene Expression Profiling Identifies Clinically Relevant Subclasses of Human Preeclampsia. Hypertension 2016, 68, 137–147.
- Benton, S.J.; Leavey, K.; Grynspan, D.; Cox, B.J.; Bainbridge, S.A. The clinical heterogeneity of preeclampsia is related to both placental gene expression and placental histopathology. Am. J. Obstet. Gynecol. 2018, 219, 604.e1–604.e25.
- Rana, S.; Lemoine, E.; Granger, J.P.; Karumanchi, S.A. Preeclampsia: Pathophysiology, Challenges, and Perspectives. Circ. Res. 2019, 124, 1094–1112.
- Licini, C.; Avellini, C.; Picchiassi, E.; Mensa, E.; Fantone, S.; Ramini, D.; Tersigni, C.; Tossetta, G.; Castellucci, C.; Tarquini, F.; et al. Pre-eclampsia predictive ability of maternal miR-125b: A clinical and experimental study. Transl. Res. 2021, 228, 13–27.
- Liu, Z.; Pei, J.; Zhang, X.; Wang, C.; Tang, Y.; Liu, H.; Yu, Y.; Luo, S.; Gu, W. APOA1 Is a Novel Marker for Preeclampsia. Int. J. Mol. Sci. 2023, 24, 16363.
- Piani, F.; Agnoletti, D.; Baracchi, A.; Scarduelli, S.; Verde, C.; Tossetta, G.; Montaguti, E.; Simonazzi, G.; Degli Esposti, D.; Borghi, C.; et al. Serum uric acid to creatinine ratio and risk of preeclampsia and adverse pregnancy outcomes. J. Hypertens. 2023, 41, 1333–1338.
- Soobryan, N.; Reddy, K.; Ibrahim, U.H.; Moodley, J.; Kumar, A.; Mackraj, I. Identification of gene signature markers in gestational hypertension and early-onset pre-eclampsia. Placenta 2023, 145, 1–8.
- Piani, F.; Tossetta, G.; Fantone, S.; Agostinis, C.; Di Simone, N.; Mandala, M.; Bulla, R.; Marzioni, D.; Borghi, C. First Trimester CD93 as a Novel Marker of Preeclampsia and Its Complications: A Pilot Study. High. Blood Press. Cardiovasc. Prev. 2023, 30, 591–594.
- Qin, S.; Sun, N.; Xu, L.; Xu, Y.; Tang, Q.; Tan, L.; Chen, A.; Zhang, L.; Liu, S. The Value of Circulating microRNAs for Diagnosis and Prediction of Preeclampsia: A Meta-analysis and Systematic Review. Reprod. Sci. 2022, 29, 3078–3090.
- Ping, Z.; Feng, Y.; Lu, Y.; Ai, L.; Jiang, H. Integrated analysis of microRNA and mRNA expression profiles in Preeclampsia. BMC Med. Genom. 2023, 16, 309.
- Kimura, T. Non-coding Natural Antisense RNA: Mechanisms of Action in the Regulation of Target Gene Expression and Its Clinical Implications. Yakugaku Zasshi 2020, 140, 687–700.
- Zeng, Q.; Wan, H.; Zhao, S.; Xu, H.; Tang, T.; Oware, K.A.; Qu, S. Role of PIWI-interacting RNAs on cell survival: Proliferation, apoptosis, and cycle. IUBMB Life 2020, 72, 1870–1878.
- Wang, N.; Yu, Y.; Xu, B.; Zhang, M.; Li, Q.; Miao, L. Pivotal prognostic and diagnostic role of the long non-coding RNA colon cancer-associated transcript 1 expression in human cancer (Review). Mol. Med. Rep. 2019, 19, 771–782.
- Zhao, W.; An, Y.; Liang, Y.; Xie, X.W. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1930–1936.
- Shields, E.J.; Petracovici, A.F.; Bonasio, R. lncRedibly versatile: Biochemical and biological functions of long noncoding RNAs. Biochem. J. 2019, 476, 1083–1104.
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655.
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561.
- Schwarzenbach, H.; Gahan, P.B. MicroRNA Shuttle from Cell-To-Cell by Exosomes and Its Impact in Cancer. Noncoding RNA 2019, 5, 28.
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234.
