Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ye Zhang | -- | 2676 | 2024-01-30 04:08:43 | | | |
| 2 | Sirius Huang | Meta information modification | 2676 | 2024-01-31 01:25:47 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Zhang, Y.; Bhat, J.A.; Zhang, Y.; Yang, S. Molecular Regulatory Networks of Seed Size in Soybean. Encyclopedia. Available online: https://encyclopedia.pub/entry/54509 (accessed on 07 February 2026).
Zhang Y, Bhat JA, Zhang Y, Yang S. Molecular Regulatory Networks of Seed Size in Soybean. Encyclopedia. Available at: https://encyclopedia.pub/entry/54509. Accessed February 07, 2026.
Zhang, Ye, Javaid Akhter Bhat, Yaohua Zhang, Suxin Yang. "Molecular Regulatory Networks of Seed Size in Soybean" Encyclopedia, https://encyclopedia.pub/entry/54509 (accessed February 07, 2026).
Zhang, Y., Bhat, J.A., Zhang, Y., & Yang, S. (2024, January 30). Molecular Regulatory Networks of Seed Size in Soybean. In Encyclopedia. https://encyclopedia.pub/entry/54509
Zhang, Ye, et al. "Molecular Regulatory Networks of Seed Size in Soybean." Encyclopedia. Web. 30 January, 2024.
Copy Citation
Soybean being a major cash crop provides half of the vegetable oil and a quarter of the plant proteins to the global population. Seed size traits are the most important agronomic traits determining the soybean yield. These are complex traits governed by polygenes with low heritability as well as are highly influenced by the environment as well as by genotype x environment interactions. Extensive efforts have been made to unravel the genetic basis and molecular mechanism of seed size in soybean.
soybean
seed size
regulatory mechanism
genes
yield
1. Introduction
Soybean [Glycine max (Linn.) Merr.] is an important oilseed crop that supplies half of the world’s vegetable oil and a quarter of the plant protein to the world population [1]. Throughout the history of crop breeding, substantial yield increases have been achieved for major grain crops such as rice, wheat, and maize; however, the yield gains of other important crops such as soybean have remained relatively stable or experienced slow growth. For example, in the highest soybean-consuming countries such as China, the past five decades have seen that soybean yield improvement efforts have been almost stagnant [2]. Hence, there is a great need for these countries as well as the world to increase domestic production to make self-sufficiency in soybean production. Breeders target different yield-related traits to increase soybean production. In this regard, seed size traits are an important trait that is directly related to the yield of soybeans [3]. Nitrogen fixation is vital for plant growth and yield, especially in legumes (such as soybeans) with their unique biological nitrogen-fixing ability [4]. Current research indicates that soybeans, through symbiosis with rhizobia, convert atmospheric nitrogen into ammonium [5], which crucially influences seed size and crop yield [6][7]. Optimizing this symbiotic advantage presents a promising avenue for agricultural yield improvement. Overall, the above results highlight the urgent need for the efficient and effective enhancement of soybean productivity [8].
2. The Seed Size Diversity of Soybean Makes It an Ideal Model Plant for Study
Besides the importance of genetic diversity for trait improvement in plant breeding, it also has great significance for the study of genetic evolution as well as the molecular regulatory mechanism. Hence, species possessing larger genetic diversity are serving as the model species in the study of crop evolution and genetic mechanisms. Among the flowering plants, leguminous plants constitute one of the largest and most diverse plant families, with approximately 20,000 species. The Fabaceae family is the largest subfamily within legumes and it encompasses a majority of model species such as soybean [9]. The soybean plants possess a unique advantage in the study of seed morphology. They exhibit a diverse range of seed sizes [10], and their embryos display diverse morphological forms (Figure 1).

Figure 1. Seed size variation across the soybean germplasm. Soybeans exhibit abundant genetic diversity, often displaying significant variations in seed size and shape among different cultivars. This makes them an ideal material for investigating seed traits. Scale bar = 1 cm.
