Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Brock Hewitt | -- | 4213 | 2024-01-24 15:45:28 | | | |
| 2 | Camila Xu | Meta information modification | 4213 | 2024-01-25 04:01:21 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Larrain, C.; Torres-Hernandez, A.; Hewitt, D.B. Artificial Intelligence in the Management of Hepatocellular Carcinoma. Encyclopedia. Available online: https://encyclopedia.pub/entry/54300 (accessed on 08 February 2026).
Larrain C, Torres-Hernandez A, Hewitt DB. Artificial Intelligence in the Management of Hepatocellular Carcinoma. Encyclopedia. Available at: https://encyclopedia.pub/entry/54300. Accessed February 08, 2026.
Larrain, Carolina, Alejandro Torres-Hernandez, Daniel Brock Hewitt. "Artificial Intelligence in the Management of Hepatocellular Carcinoma" Encyclopedia, https://encyclopedia.pub/entry/54300 (accessed February 08, 2026).
Larrain, C., Torres-Hernandez, A., & Hewitt, D.B. (2024, January 24). Artificial Intelligence in the Management of Hepatocellular Carcinoma. In Encyclopedia. https://encyclopedia.pub/entry/54300
Larrain, Carolina, et al. "Artificial Intelligence in the Management of Hepatocellular Carcinoma." Encyclopedia. Web. 24 January, 2024.
Copy Citation
Artificial Intelligence (AI) can be a useful tool in the management of disease processes such as hepatocellular carcinoma (HCC) as treatment decisions are often complex and multifaceted. AI applications in medicine are expanding with the ongoing advances in AI including more sophisticated machine learning and deep learning processes.
artificial intelligence
machine learning
deep learning
radiomics
hepatocellular carcinoma
1. Introduction
Primary liver cancer is the sixth most commonly diagnosed cancer worldwide and hepatocellular carcinoma (HCC) accounts for over 80% of newly diagnosed cases [1]. Due to the lack of early diagnostic markers, absence of specific symptoms in early disease, and inadequate screening programs in most countries, nearly two out of every three patients diagnosed with HCC have intermediate or advanced disease upon diagnosis [2][3]. Unfortunately, these patients often have a poor prognosis as their disease may not be amenable to curative interventions, such as surgical resection, liver transplantation, or ablation, and, until recently, effective systemic options were limited. As a result, HCC is the fourth most common cause of cancer-related death worldwide with a relative five-year survival rate of 18% [4][5]. However, the recent inclusion of immunotherapy into the HCC treatment paradigm and the expansion of downstaging/bridging protocols to liver transplantation have improved overall survival and established a new standard of care for patients with HCC [6][7]. In addition to surgical and systemic therapeutic options, a number of liver-directed therapies (e.g., bland transarterial embolization (TAE), transarterial chemoembolization (TACE), radioembolization (RE)) are available depending on the extent of cirrhosis and the stage of the disease [5]. With the expanding therapeutic armamentarium for patients with HCC, novel tools are needed to effectively stratify patients to maximize therapeutic benefit.
Artificial intelligence (AI) has recently emerged as a viable clinical tool with growing utility in the management of HCC. Broadly, AI is a subdivision of data science which describes the theory and development of computer systems that perform tasks requiring human-level intelligence such as visual perception or decision making. AI was originally conceptualized in the 1950s by the mathematician Alan Turing and the field has greatly expanded since its original conception. As technology continues to evolve, new AI techniques have been developed to address more complex and sophisticated problems. Machine learning and deep learning are two such subfields of AI (Figure 1).
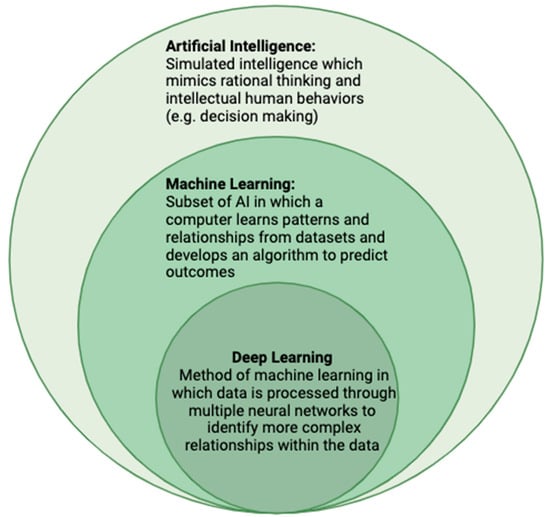
Figure 1. The Relationship between artificial intelligence, machine learning, and deep learning.