- Mayya, V.K.; Duchaine, T.F. Ciphers and Executioners: How 3′-Untranslated Regions Determine the Fate of Messenger RNAs. Front. Genet. 2019, 10, 6.
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333.
- Chen, Y.M.; Zheng, Y.L.; Su, X.; Wang, X.Q. Crosstalk Between MicroRNAs and Circular RNAs in Human Diseases: A Bibliographic Study. Front. Cell Dev. Biol. 2021, 9, 754880.
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518.
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008.
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259.
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433.
- Ortiz-Quintero, B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif. 2016, 49, 281–303.
- Axmann, M.; Meier, S.M.; Karner, A.; Strobl, W.; Stangl, H.; Plochberger, B. Serum and Lipoprotein Particle miRNA Profile in Uremia Patients. Genes 2018, 9, 533.
- Watts, G.F.; Barrett, P.H. High-density lipoprotein metabolism in familial hypercholesterolaemia: Significance, mechanisms, therapy. Nutr. Metab. Cardiovasc. Dis. 2002, 12, 36–41.
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659.
- Sabanovic, B.; Piva, F.; Cecati, M.; Giulietti, M. Promising Extracellular Vesicle-Based Vaccines against Viruses, Including SARS-CoV-2. Biology 2021, 10, 94.
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980.
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24.
- Rekker, K.; Altmae, S.; Suhorutshenko, M.; Peters, M.; Martinez-Blanch, J.F.; Codoner, F.M.; Vilella, F.; Simon, C.; Salumets, A.; Velthut-Meikas, A. A Two-Cohort RNA-seq Study Reveals Changes in Endometrial and Blood miRNome in Fertile and Infertile Women. Genes 2018, 9, 574.
- Aplin, J.D.; Kimber, S.J. Trophoblast-uterine interactions at implantation. Reprod. Biol. Endocrinol. 2004, 2, 48.
- Desrochers, L.M.; Bordeleau, F.; Reinhart-King, C.A.; Cerione, R.A.; Antonyak, M.A. Microvesicles provide a mechanism for intercellular communication by embryonic stem cells during embryo implantation. Nat. Commun. 2016, 7, 11958.
- Cretoiu, D.; Xu, J.; Xiao, J.; Suciu, N.; Cretoiu, S.M. Circulating MicroRNAs as Potential Molecular Biomarkers in Pathophysiological Evolution of Pregnancy. Dis. Markers 2016, 2016, 3851054.
- Liu, L.; Guo, J.; Gao, W.; Gao, M.; Ma, X. Research progress in the role of non-coding RNAs and embryo implantation. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2023, 48, 1377–1387.
- Gurung, S.; Greening, D.W.; Catt, S.; Salamonsen, L.; Evans, J. Exosomes and soluble secretome from hormone-treated endometrial epithelial cells direct embryo implantation. Mol. Hum. Reprod. 2020, 26, 510–520.
- Das, M.; Kale, V. Extracellular vesicles: Mediators of embryo-maternal crosstalk during pregnancy and a new weapon to fight against infertility. Eur. J. Cell Biol. 2020, 99, 151125.
- Atay, S.; Gercel-Taylor, C.; Suttles, J.; Mor, G.; Taylor, D.D. Trophoblast-derived exosomes mediate monocyte recruitment and differentiation. Am. J. Reprod. Immunol. 2011, 65, 65–77.
- Whitley, G.S.; Dash, P.R.; Ayling, L.J.; Prefumo, F.; Thilaganathan, B.; Cartwright, J.E. Increased apoptosis in first trimester extravillous trophoblasts from pregnancies at higher risk of developing preeclampsia. Am. J. Pathol. 2007, 170, 1903–1909.
- Czernek, L.; Duchler, M. Exosomes as Messengers Between Mother and Fetus in Pregnancy. Int. J. Mol. Sci. 2020, 21, 4264.
- Truong, G.; Guanzon, D.; Kinhal, V.; Elfeky, O.; Lai, A.; Longo, S.; Nuzhat, Z.; Palma, C.; Scholz-Romero, K.; Menon, R.; et al. Oxygen tension regulates the miRNA profile and bioactivity of exosomes released from extravillous trophoblast cells—Liquid biopsies for monitoring complications of pregnancy. PLoS ONE 2017, 12, e0174514.