As early as around 5000 BC mankind started to cultivate soybeans, and conserved as well as managed the rich source of soybean germplasm resources. The abundance of genetic diversity allows soybean breeders and researchers to conduct genetic analysis as well as functional studies using different soybean varieties and mutants. By using recent genetic and omics techniques combined with artificial intelligence (AI) based models such as machine learning (ML) and deep learning (DL) the soybean researchers will be able to unravel regulatory molecular mechanisms that influence seed size. Moreover, soybean exhibits extensive genetic variation in traits such as seed size (Figure 1). The seed size differences among different soybean varieties can exceed fivefold, hence it makes the soybean an ideal model plant for studying seed size.
3. Molecular Regulatory Network Underlying Seed Size in Soybean
The molecular regulatory network controlling seed size in soybeans is a complex system involving interactions at multiple levels, such as gene regulation, hormone regulation, nutrient regulation, and signal transduction pathways (Table 1). These regulatory factors collectively modulate the development of the embryo and endosperm, ultimately influencing seed size traits. Furthermore, previous studies on seed size regulatory mechanisms have mostly focused on internal factors, neglecting the profound impact of environmental factors on soybean seed development. For instance, soybean is highly sensitive to photoperiod, and the duration as well as intensity of light exposure can affect the process of seed development. In recent years, the influence of environmental factors on soybean seed development has gained widespread attention among researchers [11][12].
Table 1. Genes associated with the development of soybean seed size/weight.
| Position | Protein Name | Protein Category | Accession Number | Positive/Negative | Molecular Mechanism | Reference |
|---|---|---|---|---|---|---|
| Transcriptional regulators | BBM | AP2 family transcription factors | Glyma.09G248200 | positive | Positively regulates the process of seed embryogenesis and development and plays a decisive role in seed maturation | Ouakfaoui, 2010 [13] |
| GmAP2-1 | AP2 family transcription factors | Glyma.01G188400 | positive | All three together regulate seed size and grain weight by affecting seed length, width, and area | Jiang, 2020 [14] | |
| GmAP2-4 | AP2 family transcription factors | Glyma.13G329700 | positive | Jiang, 2020 [14] | ||
| GmAP2-6 | AP2 family transcription factors | Glyma.08G279000 | positive | Jiang, 2020 [14] | ||
| PP2C-1 | Phosphatase 2C-1 | Glyma.17G221100 | positive | Increases bead cell size and activates seed development-related genes; interacts with GmBZR1 | Lu, 2017 [15] | |
| GmBS1 | Tify domain | Glyma.10G244400 | negative | Negative regulation of primary cell proliferation by suppressing the expression of GIF1 and GRF5 | Ge,2016 [16] | |
| GmBS2 | Glyma.20G150000 | negative | ||||
| GmCYP78A5 | cytochrome P450 family protein CYP78A10 | Glyma.05G00220 | positive | Prediction by RNA-seq data analysis may correlate with seed size | Du, 2017 [17] | |
| CYP78A10 | Cytochrome P450 | Glyma.05G019200 | positive | Predicted by evolutionary correlation analysis to be extremely correlated with seed size, but leads to lower pod numbers | Wang, 2015 [18] | |
| CYP78A72 | Cytochrome P450 | Glyma.19G240800 | positive | Overexpression increases seed size and is functionally redundant with two other homologous genes | Zhao, 2016 [19] | |
| GmWRKY15a | WRKY family transcription factor | Glyma.05G096500 | positive | Differential expression in soybean pods is significantly associated with CT repeat variation during soybean domestication | Gu, 2017 [20] | |
| Phytohormone signalling and homeostasis | GmBZR1 | transcription factor of the BR signalling pathway | Glyma.17G248900 | positive | Conserved regulation by BR signalling promotes phosphorylation/dephosphorylation ratios | Zhang, 2016 [21] |
| GmGA20OX | gibberellin biosynthetic enzymes | Glyma.07G081700 | positive | Highly selected during domestication to increase the rate of GA biosynthesis | Lu, 2016 [22] | |