In machine learning, computer systems learn and adapt by using algorithms and statistical models to draw inferences from patterns present in data [8]. Unlike traditional statistical programming where the data and algorithms are provided and the output is produced, machine learning systems are provided the data and then independently “develop” an algorithm to process the data. Algorithm accuracy improves overtime as the system “learns” from additional data/output cycles. Two common learning methods are supervised and unsupervised learning. In supervised learning, the computer system is provided human-labeled datasets including desired inputs and outputs. The operator knows the desired output; the algorithm “learns” from the observations to identify patterns in the data and make predictions. Examples of supervised learning include classification, regression, and forecasting. In contrast, unsupervised machine learning systems do not receive output information. The system identifies associations and relationships to group the data in a more organized way without an “answer key”. Clustering and dimension reduction are two examples of unsupervised learning [9].
The datasets used to develop machine learning models are typically divided into training, validation, and testing datasets. Once an algorithm has been developed from a training dataset, it is further optimized and tuned with a validation set. Upon completion of the training phase, its performance will be evaluated with a test dataset comprising new data which the system has not yet encountered. Ideally, the algorithm will have a similar predictive power with the test dataset as with the one it was trained through, implying generalizability of the model. It is important to note the generalizability of an algorithm is largely based upon the characteristics of its training and the validation dataset. Any biases present within the dataset, such as homogeneity due to sampling error, will become inherent to the model. This will ultimately lead to decreased generalizability and a poorer predictive performance of the model [9].
Deep learning is a subset of machine learning and more closely mimics human intelligence. The system is built upon artificial neural networks that are modeled on the biologic neural networks of the brain [10]. Deep learning models have the ability to process more complex data, such as images, text or sounds, and are the basis of complex models such as speech recognition or large-scale image analysis. This method of AI processes data through multiple neural layers, progressively extracting higher-level features to produce a complete learned result. These networks can comprise millions of neural layers each of which receives data from the previous layer, transforms the data, and sends them to connected neurons in succeeding layers (Figure 2). Each of these connections has a different weighted value based upon which characteristics are most important and predictive in achieving an accurate output. These networks of neurons—in addition to the initial input and final output layers—are known as hidden layers. One major limitation of deep learning methods is that it is not always understood how the model has reached the final output, a phenomenon known as the “black box” of deep learning. It is often unclear how the model has transformed the data to reach the output. Furthermore, data are processed in such a granular fashion and through countless amounts of hidden neural layers that these connections are often not comprehensible or meaningful to the human brain [11]. Another major limitation of deep learning is that these models require sizable training datasets in order to extrapolate meaningful relationships within the data. This can be a deterrent to developing functional models for entities with few observations as is frequently the case when evaluating rare diseases [12].
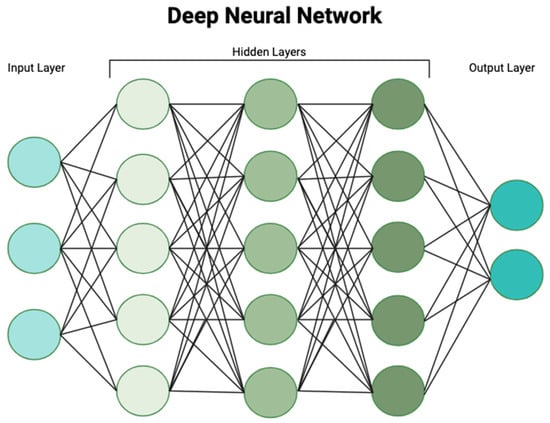
Figure 2. Deep learning processes and transforms data through multiple artificial neural networks to generate a predictive algorithm based on complex patterns within the data.
AI has a growing presence in medicine; however, ethical considerations and technical constraints initially limited broader adoption and the implementation of AI within the medical field. While many ethical considerations remain, recent advances in AI methodologies have overcome some of the technical constraints. For example, Jiang et al. reported on the superiority of a self-supervised large language model that was trained on unstructured clinical notes from an electronic health record to predict 30-day all-cause readmission, in-hospital mortality, comorbidity indices, lengths of stay, and insurance denial when compared with traditional models [13]. Furthermore, an AI model using deep network automatic segmentation outperformed a clinical model and a radiomic model in discriminating between patients with pancreatic cancer with and without lymph node metastasis [14]. These advances have improved the accuracy and expanded the clinical utility of AI systems in the field of medicine.