- Yang, X.W.; Shen, G.Z.; Cao, L.Q.; Jiang, X.F.; Peng, H.P.; Shen, G.; Chen, D.; Xue, P. MicroRNA-1269 promotes proliferation in human hepatocellular carcinoma via downregulation of FOXO1. BMC Cancer 2014, 14, 909.
- Gunel, T.; Kamali, N.; Hosseini, M.K.; Gumusoglu, E.; Benian, A.; Aydinli, K. Regulatory effect of miR-195 in the placental dysfunction of preeclampsia. J. Matern. Fetal Neonatal Med. 2020, 33, 901–908.
- Whigham, C.A.; MacDonald, T.M.; Walker, S.P.; Hiscock, R.; Hannan, N.J.; Pritchard, N.; Cannon, P.; Nguyen, T.V.; Miranda, M.; Tong, S.; et al. MicroRNAs 363 and 149 are differentially expressed in the maternal circulation preceding a diagnosis of preeclampsia. Sci. Rep. 2020, 10, 18077.
- Kolkova, Z.; Holubekova, V.; Grendar, M.; Nachajova, M.; Zubor, P.; Pribulova, T.; Loderer, D.; Zigo, I.; Biringer, K.; Hornakova, A. Association of Circulating miRNA Expression with Preeclampsia, Its Onset, and Severity. Diagnostics 2021, 11, 476.
- Zhang, Y.; Huang, G.; Zhang, Y.; Yang, H.; Long, Y.; Liang, Q.; Zheng, Z. MiR-942 decreased before 20 weeks gestation in women with preeclampsia and was associated with the pathophysiology of preeclampsia in vitro. Clin. Exp. Hypertens. 2017, 39, 108–113.
- Pan, M.; Ge, Q.; Li, H.; Yang, Q.; Lu, J.; Zhang, D.; Lu, Z. Sequencing the miRNAs in maternal plasma from women before and after parturition. J. Nanosci. Nanotechnol. 2012, 12, 4035–4043.
- Sekar, D.; Lakshmanan, G.; Mani, P.; Biruntha, M. Methylation-dependent circulating microRNA 510 in preeclampsia patients. Hypertens. Res. 2019, 42, 1647–1648.
- Whigham, C.A.; MacDonald, T.M.; Walker, S.P.; Pritchard, N.; Hannan, N.J.; Cannon, P.; Nguyen, T.V.; Hastie, R.; Tong, S.; Kaitu’u-Lino, T.J. Circulating GATA2 mRNA is decreased among women destined to develop preeclampsia and may be of endothelial origin. Sci. Rep. 2019, 9, 235.
- Jairajpuri, D.S.; Malalla, Z.H.; Mahmood, N.; Almawi, W.Y. Circulating microRNA expression as predictor of preeclampsia and its severity. Gene 2017, 627, 543–548.
- Li, Q.; Han, Y.; Xu, P.; Yin, L.; Si, Y.; Zhang, C.; Meng, Y.; Feng, W.; Pan, Z.; Gao, Z.; et al. Elevated microRNA-125b inhibits cytotrophoblast invasion and impairs endothelial cell function in preeclampsia. Cell Death Discov. 2020, 6, 35.
- Xu, P.; Zhao, Y.; Liu, M.; Wang, Y.; Wang, H.; Li, Y.X.; Zhu, X.; Yao, Y.; Wang, H.; Qiao, J.; et al. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension 2014, 63, 1276–1284.
- Tan, M.Y.; Wright, D.; Syngelaki, A.; Akolekar, R.; Cicero, S.; Janga, D.; Singh, M.; Greco, E.; Wright, A.; Maclagan, K.; et al. Comparison of diagnostic accuracy of early screening for pre-eclampsia by NICE guidelines and a method combining maternal factors and biomarkers: Results of SPREE. Ultrasound Obstet. Gynecol. 2018, 51, 743–750.
- Hromadnikova, I.; Kotlabova, K.; Ondrackova, M.; Kestlerova, A.; Novotna, V.; Hympanova, L.; Doucha, J.; Krofta, L. Circulating C19MC microRNAs in preeclampsia, gestational hypertension, and fetal growth restriction. Mediat. Inflamm. 2013, 2013, 186041.