| GmJAZ3 | protein TIFY 6a-related | Glyma.09G123600 | positive | Promotes seed size/weight and other organ sizes in stable transgenic soybean plants by increasing cell proliferation | Hu, 2023 [23] | |
| SGF14f | protein SGF14f | Glyma.02G115900 | unknown | May regulate the balance of GA and ABA signalling and determine embryogenesis and seed germination | Schooheim, 2009 [24] | |
| GmFAD3 | microsomal omega-3 fatty acid desaturase | Glyma.18G062000 | negative | Associated with fatty acid synthesis, which inhibits jasmonic acid accumulation, and silencing leads to greater seed | Singh, 2020 [25] | |
| unnamed | ethylene-response factor C3 | Glyma.19G163900 | positive | AHP can promote cytokinin signaling with multiple functionally redundant genes | Claire, 2019 [26] | |
| unnamed | Hpt domain | Glyma.19G151900 | positive | Possible involvement in cytokinin-mediated seed size and weight regulatory networks | Assefa1, 2019 [27] | |
| GmPSKγ1 | PSK-like peptide | Glyma.02G126200 | positive | Novel peptide hormone whose tyrosine is sulfated induces embryonic cell expansion for seed growth | Yu, 2019 [28] | |
| Metabolic pathway | GmST01 | UDP-galactose4-epimerase; domain 1 | Glyma.08G109100 | positive | Regulation of cell division and amplification patterns in soybean to determine seed shape | Li, 2022 [29] |
| GmST05 | Phosphatidylethanolamine-binding Protein | Glyma.05G244100 | positive | regulates seed size and affects oil and protein content, which may affect the transcription of GmSWEET10a | Duan, 2022 [30] | |
| GmSWEET10a | sugar efflux transporter SWEET39 | Glyma.15G049200 | positive | The former is strongly selected in domestication and the latter is being selected, both contribute to the sugar distribution from the seed coat to the embryo by transporting sucrose and hexose, thus increasing oil content and seed size | Wang, 2020 [31] | |
| GmSWEET10b | sugar efflux transporter SWEET24 | Glyma.08G183500 | positive | |||
| CIF1 | Cell wall invertase inhibitors | Glyma.17G036300 | negative | Coordination of seed maturation by post-translational fine-tuning of sucrose metabolism and library strength by CWI | Tang, 2017 [32] | |
| GmDREBL | Caskin/Ankyrin repeat-containing protein | Glyma.12G11150 | positive | Involved in the regulation of fatty acid accumulation by controlling the expression of WRI1 and its downstream genes | Zhang, 2016 [33] | |
| GmNSS | PEPTIDASE-C1 DOMAIN-CONTAINING PROTEIN | Glyma.08G309000 | positive | Regulation of the size of the outer bead cover cell | Zhang, 2023 [34] | |
| Other regulators | GmDof4 | DOF ZINC FINGER PROTEIN DOF1.1-RELATED | Glyma.17G081800 | positive | Increasing lipid content in soybean seeds by upregulating genes related to fatty acid biosynthesis and direct binding downregulation of the storage protein gene CRA1 by the cis-DNA element in its promoter region | Wang, 2007 [35] |
| GmDof11 | DOF ZINC FINGER PROTEIN DOF1.1-RELATED | Glyma.13G329000 | positive | |||
| unnamed | RING FINGER AND CHY ZINC FINGER DOMAIN-CONTAINING PROTEIN 1 | Glyma.17G202700 | unknown | Prediction by GWAS analysis may correlate with seed size | Assefa1, 2019 [27] | |
| GmSSS1 | tetratricopeptide repeat protein, TPR | Glyma.19G196000 | positive | Exerts a positive impact on cell expansion and cell division, thus regulating the ultimate size of soybean seeds | Zhu, 2022 [36] |
3.1. The Ubiquitin-Proteasome Pathway
Protein ubiquitination controls many aspects of cellular processes by affecting protein stability, activity, and localization [37]. The ubiquitination process involves a series of specialized enzymes such as ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s) [38]. Ubiquitin or ubiquitin chains can be removed by deubiquitinating enzymes. Recent studies have revealed the important role of the ubiquitin-proteasome pathway in the regulation of seed size. For example, DA1 is a ubiquitin receptor in A. thaliana that contains two ubiquitin-interacting motif (UIM) domains viz., a LIM domain and a C-terminal peptidase domain [39]. DA1 negatively regulates seed and organ growth by modulating cell proliferation in the ovule integument [40]. Additionally, DA1 can interact with its closest homolog, DAR, to control seed size.