2. Screening and Detection
Since many patients diagnosed with HCC have advanced disease and limited therapeutic options, improvements in screening and early detection are necessary to improve outcomes. For example, a review of country-level HCC surveillance programs demonstrated that countries with established and effective HCC surveillance programs detected HCC at significantly earlier stages and had lower overall mortality [15]. Effective programs identify and stratify patients at high-risk of developing HCC and enroll them into regular surveillance protocols such as biannual evaluation with liver ultrasound and liquid tumor markers. Ideally, healthcare professionals intervene prior to the development of irreversible HCC risk factors (i.e., cirrhosis). However, cost/benefit restraints and high false-positive rates limit the utility of screening low-prevalence populations. AI techniques may help us to overcome these limitations (Table 1). In a 2022 study, Blanes-Vidal et al. evaluated asymptomatic patients from a primary care population without a prior diagnosis of liver disease [16]. The authors tested the diagnostic performance of ensemble models, a machine learning approach, to detect liver fibrosis and then compared the performance with standard blood-based scoring systems. The ensemble models included data readily available during a primary care visit. In a subset of 463 patients that received a liver biopsy, the ensemble learning models significantly outperformed standard blood-based indices to detect liver stiffness (>8 kPa) and fibrosis (Kleiner biopsy stage F2 to F4) with AUCs of 0.86–0.94 vs. 0.60–0.76. Furthermore, all the ensemble models had a ≥98% negative predictive value. Similarly, other AI models have demonstrated superiority to standard models in predicting the development of HCC [17][18].
Radiomics, a quantitative method to extract features (e.g., shape, intensity, texture) from medical imaging, can significantly improve the diagnostic yield of imaging modalities, especially when combined with other AI techniques such as deep learning. While established criteria exist for the radiologic diagnosis of HCC in high-risk patients (i.e., LI-RADS), the true proportion of patients with LI-RADS 5 lesions (diagnostic of HCC) is unclear. Additionally, this classification system only applies to patients at high risk of HCC (e.g., cirrhosis, HBV infection) [19]. The diagnostic uncertainty of many newly identified liver lesions may lead to unnecessary serial imaging, invasive procedures (e.g., biopsy), and undue psychological stress on the patient. For these reasons, much energy has been devoted to building radiomic-based models that can accurately detect and diagnose HCC (Table 1). Yasaka et al. developed a deep learning model using a convolutional neural network (CNN) to differentiate malignant liver lesions from non-malignant lesions [20]. In this retrospective study, the authors built a CNN using imaging sets from 460 patients who were found to have liver lesions on triple-phase CT. The lesions were classified into five categories as follows: classic HCC (A), malignant tumors other than HCC (B), indeterminate masses and rare benign liver masses (C), hemangiomas (D), or cysts €. The CNN accurately identified malignant lesions with the median AUC of 0.92 for differentiating categories A–B from categories C–E. Furthermore, Mokrane et al. used a radiomic model to improve the diagnostic accuracy of HCC in cirrhotic patients with indeterminate liver nodules [21]. In this multicenter retrospective study, multiphasic CT scans of 178 cirrhotic patients who had undergone the biopsy of indeterminate liver nodules were included. Nearly 14,000 quantitative features were extracted from imaging sets. With machine learning algorithms, a radiomic signature was created and validated to classify these indeterminate liver nodules as HCC or non-HCC. While the LI-RADS scores were similar between the HCC and non-HCC groups, the radiomic signature reached an AUC of 0.70 in the discovery cohort and 0.66 in the validation cohort. AI has also been used to augment the diagnostic capabilities of other imaging modalities including ultrasound and magnetic resonance imaging (MRI) [22][23][24].
