
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Gyöngyvér Mara | -- | 4478 | 2024-01-25 15:55:49 | | | |
| 2 | Lindsay Dong | Meta information modification | 4478 | 2024-01-29 01:37:12 | | |
Video Upload Options
The soil microbiome plays an important role in maintaining soil health, plant productivity, and soil ecosystem services. Molecular-based studies have shed light on the fact that the soil microbiome has been quantitatively underestimated. In addition to metagenomic studies, metaproteomics and metatranscriptomic studies that target the functional part of the microbiome are becoming more common. These are important for a better understanding of the functional role of the microbiome and for deciphering plant-microbe interactions. Free-living beneficial bacteria that promote plant growth by colonizing plant roots are called plant growth-promoting rhizobacteria (PGPRs). They exert their beneficial effects in different ways, either by facilitating the uptake of nutrients and synthesizing particular compounds for plants or by preventing and protecting plants from diseases.
1. Introduction
2. Soil Microbiome
2.1. Spatial Distribution of Soil Microbiome
2.2. Soil Microbiome Taxonomic Diversity: Structure and Function
2.2.1. Taxonomic Diversity
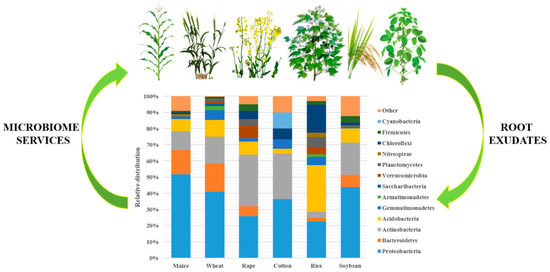
2.2.2. Factors Affecting Diversity
2.2.3. Ecological Function
2.3. Beneficial Plant–Microbe Interactions
2.3.1. Biostimulant Microbes
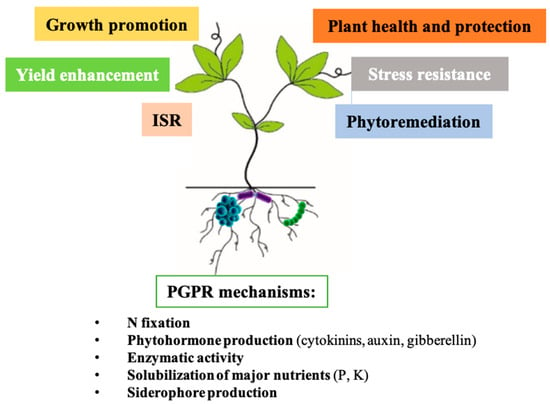
Biological Nitrogen Fixation
Phytohormone Production
Enzymatic Activity
Solubilization of Major Nutrients
Solubilization of Iron with Siderophore Production
2.3.2. Biocontrol Activity of Microbes
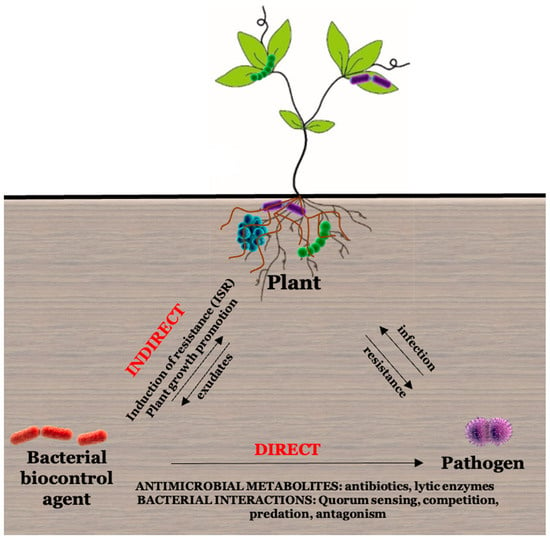
Antibiotics
Interference of Quorum Sensing with Virulence
Lytic Enzymes
Induced Systemic Resistance (ISR)
2.4. Plant-Beneficial Function Encoding Gene Clusters and Mobile Genetic Elements
2.5. Synergistic Microbial Processes
2.6. Innovations in Carrier Materials for Bioinoculants
2.7. Engineering Microbiome
References
- De Corato, U. Towards New Soil Management Strategies for Improving Soil Quality and Ecosystem Services in Sustainable Agriculture: Editorial Overview. Sustainability 2020, 12, 9398.
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511.