In soybean, a gene encoding an E3 ligase zinc finger protein (Glyma.17G202700) is highly expressed in the seed coat of large seeds. Additionally, a gene encoding an E2 conjugating enzyme phosphatase 2 (Glyma.07G196500) is expressed at seven-fold higher levels in the seed coat of large seeds compared to small seeds. These findings suggest the key role of the ubiquitin-mediated protein degradation pathway in the regulation of seed size in soybean [17].
3.2. Plant Hormone Signaling Pathways
Plant hormones have been documented to play a crucial role in the regulation of seed development. Different hormones exert their specific effects on various aspects of seed development, including seed size, embryo development, endosperm development, and seed dormancy.
3.3. Regulation of Seed Size by Transcription Factors
Transcriptional regulation is crucial for plant growth and development, and several transcription factors have been identified as the key regulators of seed size. These transcription factors, along with transcriptional co-activators and regulators involved in chromatin modification, play important roles in determining seed size in plants.
In soybeans, several transcription factors have been reported to be involved in seed size regulation. This includes important gene families such as WRKY and CP450 [17]. These transcription factors likely control seed size by regulating the expression of genes that are involved in cell proliferation, cell expansion, and other processes during seed development. Their precise mechanisms of action and specific target genes are still being investigated to gain a better understanding of their roles in the regulation of seed size in soybeans.
3.4. Photoperiod Regulation of Soybean Seed Size
The photoperiod i.e., the duration of light and dark periods in a day, can have an impact on seed size in plants [41]. Different plant species showed varied responses to photoperiod, and it can influence different aspects of seed development and growth. Plants have developed effective strategies to adapt to environmental changes throughout their evolution. For instance, plants perceive variations in daylight duration, allowing them to undergo the transition to flowering during seasons most conducive to successful reproduction [42]. Flowering time is a critical trait in domestication, and increased knowledge of flowering regarding its genetic basis and molecular mechanism will greatly enhance crop plants’ adaptation to the environment [43].
CONSTANS (CO) acts as a central regulator in the photoperiodic flowering pathway, and CO orthologs have been identified in various plant species [44][45][46][47]. Yu et al. [43] discovered that under favourable conditions of reproductive growth, plants tend to suppress the transcription of APETALA2 (AP2) through CO, thereby regulating the proliferation of seed coat epidermal cells and promoting larger seed production [48]. Under conditions favouring vegetative growth, CO protein becomes unstable, leading the plants to prioritize nutrient acquisition and generate smaller seeds by attenuating the inhibitory effect of CO on AP2. When CO is mutated, the seed size of the plant becomes unresponsive to photoperiod. Thus providing the first insight into the core regulatory module governing seed size in response to photoperiodic cues.
3.5. Other Regulatory Pathways of Soybean Seed Size
Nguyen et al. [49] reported and characterized the GmKIX8-1, a gene that encodes a nuclear protein regulating organ size in soybeans. GmKIX8-1 encodes a conserved KIX domain protein that is homologous to AtKIX8 in A. thaliana, which limits organ growth by modulating cell proliferation in meristematic tissues [49]. Previous studies have revealed that the KIX-PPD-MYC-GIF1 module controls seed size in A. thaliana by inhibiting cell proliferation in the outer integument during the development of embryo sac and early seed [50], this regulatory mechanism may also exist during seed development in soybean.