While the generalizability of screening and diagnostic models derived from AI techniques requires further evaluation, these models offer non-invasive and resource-efficient means to reliably screen and detect HCC in select populations. This technology can be especially useful in low resource centers where specialized liver radiologists may not be readily available to aid in the diagnosis of complex liver lesions. Furthermore, these models may help institutions and governments to more efficiently allocate scarce resources to HCC screening and intervention due to improved risk stratification. For example, using a deep learning recurrent neural network model to generate HCC risk scores, Ioannou et al. found that 80% of HCC cases diagnosed in the subsequent three years occurred in the highest 51% of risk scores [18]. Risk-based screening supported by AI algorithms may increase diagnostic yield, optimize resource utilization, and help us to overcome the suboptimal performance of existing tools [25].
Table 1. Select studies utilizing AI in screening and diagnosis of HCC.
| Author, Year | Model Design | Population | AI Methodology | Accuracy |
|---|---|---|---|---|
| Blanes-Vidal et al. (2022) [16] | Prediction of liver fibrosis using clinical data readily available to primary care physicians | Low-prevalence primary care population | Ensemble learning model | AUC: 0.86–0.94 |
| Ioannou et al. (2020) [18] | Identification of patients at high risk of developing HCC by extracting data from electronic medical records | Patients with known Hepatitis C Virus and cirrhosis | Recurrent neural network | AUROC: 0.759 |
| Yasaka et al. (2018) [20] | Differentiation of liver masses on CT, with categorization into HCC, other liver tumors, hemangiomas, or cysts | Patients who had undergone dynamic contrast-enhanced CT for evaluation of liver lesions | Convolutional neural network | AUROC: 0.92 |
| Mokrane et al. (2020) [21] | Diagnosis of liver nodules as HCC vs. non-HCC based on quantitative features extracted from triphasic CT | Patients with cirrhosis and biopsy-proven indeterminate liver nodules | Machine learning-based radiomic signature | AUROC: 0.66 |
| Schmauch et al. (2019) [22] | Detection and characterization of focal liver lesions as benign- vs. malignant-based on ultrasound characteristics | Patients with known liver nodules | Residual neural network | AUROC: 0.935 |
3. Prognosis and Treatment
3.1. HCC Prognosis and Risk of Recurrence
Recent multi-omic investigations have advanced the understanding of the carcinogenic mechanisms responsible for HCC, revealing many potential biomarkers [26]. Evaluating these large data sets with AI methods may improve current prognostic ability by identifying more aggressive subtypes and patients at high risk of recurrence (Table 2). In an early study using a deep learning framework to integrate multi-omic data in patients with HCC, Chaudhary et al. developed a deep learning model from 360 patients with HCC using RNA sequencing, miRNA sequencing, and methylation data from The Cancer Genome Atlas [27]. The model identified a more aggressive subtype with worse survival characterized through frequent TP53 mutations; a higher expression of KRT19, EPCAM, and BIRC5; and activated Wnt and Akt signaling pathways. The investigators then validated the model on five external data sets with acceptable results.
In addition to detecting relationships within large multi-omic data sets to improve prognostication, AI techniques can help us identify biomarkers in the preoperative setting typically only identified through pathologic evaluation such as microvascular invasion (MVI) [28]. For example, multiple studies have shown the feasibility of using machine learning algorithms to accurately predict the presence of MVI based on preoperative axial imaging characteristics [29][30][31]. Chong et al. built a radiomic-based nomogram to assess the risk of MVI [32]. This retrospective study analyzed preoperative MRIs from 356 patients with pathologically confirmed solitary HCC less than 5 cm. The nomogram, built by extracting radiomic features from images containing tumors, peritumoral tissue, and non-tumoral liver parenchyma, accurately predicted the risk of MVI as well as recurrence-free survival.
Similarly, other groups have identified biomarkers of HCC recurrence using machine learning methods (Table 2) [33]. Yan et al. created a deep learning MR signature derived from imaging characteristics including tumor size, arterial phase enhancement type, capsular appearance, presence of a hypointense halo, intratumoral necrosis, satellite nodules, and peritumoral hypointensity [34]. When combined with clinical factors such as MVI and tumor number, the MR signature predicted early recurrence better than clinical data alone. Another model created by Ji et al. extracted radiomic features from preoperative CT scans to build a radiomic signature that, when integrating clinical data such as MVI, AFP level, and tumor number, accurately predicted the risk of recurrence in patients after resection of early-stage HCC [35]. Improved prognostication and risk stratification with AI techniques may better inform management decisions for patients with HCC.