- Dubey, D.-A.; Malla, M.; Khan, F.; Chowdhary, K.; Yadav, S.; Kumar, A.; Sharma, S.; Khare, P.K.; Khan, M.; Khan, M. Soil microbiome: A key player for conservation of soil health under changing climate. Biodivers. Conserv. 2019, 28, 2405–2429.
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663.
- O’Brien, S.L.; Gibbons, S.M.; Owens, S.M.; Hampton-Marcell, J.; Johnston, E.R.; Jastrow, J.D.; Gilbert, J.A.; Meyer, F.; Antonopoulos, D.A. Spatial scale drives patterns in soil bacterial diversity. Environ. Microbiol. 2016, 18, 2039–2051.
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46.
- Thakur, M.P.; Geisen, S. Trophic Regulations of the Soil Microbiome. Trends Microbiol. 2019, 27, 771–780.
- Fan, K.; Cardona, C.; Li, Y.; Shi, Y.; Xiang, X.; Shen, C.; Wang, H.; Jack, G.; Chu, H. Rhizosphere-associated bacterial network structure and spatial distribution differ significantly from bulk soil in wheat crop fields. Soil Biol. Biochem. 2017, 113, 275–284.
- Bakker, P.; Berendsen, R.; Doornbos, R.; Wintermans, P.; Pieterse, C. The rhizosphere revisited: Root microbiomics. Front. Plant Sci. 2013, 4, 165.
- Saleem, M.; Hu, J.; Jousset, A. More than the Sum of Its Parts: Microbiome Biodiversity as a Driver of Plant Growth and Soil Health. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 145–168.
- Ettema, C.H.; Wardle, D.A. Spatial soil ecology. Trends Ecol. Evol. 2002, 17, 177–183.
- Bakker, M.G.; Chaparro, J.M.; Manter, D.K.; Vivanco, J.M. Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays. Plant Soil 2015, 392, 115–126.
- Li, X.; Rui, J.; Mao, Y.; Yannarell, A.; Mackie, R. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 2014, 68, 392–401.
- Yadav, A.N.; Kumar, V.; Dhaliwal, H.; Prasad, R.; Saxena, A. Microbiome in Crops: Diversity, Distribution, and Potential Role in Crop Improvement. In New and Future Developments in Microbial Biotechnology and Bioengineering: Crop Improvement through Microbial Biotechnology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 305–332. ISBN 978-0-444-63987-5.
- Mahoney, A.K.; Yin, C.; Hulbert, S.H. Community Structure, Species Variation, and Potential Functions of Rhizosphere-Associated Bacteria of Different Winter Wheat (Triticum aestivum) Cultivars. Front. Plant Sci. 2017, 8, 132.
- Rathore, R.; Dowling, D.N.; Forristal, P.D.; Spink, J.; Cotter, P.D.; Bulgarelli, D.; Germaine, K.J. Crop Establishment Practices Are a Driver of the Plant Microbiota in Winter Oilseed Rape (Brassica napus). Front. Microbiol. 2017, 8, 1489.
- Ullah, A.; Akbar, A.; Luo, Q.; Khan, A.H.; Manghwar, H.; Shaban, M.; Yang, X. Microbiome Diversity in Cotton Rhizosphere Under Normal and Drought Conditions. Microb. Ecol. 2019, 77, 429–439.
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920.
- Sugiyama, A.; Ueda, Y.; Zushi, T.; Takase, H.; Yazaki, K. Changes in the Bacterial Community of Soybean Rhizospheres during Growth in the Field. PLoS ONE 2014, 9, e100709.
- Praeg, N.; Pauli, H.; Illmer, P. Microbial Diversity in Bulk and Rhizosphere Soil of Ranunculus glacialis Along a High-Alpine Altitudinal Gradient. Front. Microbiol. 2019, 10, 1429. Available online: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01429 (accessed on 24 August 2023).
- Lakshmanan, V.; Selvaraj, G.; Bais, H.P. Functional soil microbiome: Belowground solutions to an aboveground problem. Plant Physiol. 2014, 166, 689–700.
- Aira, M.; Gómez-Brandón, M.; Lazcano, C.; Bååth, E. Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 2010, 42, 2276–2281.
- Geller, A.M.; Levy, A. “What I cannot create, I do not understand”: Elucidating microbe-microbe interactions to facilitate plant microbiome engineering. Curr. Opin. Microbiol. 2023, 72, 102283.
- Khan, A.A.H. Plant-Bacterial Association and Their Role as Growth Promoters and Biocontrol Agents. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Cham, Switzerland, 2019; pp. 389–419. ISBN 9789811369858.