Flavonoids viz., anthocyanins, proanthocyanidins, flavonols, and isoflavones, are the most important components affecting seed coat colour [51]. Zhang et al. [34] identified a novel gene, Glyma.08G309000, from the mutant S006, which was named Novel Seed Size (NSS) [34]. This gene is associated with brown and small-seeded phenotypes. This study indicated a significant increase in anthocyanin accumulation in the S006 mutant leading to pigmentation in the seed coat. Moreover, the outer integument cell area was significantly reduced in the S006 mutant, thus negatively impacting seed size in soybeans. This study provides evidence that the brown seed coat colour might be attributed to elevated expression of chalcone synthase 7/8 genes, while decreased expression of NSS contributes to reduced seed size. These findings suggest that the NSS gene represents a novel regulator of seed development. The NSS gene encodes a protein of unknown function, but it contains a peptidase-c1 domain resembling a potential DNA helicase RuvA subunit, indicating a possible involvement in apoptosis.
The plant cell wall is a highly complex structure composed of structural proteins, enzymes, and various polysaccharides as well as pectin [52], which plays a crucial role in the primary cell wall of plants [53]. Li et al. [29] reported a semi-dominant locus named ST1 (Seed Thickness 1) [29] and successfully identified the underlying major functional gene of ST1 as Glyma.08G109100. This gene was documented to regulate soybean seed thickness and encodes a UDP-D-glucuronic acid 4-epimerase, which regulates the production of UDP-rhamnose and promotes pectin biosynthesis. Thus, it may determine seed shape by modulating cell division and expansion patterns in soybeans. Interestingly, this morphological variation simultaneously increases seed oil content. Together with the upregulation of sugar metabolism regulated by ST1, soybean has become a major oilseed crop, suggesting a strong selection for this gene during domestication. Additionally, Tang et al. [32] discovered that the soybean transformation inhibitor GmCIF1 participates in controlling seed maturation by specifically inhibiting the activity of cell wall invertase (CWI) [32]. Silencing of GmCIF1 expression elevates the CWI post-translational levels to finely tune sucrose metabolism and sink strength, thereby coordinating the process of seed maturation and increasing seed weight, as well as accumulating hexoses, starch, and proteins in mature seeds.
The 100-seed weight is considered one of the pivotal domestication traits that greatly influences soybean yield. Nevertheless, its elusive genetic foundation remains enigmatic. Zhu et al. [36] elucidated a soybean seed size 1 (sss1) mutant with enlarged seeds in comparison to its wild-type counterpart [36]. These authors showed that candidate gene GmSSS1 (SOYBEAN SEED SIZE 1) encodes a SPINDLY homolog that resides within a well-defined quantitative trait locus (QTL) hotspot on chromosome 19, which underwent intense selection during the cultivation of soybeans. Deletion of GmSSS1 causes reduced seed size, while its overexpression results the larger seeds, subsequently augmenting the 100-seed weight of a soybean. Further investigations indicate that GmSSS1 exerts a positive effect on cell expansion and cell division, thus regulating the ultimate size of soybean seeds (Figure 2).
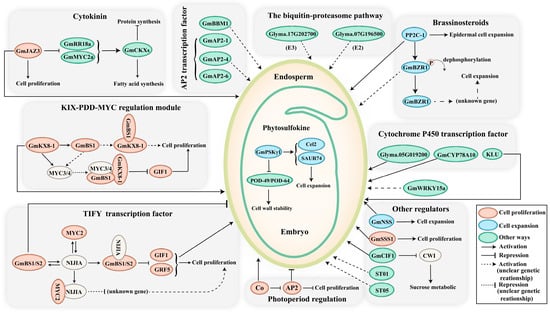
Figure 2. The major signalling pathways involved in soybean seed size control. The size of seeds is regulated through a complex network of signals, involving genes, hormones, transcription factors, and other intricate mechanisms. Dashed lines represent uncertain genetic relationships. Modulators that regulate seed size by influencing cell proliferation, cell expansion, and other regulatory processes are depicted in red, blue, and green, respectively. Abbreviations: BS, big seed; NIJIA, novel interactors of JAZ; NSS, novel seed size; SSS, soybean seed size; CWI, cell wall invertase; ST, seed thickness.
References
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183.
- Bhat, J.A.; Yu, D. High-Throughput NGS-Based Genotyping and Phenotyping: Role in Genomic-Assisted Breeding for Soybean Improvement. Legume Sci. 2021, 3, e81.