Table 2. Key studies utilizing AI to predict prognosis and risk of recurrence of HCC.
| Author, Year | Model Design | Pertinent Risk Factors | Population | AI Methodology | Accuracy |
|---|---|---|---|---|---|
| Chaudhary et al. (2018) [27] | Predictive model for HCC prognosis based on molecular signature and multi-omic data |
|
HCC patients within the Genome Cancer Atlas (TCGA) | Deep learning | C-index: 0.68 |
| Liu et al. (2021) [30] | Prediction of MVI preoperatively based on CT imaging characteristics and patient clinical factors |
|
Patients with HCC | Residual Neural Network | AUC: 0.845 |
| Chong et al. (2021) [32] | Creation of radiomic-based nomogram to preoperatively predict risk of MVI and recurrence-free survival, based. on MRI characteristics and clinical data |
|
Patients with solitary HCC smaller than 5cm | Random Forrest | AUC: 0.92 |
| Ji et al. (2020) [35] | Creation of radiomic signature with pre- and post-resection features to predict recurrence for early-stage HCC |
|
Patients with HCC that met the Milan Criteria and underwent curative intent resection | Machine learning-based radiomic signature | C-index: 0.77 |
3.2. Pathologic Assessment
AI techniques have also been applied to evaluate associations between histologic features and outcomes in numerous disease processes, including HCC (Table 3) [36][37][38][39]. Using whole-slide imaging, Yamashita et al. developed a deep learning-based system to predict a recurrence-free disease interval after curative-intent hepatectomy in patients with HCC. Their model stratified patients into low-risk and high-risk subgroups and outperformed the standard tumor–node–metastasis (TNM) staging system [40].
Chen et al. built a neural network that was able to assist in the prognostication of HCC based on histologic whole-slide imaging. The model used hematoxylin and eosin slides from a genomic database to train a neural network to classify liver lesions as malignant with 96.0% accuracy and predict lesion histopathological grade with 89.6% accuracy. Furthermore, the model also predicted select gene mutations including CTNNB1, FMN2, TP53, and XFZ4 [41]. AI-based pathology models can also predict the activation of immune signatures in HCC. Zeng et al. used deep learning approaches on whole-slide histologic images and gene expression profiling, derived from the Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) public dataset, to develop models that predicted the activation of six key immune signatures that, when overexpressed, correlated with the response to immunotherapy [42]. While prospective validation studies are needed, these data demonstrate the potential utility of AI to select patients who will have a greater response to immunotherapy and may inform adjuvant therapy decisions.
Table 3. Key studies demonstrating the use of AI on whole-slide imaging.
| Author, Year | Model Design | Population | AI Methodology | Accuracy |
|---|---|---|---|---|
| Qu et al. (2022) [36] | Creation of histological score using whole-slide imaging to predict HCC recurrence | Patients with early-stage HCC who had undergone surgical resection in a single institutional dataset and the TCGA dataset | Convolutional neural network | C-index: 0.804 |
| Saillard et al. (2020) [37] | Use of whole-slide imaging to predict risk of HCC recurrence and stratifying it into low- and high-risk subgroups | Patients with HCC who had undergone surgical resection in a single institutional dataset and the TCGA dataset | Convolutional neural network | C-index: 0.72 |
| Yamashita et al. (2021) [40] | Use of whole-slide imaging to formulate a risk score predictive of HCC recurrence | Patients with HCC in the TCGA and Stanford-HCC dataset | Convolutional neural network | C-index: 0.724 |
| Zeng et al. (2022) [42] | Prediction of activation of immune gene signatures based on whole-slide imaging | Patients with HCC who had undergone surgical resection in the TCGA dataset | Clustering-constrained attention multiple instance learning | AUROC: 0.78–0.91 |
3.3. Locoregional Therapies
AI-based models may help to select patients who are good candidates for locoregional therapies such as radiofrequency ablation (RFA) (Table 4). Wu et al. built an artificial neural network based on fifteen clinical variables from HCC patients who had undergone CT-guided RFA [43]. Variables included patient characteristics, tumor size, tumor number, and laboratory values (e.g., AFP). Ultimately, the artificial neural network model predicted one-year disease-free survival with an AUC of 0.84 and one-year disease-free survival with an AUC of 0.75. Lui et al. created a deep learning radiomic-based model of preoperative contrast-enhanced liver ultrasound images and predicted the response to the first TACE session in patients with HCC [44]. In another study, investigators trained and validated a CNN to automatically assess splenic volume. Patients with higher splenic volumes, as automatically determined by the CNN, had significantly higher risk of hepatic decompensation and lower overall survival after TACE. Conversely, axial and craniocaudal splenic diameter did not correlate with outcomes [45].