- Eva, L.; Gyongyver, M. Is PGPR an Alternative for NPK Fertilizers in Sustainable Agriculture? In Microbial Interventions in Agriculture and Environment; Springer: Cham, Switzerland, 2019; pp. 51–62. ISBN 9789811383908.
- Turan, M.; Topcuoğlu, B.; Kıtır, N.; Alkaya, Ü.; Erçelik, F.; Nikerel, E.; Güneş, A.; Turan, M.; Topcuoğlu, B.; Kıtır, N.; et al. Plant Growth Promoting Rhizobacteria’s (PGPRS) Enzyme Dynamics in Soil Remediation. In Soil Contamination—Current Consequences and Further Solutions; IntechOpen: London, UK, 2016; ISBN 978-953-51-2816-8.
- Fowler, D.; Steadman, C.E.; Stevenson, D.; Coyle, M.; Rees, R.M.; Skiba, U.M.; Sutton, M.A.; Cape, J.N.; Dore, A.J.; Vieno, M.; et al. Effects of global change during the 21st century on the nitrogen cycle. Atmos. Chem. Phys. 2015, 15, 13849–13893.
- Ladha, J.K.; Peoples, M.B.; Reddy, P.M.; Biswas, J.C.; Bennett, A.; Jat, M.L.; Krupnik, T.J. Biological nitrogen fixation and prospects for ecological intensification in cereal-based cropping systems. Field Crops Res. 2022, 283, 108541.
- Matilla, M.; Krell, T. Plant Growth Promotion and Biocontrol Mediated by Plant-Associated Bacteria. In Plant Microbiome: Stress Response; Springer: Cham, Switzerland, 2018; pp. 45–80. ISBN 978-981-10-5513-3.
- Ahmad, M.; Nadeem, S.M.; Zahir, Z.A. Plant-Microbiome Interactions in Agroecosystem: An Application. In Microbiome in Plant Health and Disease; Springer: Singapore, 2019; pp. 251–291.
- Salazar-Cerezo, S.; Martínez-Montiel, N.; García-Sánchez, J.; Pérez-Y-Terrón, R.; Martínez-Contreras, R.D. Gibberellin biosynthesis and metabolism: A convergent route for plants, fungi and bacteria. Microbiol. Res. 2018, 208, 85–98.
- de Garcia Salamone, I.E.; Hynes, R.K.; Nelson, L.M. Role of Cytokinins in Plant Growth Promotion by Rhizosphere Bacteria. In PGPR: Biocontrol and Biofertilization; Siddiqui, Z.A., Ed.; Springer: Dordrecht, The Netherlands, 2006; pp. 173–195. ISBN 978-1-4020-4152-5.
- Ramadan, E.; Abdelhafez, A.; Enas, A.; Saber, F. Plant growth promoting rhizobacteria and their potential for biocontrol of phytopathogens. Afr. J. Microbiol. Res. 2016, 10, 486–504.
- Zhang, S.; Gan, Y.; Xu, B. Mechanisms of the IAA and ACC-deaminase producing strain of Trichoderma longibrachiatum T6 in enhancing wheat seedling tolerance to NaCl stress. BMC Plant Biol. 2019, 19, 22.
- Sakure, S.; Bhosale, S. Actinobacteria for Biotic Stress Management. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Cham, Switzerland, 2019; pp. 363–378. ISBN 9789811369858.
- Mathur, A.; Koul, A.; Hattewar, J. Plant Growth-Promoting Rhizobacteria (PGPRs): Significant Revolutionary Tools for Achieving Long-Term Sustainability and Combating the Biotic Stress Caused by the Attack of Pathogens Affecting Crops in Agriculture. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Cham, Switzerland, 2019; pp. 379–388. ISBN 9789811369858.
- Tandon, S.; Vats, S. Microbial Biosynthesis of Cadmium Sulfide (CDS) Nanoparticles and their Characterization. Eur. J. Pharm. Med. Res. 2016, 3, 545–550.
- Vats, S.; Bhargava, P. Alternate Energy: Fuel for “Modi’s India” and “Smart cities”. Int. J. Curr. Res. 2017, 9, 49090–49097.
- Saxena, P.; Srivastava, J.; Pandey, S.; Srivastava, S.; Maurya, N.; Chand, K.; Mishra, S.; Asthana, G.; Bhargava, P.; Kumar, R.; et al. Plants for Biocontrol and Biological Control of Plant Pathogens. In Plant Biotic Interactions; Springer: Cham, Switzerland, 2019; pp. 147–179. ISBN 978-3-030-26656-1.