- Hina, A.; Cao, Y.; Song, S.; Li, S.; Sharmin, R.A.; Elattar, M.A.; Bhat, J.A.; Zhao, T. High-Resolution Mapping in Two RIL Populations Refines Major “QTL Hotspot” Regions for Seed Size and Shape in Soybean (Glycine max L.). Int. J. Mol. Sci. 2020, 21, 1040.
- Li, X.; Zhao, J.; Tan, Z.; Zeng, R.; Liao, H. GmEXPB2, a Cell Wall β-Expansin, Affects Soybean Nodulation through Modifying Root Architecture and Promoting Nodule Formation and Development. Plant Physiol. 2015, 169, 2640–2653.
- Taylor, B.N.; Menge, D.N.L. Light regulates tropical symbiotic nitrogen fixation more strongly than soil nitrogen. Nat. Plants 2018, 4, 655–661.
- Chen, L.; Qin, L.; Zhou, L.; Li, X.; Chen, Z.; Sun, L.; Wang, W.; Lin, Z.; Zhao, J.; Yamaji, N.; et al. A nodule-localized phosphate transporter GmPT7 plays an important role in enhancing symbiotic N2 fixation and yield in soybean. New Phytol. 2019, 221, 2013–2025.
- Yang, Z.; Du, H.; Xing, X.; Li, W.; Kong, Y.; Li, X.; Zhang, C. A small heat shock protein, GmHSP17.9, from nodule confers symbiotic nitrogen fixation and seed yield in soybean. Plant Biotechnol. J. 2022, 20, 103–115.
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “Green Revolution” for Soybean. Mol. Plant 2020, 13, 688–697.
- Doyle, J.J.; Luckow, M.A. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 2003, 131, 900–910.
- Vaughan, J.; Geissler, C.; Nicholson, B.; Dowle, E.; Rice, E. The New Oxford Book of Food Plants; OUP Oxford: Oxford, UK, 1997.
- Luo, S.; Jia, J.; Liu, R.; Wei, R.; Guo, Z.; Cai, Z.; Chen, B.; Liang, F.; Xia, Q.; Nian, H.; et al. Identification of major QTLs for soybean seed size and seed weight traits using a RIL population in different environments. Front. Plant Sci. 2022, 13, 1094112.
- Krause, M.D.; Piepho, H.-P.; Dias, K.O.G.; Singh, A.K.; Beavis, W.D. Models to estimate genetic gain of soybean seed yield from annual multi-environment field trials. TAG Theor. Appl. Genet. Theor. Und Angew. Genet. 2023, 136, 252.
- El Ouakfaoui, S.; Schnell, J.; Abdeen, A.; Colville, A.; Labbé, H.; Han, S.; Baum, B.; Laberge, S.; Miki, B. Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol. Biol. 2010, 74, 313–326.
- Jiang, W.; Zhang, X.; Song, X.; Yang, J.; Pang, Y. Genome-Wide Identification and Characterization of APETALA2/Ethylene-Responsive Element Binding Factor Superfamily Genes in Soybean Seed Development. Front. Plant Sci. 2020, 11, 566647.
- Lu, X.; Xiong, Q.; Cheng, T.; Li, Q.T.; Liu, X.L.; Bi, Y.D.; Li, W.; Zhang, W.K.; Ma, B.; Lai, Y.C.; et al. A PP2C-1 Allele Underlying a Quantitative Trait Locus Enhances Soybean 100-Seed Weight. Mol. Plant 2017, 10, 670–684.
- Ge, L.; Yu, J.; Wang, H.; Luth, D.; Bai, G.; Wang, K.; Chen, R. Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc. Natl. Acad. Sci. USA 2016, 113, 12414–12419.
- Du, J.; Wang, S.; He, C.; Zhou, B.; Ruan, Y.-L.; Shou, H. Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J. Exp. Bot. 2017, 68, 1955–1972.
- Wang, X.; Li, Y.; Zhang, H.; Sun, G.; Zhang, W.; Qiu, L. Evolution and association analysis of GmCYP78A10 gene with seed size/weight and pod number in soybean. Mol. Biol. Rep. 2015, 42, 489–496.