3.4. Automatic Methods for Liver and Tumor Segmentation
Automatic liver and tumor segmentation has numerous clinical applications which may aid in the management of HCC such as the preoperative volumetric quantification and assessment of treatment responses to locoregional therapies (Table 4). The use of manual and semi-automatic liver segmentation methods can lead to inconsistencies due to user variability. Furthermore, segmentation is often a slow, labor-intensive process. With advancements in deep learning techniques and CNNs, completely automatic liver segmentation is now feasible [46][47][48]. However, automatic algorithms do have their own limitations including the need for large training datasets to develop accurate algorithms. Automatic tumor segmentation can be even more challenging as tumor characteristics, such as variability in size, variability in location, and indiscrete borders between healthy liver parenchyma and tumors, can decrease the accuracy of models. Regardless, early studies demonstrate that automatic segmentations outperform semi-automatic methods with regard to the accuracy and repeatability of segmentation [49]. Meng et al. used a three-dimensional (3D) dual-path multiscale CNN to build a liver and liver tumor segmentation algorithm based on abdominal CT images. The dual path multiscale 3D network architecture allowed the model to capture large scale global features through one path while capturing more granular local features through the second path. Notably, their algorithm performed best on large tumors [50]. Zheng et al. built a four-dimensional (4D) deep learning model to segment HCC lesions based on dynamic contrast-enhanced MRIs. In an attempt to avoid the “black box” learning that occurs with many automatic segmentation models, the investigators first build a 3D CNN to separately extract imaging features from each individual phase of imaging. From there, these data were fed into a convolutional long short-term memory (C-LSTM) network module in order to extract higher-level features, including temporal information and dynamic features, that varied through the multi-phase imaging. This allows us to use dynamic features that are not only characteristic of HCC but critical to diagnosis, such as arterial phase hyperenhancement and portal venous or delayed-phase washout. Their model achieved a Dice score of 0.825 ± 0.077 for HCC tumor segmentation in an internal test set and 0.786 ± 0.073 in an external set, implying good generalizability of this model [51].
3.5. Surgical Complications
Many HCC patients have some level of hepatic dysfunction upon presentation, increasing the risk of post-hepatectomy liver failure (PHLF). PHLF remains the most frequent cause of postoperative mortality in patients following hepatectomy, highlighting the importance of identifying patients at high risk of PHLF prior to resection to more effectively select patients for adjuncts such as portal vein embolization. Wang et al. constructed a machine learning clinical model using laboratory values, tumor characteristics, and surgical variables (e.g., surgical approach, extent of resection, intraoperative blood loss) to predict the risk of PHLF. The model outperformed traditional models such as MELD, Child-Turcotte-Pugh, or albumin-bilirubin grade when predicting PHLF [52]. AI-derived algorithms have successfully predicted other surgical complications. Laino et al. predicted the risk of a postoperative bile leak following hepatectomy using a combined clinical–radiomic model in 378 patients with a preoperative CT. Radiomic data was extracted from a virtual liver biopsy obtained from a 2 cm cylinder of non-tumoral liver parenchyma on the portal phase of preoperative CT. Their combined model (AUC 0.74) outperformed a model using clinical data alone (AUC 0.61) to predict the risk of a bile leak [53].
Table 4. Key studies demonstrating the use of AI in management of HCC.