- Arseneault, T.; Filion, M. Biocontrol through antibiosis: Exploring the role played by subinhibitory concentrations of antibiotics in soil and their impact on plant pathogens. Can. J. Plant Pathol. 2017, 39, 267–274.
- Ram, R.M.; Keswani, C.; Bisen, K.; Tripathi, R.; Singh, S.P.; Singh, H.B. Biocontrol Technology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 177–190.
- Quiñones, B.; Dulla, G.; Lindow, S.E. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 2005, 18, 682–693.
- Koutsoudis, M.D.; Tsaltas, D.; Minogue, T.D.; von Bodman, S.B. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. USA 2006, 103, 5983–5988.
- Hussain, M.B.B.M.; Zhang, H.-B.; Xu, J.-L.; Liu, Q.; Jiang, Z.; Zhang, L.-H. The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J. Bacteriol. 2008, 190, 1045–1053.
- Kang, B.R.; Anderson, A.J.; Kim, Y.C. Hydrogen Cyanide Produced by Pseudomonas chlororaphis O6 Exhibits Nematicidal Activity against Meloidogyne hapla. Plant Pathol. J. 2018, 34, 35–43.
- Yu, Y.; Gui, Y.; Li, Z.; Jiang, C.; Guo, J.; Niu, D. Induced Systemic Resistance for Improving Plant Immunity by Beneficial Microbes. Plants 2022, 11, 386.
- Lekired, A.; Cherif-Silini, H.; Silini, A.; Ben Yahia, H.; Ouzari, H.-I. Comparative genomics reveals the acquisition of mobile genetic elements by the plant growth-promoting Pantoea eucrina OB49 in polluted environments. Genomics 2023, 115, 110579.
- Maheshwari, M.; Abulreesh, H.H.; Khan, M.S.; Ahmad, I.; Pichtel, J. Horizontal Gene Transfer in Soil and the Rhizosphere: Impact on Ecological Fitness of Bacteria. In Agriculturally Important Microbes for Sustainable Agriculture: Volume I: Plant-Soil-Microbe Nexus; Meena, V.S., Mishra, P.K., Bisht, J.K., Pattanayak, A., Eds.; Springer: Singapore, 2017; pp. 111–130. ISBN 978-981-10-5589-8.
- Sánchez-Salazar, A.M.; Taparia, T.; Olesen, A.K.; Acuña, J.J.; Sørensen, S.J.; Jorquera, M.A. An overview of plasmid transfer in the plant microbiome. Plasmid 2023, 127, 102695.
- Ku, Y.-S.; Wang, Z.; Duan, S.; Lam, H.-M. Rhizospheric Communication through Mobile Genetic Element Transfers for the Regulation of Microbe–Plant Interactions. Biology 2021, 10, 477.
- Popowska, M.; Krawczyk-Balska, A. Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 2013, 4, 44.
- Ma, W.; Charles, T.C.; Glick, B.R. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl. Environ. Microbiol. 2004, 70, 5891–5897.
- Serova, T.A.; Tikhonovich, I.A.; Tsyganov, V.E. Analysis of nodule senescence in pea (Pisum sativum L.) using laser microdissection, real-time PCR, and ACC immunolocalization. J. Plant Physiol. 2017, 212, 29–44.
- Camerini, S.; Senatore, B.; Lonardo, E.; Imperlini, E.; Bianco, C.; Moschetti, G.; Rotino, G.L.; Campion, B.; Defez, R. Introduction of a novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch. Microbiol. 2008, 190, 67–77.
- Defez, R.; Esposito, R.; Angelini, C.; Bianco, C. Overproduction of Indole-3-Acetic Acid in Free-Living Rhizobia Induces Transcriptional Changes Resembling Those Occurring in Nodule Bacteroids. Mol. Plant Microbe Interact. 2016, 29, 484–495.
- Defez, R.; Andreozzi, A.; Romano, S.; Pocsfalvi, G.; Fiume, I.; Esposito, R.; Angelini, C.; Bianco, C. Bacterial IAA-Delivery into Medicago Root Nodules Triggers a Balanced Stimulation of C and N Metabolism Leading to a Biomass Increase. Microorganisms 2019, 7, 403.
- Nascimento, F.X.; Brígido, C.; Glick, B.R.; Oliveira, S.; Alho, L. Mesorhizobium ciceri LMS-1 expressing an exogenous 1-aminocyclopropane-1-carboxylate (ACC) deaminase increases its nodulation abilities and chickpea plant resistance to soil constraints. Lett. Appl. Microbiol. 2012, 55, 15–21.