- Zhao, B.; Dai, A.; Wei, H.; Yang, S.; Wang, B.; Jiang, N.; Feng, X. Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 2016, 90, 33–47.
- Gu, Y.; Li, W.; Jiang, H.; Wang, Y.; Gao, H.; Liu, M.; Chen, Q.; Lai, Y.; He, C. Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size. J. Exp. Bot. 2017, 68, 2717–2729.
- Zhang, Y.; Zhang, Y.-J.; Yang, B.-J.; Yu, X.-X.; Wang, D.; Zu, S.-H.; Xue, H.-W.; Lin, W.-H. Functional characterization of GmBZL2 (AtBZR1 like gene) reveals the conserved BR signaling regulation in Glycine max. Sci. Rep. 2016, 6, 31134.
- Lu, X.; Li, Q.-T.; Xiong, Q.; Li, W.; Bi, Y.-D.; Lai, Y.-C.; Liu, X.-L.; Man, W.-Q.; Zhang, W.-K.; Ma, B.; et al. The transcriptomic signature of developing soybean seeds reveals the genetic basis of seed trait adaptation during domestication. Plant J. Cell Mol. Biol. 2016, 86, 530–544.
- Hu, Y.; Liu, Y.; Tao, J.J.; Lu, L.; Jiang, Z.H.; Wei, J.J.; Wu, C.M.; Yin, C.C.; Li, W.; Bi, Y.D.; et al. GmJAZ3 interacts with GmRR18a and GmMYC2a to regulate seed traits in soybean. J. Integr. Plant Biol. 2023, 65, 1983–2000.
- Schoonheim, P.J.; Costa Pereira, D.D.A.; De Boer, A.H. Dual role for 14-3-3 proteins and ABF transcription factors in gibberellic acid and abscisic acid signalling in barley (Hordeum vulgare) aleurone cells. Plant Cell Environ. 2009, 32, 439–447.
- Singh, A.K.; Fu, D.-Q.; El-Habbak, M.; Navarre, D.; Ghabrial, S.; Kachroo, A. Silencing genes encoding omega-3 fatty acid desaturase alters seed size and accumulation of Bean pod mottle virus in soybean. Mol. Plant Microbe Interact. 2011, 24, 506–515.
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006, 18, 3073–3087.
- Assefa, T.; Otyama, P.I.; Brown, A.V.; Kalberer, S.R.; Kulkarni, R.S.; Cannon, S.B. Genome-wide associations and epistatic interactions for internode number, plant height, seed weight and seed yield in soybean. BMC Genom. 2019, 20, 527.
- Yu, L.; Liu, Y.; Zeng, S.; Yan, J.; Wang, E.; Luo, L. Expression of a novel PSK-encoding gene from soybean improves seed growth and yield in transgenic plants. Planta 2019, 249, 1239–1250.
- Li, J.; Zhang, Y.; Ma, R.; Huang, W.; Hou, J.; Fang, C.; Wang, L.; Yuan, Z.; Sun, Q.; Dong, X.; et al. Identification of ST1 reveals a selection involving hitchhiking of seed morphology and oil content during soybean domestication. Plant Biotechnol. J. 2022, 20, 1110–1121.
- Duan, Z.; Zhang, M.; Zhang, Z.; Liang, S.; Fan, L.; Yang, X.; Yuan, Y.; Pan, Y.; Zhou, G.; Liu, S.; et al. Natural allelic variation of GmST05 controlling seed size and quality in soybean. Plant Biotechnol. J. 2022, 20, 1807–1818.
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.-C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.-Q.; et al. Simultaneous changes in seed size, oil content and protein content driven by selection of homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786.
- Tang, X.; Su, T.; Han, M.; Wei, L.; Wang, W.; Yu, Z.; Xue, Y.; Wei, H.; Du, Y.; Greiner, S.; et al. Suppression of extracellular invertase inhibitor gene expression improves seed weight in soybean (Glycine max). J. Exp. Bot. 2017, 68, 469–482.