- Ntsethe, A.; Mackraj, I. An Investigation of Exosome Concentration and Exosomal microRNA (miR-155 and miR-222) Expression in Pregnant Women with Gestational Hypertension and Preeclampsia. Int. J. Womens Health 2022, 14, 1681–1689.
- Luque, A.; Farwati, A.; Crovetto, F.; Crispi, F.; Figueras, F.; Gratacos, E.; Aran, J.M. Usefulness of circulating microRNAs for the prediction of early preeclampsia at first-trimester of pregnancy. Sci. Rep. 2014, 4, 4882.
- Witvrouwen, I.; Mannaerts, D.; Ratajczak, J.; Boeren, E.; Faes, E.; Van Craenenbroeck, A.H.; Jacquemyn, Y.; Van Craenenbroeck, E.M. MicroRNAs targeting VEGF are related to vascular dysfunction in preeclampsia. Biosci. Rep. 2021, 41, BSR20210874.
- Hromadnikova, I.; Kotlabova, K.; Ivankova, K.; Krofta, L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS ONE 2017, 12, e0171756.
- Hromadnikova, I.; Dvorakova, L.; Kotlabova, K.; Krofta, L. The Prediction of Gestational Hypertension, Preeclampsia and Fetal Growth Restriction via the First Trimester Screening of Plasma Exosomal C19MC microRNAs. Int. J. Mol. Sci. 2019, 20, 2972.
- Fraser, A.; Nelson, S.M.; Macdonald-Wallis, C.; Sattar, N.; Lawlor, D.A. Hypertensive disorders of pregnancy and cardiometabolic health in adolescent offspring. Hypertension 2013, 62, 614–620.
- Apicella, C.; Ruano, C.S.M.; Mehats, C.; Miralles, F.; Vaiman, D. The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia. Int. J. Mol. Sci. 2019, 20, 2837.
- Ji, L.; Brkic, J.; Liu, M.; Fu, G.; Peng, C.; Wang, Y.L. Placental trophoblast cell differentiation: Physiological regulation and pathological relevance to preeclampsia. Mol. Aspects Med. 2013, 34, 981–1023.
- Chhabra, R. miRNA and methylation: A multifaceted liaison. Chembiochem 2015, 16, 195–203.
- Morales, S.; Monzo, M.; Navarro, A. Epigenetic regulation mechanisms of microRNA expression. Biomol. Concepts 2017, 8, 203–212.
- Wu, D.; Shi, L.; Chen, F.; Lin, Q.; Kong, J. Methylation Status of the miR-141-3p Promoter Regulates miR-141-3p Expression, Inflammasome Formation, and the Invasiveness of HTR-8/SVneo Cells. Cytogenet. Genome Res. 2021, 161, 501–513.
- Brodowski, L.; Schroder-Heurich, B.; von Hardenberg, S.; Richter, K.; von Kaisenberg, C.S.; Dittrich-Breiholz, O.; Meyer, N.; Dork, T.; von Versen-Hoynck, F. MicroRNA Profiles of Maternal and Neonatal Endothelial Progenitor Cells in Preeclampsia. Int. J. Mol. Sci. 2021, 22, 5320.
- Lin, C.; Rajakumar, A.; Plymire, D.A.; Verma, V.; Markovic, N.; Hubel, C.A. Maternal endothelial progenitor colony-forming units with macrophage characteristics are reduced in preeclampsia. Am. J. Hypertens. 2009, 22, 1014–1019.
- Suarez, Y.; Fernandez-Hernando, C.; Pober, J.S.; Sessa, W.C. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007, 100, 1164–1173.
- Suarez, Y.; Fernandez-Hernando, C.; Yu, J.; Gerber, S.A.; Harrison, K.D.; Pober, J.S.; Iruela-Arispe, M.L.; Merkenschlager, M.; Sessa, W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 14082–14087.
- Vitale, S.G.; Fulghesu, A.M.; Mikuš, M.; Watrowski, R.; D’Alterio, M.N.; Lin, L.T.; Shah, M.; Reyes-Muñoz, E.; Sathyapalan, T.; Angioni, S. The Translational Role of miRNA in Polycystic Ovary Syndrome: From Bench to Bedside-A Systematic Literature Review. Biomedicines 2022, 10, 1816.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
508
Revisions:
2 times
(View History)
Update Date:
01 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