- Nascimento, F.X.; Tavares, M.J.; Franck, J.; Ali, S.; Glick, B.R.; Rossi, M.J. ACC deaminase plays a major role in Pseudomonas fluorescens YsS6 ability to promote the nodulation of Alpha- and Betaproteobacteria rhizobial strains. Arch. Microbiol. 2019, 201, 817–822.
- Alemneh, A.A.; Zhou, Y.; Ryder, M.H.; Denton, M.D. Mechanisms in plant growth-promoting rhizobacteria that enhance legume-rhizobial symbioses. J. Appl. Microbiol. 2020, 129, 1133–1156.
- Orozco-Mosqueda, M.d.C.; Glick, B.R.; Santoyo, G. ACC deaminase in plant growth-promoting bacteria (PGPB): An efficient mechanism to counter salt stress in crops. Microbiol. Res. 2020, 235, 126439.
- Alemneh, A.A.; Zhou, Y.; Ryder, M.H.; Denton, M.D. Is phosphate solubilizing ability in plant growth-promoting rhizobacteria isolated from chickpea linked to their ability to produce ACC deaminase? J. Appl. Microbiol. 2021, 131, 2416–2432.
- Nukui, N.; Minamisawa, K.; Ayabe, S.-I.; Aoki, T. Expression of the 1-aminocyclopropane-1-carboxylic acid deaminase gene requires symbiotic nitrogen-fixing regulator gene nifA2 in Mesorhizobium loti MAFF303099. Appl. Environ. Microbiol. 2006, 72, 4964–4969.
- Balla, A.; Silini, A.; Cherif-Silini, H.; Chenari Bouket, A.; Alenezi, F.N.; Belbahri, L. Recent Advances in Encapsulation Techniques of Plant Growth-Promoting Microorganisms and Their Prospects in the Sustainable Agriculture. Appl. Sci. 2022, 12, 9020.
- Pereira, J.F.; Oliveira, A.L.M.; Sartori, D.; Yamashita, F.; Mali, S. Perspectives on the Use of Biopolymeric Matrices as Carriers for Plant-Growth Promoting Bacteria in Agricultural Systems. Microorganisms 2023, 11, 467.
- Lobo, C.B.; Juárez Tomás, M.S.; Viruel, E.; Ferrero, M.A.; Lucca, M.E. Development of low-cost formulations of plant growth-promoting bacteria to be used as inoculants in beneficial agricultural technologies. Microbiol. Res. 2019, 219, 12–25.
- Marcelino, P.R.F.; Milani, K.M.L.; Mali, S.; Santos, O.J.A.P.D.; de Oliveira, A.L.M. Formulations of polymeric biodegradable low-cost foam by melt extrusion to deliver plant growth-promoting bacteria in agricultural systems. Appl. Microbiol. Biotechnol. 2016, 100, 7323–7338.
- Panichikkal, J.; Prathap, G.; Nair, R.A.; Krishnankutty, R.E. Evaluation of plant probiotic performance of Pseudomonas sp. encapsulated in alginate supplemented with salicylic acid and zinc oxide nanoparticles. Int. J. Biol. Macromol. 2021, 166, 138–143.
- Zago, S.; Fonseca dos Santos, M.; Konrad, D.; Fiorini, A.; Rosado, F.; Missio, R.; Vendruscolo, E. Shelf Life of Azospirillum brasilense in Alginate Beads Enriched with Trehalose and Humic Acid. J. Agric. Sci. 2019, 11, 269.
- Jurić, S.; Đermić, E.; Topolovec-Pintarić, S.; Bedek, M.; Vinceković, M. Physicochemical properties and release characteristics of calcium alginate microspheres loaded with Trichoderma viride spores. J. Integr. Agric. 2019, 18, 2534–2548.
- Kumar, A.; Dubey, A. Rhizosphere microbiome: Engineering bacterial competitiveness for enhancing crop production. J. Adv. Res. 2020, 24, 337–352.
- Bano, S.; Wu, X.; Zhang, X. Towards sustainable agriculture: Rhizosphere microbiome engineering. Appl. Microbiol. Biotechnol. 2021, 105, 7141–7160.
- Haskett, T.L.; Tkacz, A.; Poole, P.S. Engineering rhizobacteria for sustainable agriculture. ISME J. 2021, 15, 949–964.