- Zhang, Y.-Q.; Lu, X.; Zhao, F.-Y.; Li, Q.-T.; Niu, S.-L.; Wei, W.; Zhang, W.-K.; Ma, B.; Chen, S.-Y.; Zhang, J.-S. Soybean GmDREBL Increases Lipid Content in Seeds of Transgenic Arabidopsis. Sci. Rep. 2016, 6, 34307.
- Zhang, M.; Dong, R.; Huang, P.; Lu, M.; Feng, X.; Fu, Y.; Zhang, X. Novel Seed Size: A Novel Seed-Developing Gene in Glycine max. Int. J. Mol. Sci. 2023, 24, 4189.
- Wang, H.-W.; Zhang, B.; Hao, Y.-J.; Huang, J.; Tian, A.-G.; Liao, Y.; Zhang, J.-S.; Chen, S.-Y. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. Cell Mol. Biol. 2007, 52, 716–729.
- Zhu, W.; Yang, C.; Yong, B.; Wang, Y.; Li, B.; Gu, Y.; Wei, S.; An, Z.; Sun, W.; Qiu, L.; et al. An enhancing effect attributed to a nonsynonymous mutation in SOYBEAN SEED SIZE 1, a SPINDLY-like gene, is exploited in soybean domestication and improvement. New Phytol. 2022, 236, 1375–1392.
- Xu, G.; Jaffrey, S.R. The new landscape of protein Ubiquitination: Proteome-wide identification of ubiquitination events reveals their functional classes and identifies substrates for ubiquitin ligases. Nat. Biotechnol. 2011, 29, 1098–1100.
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83.
- Dong, H.; Dumenil, J.; Lu, F.-H.; Na, L.; Vanhaeren, H.; Naumann, C.; Klecker, M.; Prior, R.; Smith, C.; McKenzie, N.; et al. Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in. Genes Dev. 2017, 31, 197–208.
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008, 22, 1331–1336.
- Achary, R.K.; Majee, M. CONSTANS, a key-player connecting day length to seed size. Trends Plant Sci. 2023, 28, 975–977.
- Johansson, M.; Staiger, D. Time to flower: Interplay between photoperiod and the circadian clock. J. Exp. Bot. 2015, 66, 719–730.
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 2020, 52, 428–436.
- Song, J.; Yamamoto, K.; Shomura, A.; Itadani, H.; Zhong, H.S.; Yano, M.; Sasaki, T. Isolation and mapping of a family of putative zinc-finger protein cDNAs from rice. DNA Res. 1998, 5, 95–101.
- Griffiths, S.; Dunford, R.P.; Coupland, G.; Laurie, D.A. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003, 131, 1855–1867.
- Hayama, R.; Agashe, B.; Luley, E.; King, R.; Coupland, G. A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 2007, 19, 2988–3000.
- Wong, A.C.S.; Hecht, V.F.G.; Picard, K.; Diwadkar, P.; Laurie, R.E.; Wen, J.; Mysore, K.; Macknight, R.C.; Weller, J.L. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 2014, 5, 486.
- Yu, B.; He, X.; Tang, Y.; Chen, Z.; Zhou, L.; Li, X.; Zhang, C.; Huang, X.; Yang, Y.; Zhang, W.; et al. Photoperiod controls plant seed size in a CONSTANS-dependent manner. Nat. Plants 2023, 9, 343–354.
- Nguyen, C.X.; Paddock, K.J.; Zhang, Z.; Stacey, M.G. GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol. 2021, 229, 920–934.
- Liu, Z.; Li, N.; Zhang, Y.; Li, Y. Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis. Nat. Commun. 2020, 11, 1846.
- Liu, J.; Wang, X.; Yong, H.; Kan, J.; Jin, C. Recent advances in flavonoid-grafted polysaccharides: Synthesis, structural characterization, bioactivities and potential applications. Int. J. Biol. Macromol. 2018, 116, 1011–1025.
- Keegstra, K. Plant cell walls. Plant Physiol. 2010, 154, 483–486.
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
709
Revisions:
2 times
(View History)
Update Date:
31 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