| Author, Year | Model Design | Population | AI Methodology | Accuracy |
|---|---|---|---|---|
| Wu et al. (2017) [42] | Prediction of disease-free survival after radiofrequency ablation based on clinical variables | Patients who underwent CT-guided radiofrequency ablation | Artificial neural network | AUC: 0.75–0.84 |
| Liu et al. (2020) [44] | Prediction of response to first TACE session using contrast- enhanced liver ultrasound | Patients who underwent ultrasound within one week of TACE for HCC | Radiomic-based deep learning | AUC: 0.81–0.93 |
| Meng et al. (2020) [50] | Automatic liver parenchyma and liver tumor segmentation from CT images | Multi-institutional liver tumor segmentation (LiTS) dataset | Dual path multiscale convolutional neural network | Dice: 0.689–0.965 |
| Zheng et al. (2022) [51] | Automatic segmentation of HCC lesions based on dynamic MRI | Patients with HCC who underwent dynamic contrast-enhanced MRI | Convolutional neural network and recurrence neural network | Dice: 0.825 |
| Wang et al. (2022) [52] | Prediction of post-hepatectomy liver failure based on clinical characteristics and surgical variables | Patients with HCC who underwent hepatectomy | Light gradient boosting machine learning | AUC: 0.822–0.944 |
4. Intraoperative Use of Artificial Intelligence
A thorough understanding of the vascular and biliary anatomy from adequate multiphasic abdominal imaging is necessary to perform a safe hepatectomy and minimize complications. Three-dimensional liver reconstruction technology may improve perioperative outcomes in patients undergoing a major hepatectomy by further elucidating spatial relationships between the tumor and critical vascular and biliary structures. A meta-analysis evaluating the efficacy and safety of 3D-reconstruction liver models in patients undergoing a major hepatectomy showed shorter operative times, less intraoperative blood loss, fewer hepatic inflow occlusion events, shorter hospital stays, and fewer postoperative complications when using such technology [54]. Deep learning algorithms can help us to automate the reconstruction of 3D liver models with reliable accuracy and detail [55].
As an adjunct to intraoperative liver ultrasound (IOUS), machine learning algorithms may help us to overcome some limitations of traditional ultrasounds and improve the accuracy of identifying liver lesions intraoperatively. Barash et al. trained a CNN on intraoperative ultrasound imaging to detect liver lesions. The algorithm achieved an AUC of 80.2% and an overall classification accuracy of 74.6% [56]. Furthermore, Takamoto et al. used real-time virtual sonography (RVS), an AI-assisted platform that merges preoperative CT images with real-time IOUS, to enhance IOUS with the identification of small intrahepatic lesions [57]. The median liver lesion size was 6.0 mm and RVS significantly improved surgeon confidence in lesion identification. Importantly, of the 17 lesions undetectable using fundamental IOUS, 14 were identified through RVS and ultimately treated.
However, despite the successful applications of AI technology during liver surgery, several challenges remain. This is especially true of 3D overlays, which are difficult to use during hepatectomy. The mobilization of the liver and the deformation of the parenchyma during hepatectomy make real-time overlays onto tissues more challenging and less accurate. This is in comparison with other surgical disciplines such as neurosurgery, where the target—in this case the brain—is rigid and fixed, allowing for an easy overlay of 3D reconstructions. Preliminary studies demonstrate the feasibility of a physics-based elastic augmented reality model that can provide a real-time 3D overlay during hepatectomy, allowing for the deformation and mobilization of the tissue; however, further quality improvement needs to occur prior to meaningful use during hepatectomy [58].
References
- Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; Anderson, B.O.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018, 4, 1553–1568.
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314.
- Park, J.W.; Chen, M.; Colombo, M.; Roberts, L.R.; Schwartz, M.; Chen, P.J.; Kudo, M.; Johnson, P.; Wagner, S.; Orsini, L.S.; et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: The BRIDGE Study. Liver Int. 2015, 35, 2155–2166.
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Vogel, A.; Meyer, T.; Sapisochin, G.; Salem, R.; Saborowski, A. Hepatocellular carcinoma. Lancet 2022, 400, 1345–1362.
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905.
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173.
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine learning and deep learning. Electron. Mark. 2021, 31, 685–695.
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14.
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444.
- Rudin, C. Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead. Nat. Mach. Intell. 2019, 1, 206–215.
- Banerjee, J.; Taroni, J.N.; Allaway, R.J.; Prasad, D.V.; Guinney, J.; Greene, C. Machine learning in rare disease. Nat. Methods 2023, 20, 803–814.
- Jiang, L.Y.; Liu, X.C.; Nejatian, N.P.; Nasir-Moin, M.; Wang, D.; Abidin, A.; Eaton, K.; Riina, H.A.; Laufer, I.; Punjabi, P.; et al. Health system-scale language models are all-purpose prediction engines. Nature 2023, 619, 357–362.
- Bian, Y.; Zheng, Z.; Fang, X.; Jiang, H.; Zhu, M.; Yu, J.; Zhao, H.; Zhang, L.; Yao, J.; Lu, L.; et al. Artificial Intelligence to Predict Lymph Node Metastasis at CT in Pancreatic Ductal Adenocarcinoma. Radiology 2023, 306, 160–169.
- Kudo, M. Management of Hepatocellular Carcinoma in Japan as a World-Leading Model. Liver Cancer 2018, 7, 134–147.
- Blanes-Vidal, V.; Lindvig, K.P.; Thiele, M.; Nadimi, E.S.; Krag, A. Artificial intelligence outperforms standard blood-based scores in identifying liver fibrosis patients in primary care. Sci. Rep. 2022, 12, 2914.
- Singal, A.G.; Mukherjee, A.; Elmunzer, B.J.; Higgins, P.D.; Lok, A.S.; Zhu, J.; Marrero, J.A.; Waljee, A.K. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am. J. Gastroenterol. 2013, 108, 1723–1730.
- Ioannou, G.N.; Tang, W.; Beste, L.A.; Tincopa, M.A.; Su, G.L.; Van, T.; Tapper, E.B.; Singal, A.G.; Zhu, J.; Waljee, A.K. Assessment of a Deep Learning Model to Predict Hepatocellular Carcinoma in Patients with Hepatitis C Cirrhosis. JAMA Netw. Open 2020, 3, e2015626.
- Mitchell, D.G.; Bruix, J.; Sherman, M.; Sirlin, C.B. LI-RADS (Liver Imaging Reporting and Data System): Summary, discussion, and consensus of the LI-RADS Management Working Group and future directions. Hepatology 2015, 61, 1056–1065.
- Yasaka, K.; Akai, H.; Abe, O.; Kiryu, S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology 2018, 286, 887–896.
- Mokrane, F.Z.; Lu, L.; Vavasseur, A.; Otal, P.; Peron, J.M.; Luk, L.; Yang, H.; Ammari, S.; Saenger, Y.; Rousseau, H.; et al. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur. Radiol. 2020, 30, 558–570.
- Schmauch, B.; Herent, P.; Jehanno, P.; Dehaene, O.; Saillard, C.; Aubé, C.; Luciani, A.; Lassau, N.; Jégou, S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn. Interv. Imaging 2019, 100, 227–233.
- Yang, Q.; Wei, J.; Hao, X.; Kong, D.; Yu, X.; Jiang, T.; Xi, J.; Cai, W.; Luo, Y.; Jing, X.; et al. Improving B-mode ultrasound diagnostic performance for focal liver lesions using deep learning: A multicentre study. EBioMedicine 2020, 56, 102777.
- Jansen, M.J.A.; Kuijf, H.J.; Veldhuis, W.B.; Wessels, F.J.; Viergever, M.A.; Pluim, J.P.W. Automatic classification of focal liver lesions based on MRI and risk factors. PLoS ONE 2019, 14, e0217053.
- McMahon, B.; Cohen, C.; Brown, R.S., Jr.; El-Serag, H.; Ioannou, G.N.; Lok, A.S.; Roberts, L.R.; Singal, A.G.; Block, T. Opportunities to address gaps in early detection and improve outcomes of liver cancer. JNCI Cancer Spectr. 2023, 7, pkad034.
- Liu, X.; Xiao, C.; Yue, K.; Chen, M.; Zhou, H.; Yan, X. Identification of multi-omics biomarkers and construction of the novel prognostic model for hepatocellular carcinoma. Sci. Rep. 2022, 12, 12084.
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2018, 24, 1248–1259.
- Lee, S.; Kang, T.W.; Song, K.D.; Lee, M.W.; Rhim, H.; Lim, H.K.; Kim, S.Y.; Sinn, D.H.; Kim, J.M.; Kim, K.; et al. Effect of Microvascular Invasion Risk on Early Recurrence of Hepatocellular Carcinoma after Surgery and Radiofrequency Ablation. Ann. Surg. 2021, 273, 564–571.
- Zhang, J.; Huang, S.; Xu, Y.; Wu, J. Diagnostic Accuracy of Artificial Intelligence Based on Imaging Data for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2022, 12, 763842.
- Liu, S.C.; Lai, J.; Huang, J.Y.; Cho, C.F.; Lee, P.H.; Lu, M.H.; Yeh, C.C.; Yu, J.; Lin, W.C. Predicting microvascular invasion in hepatocellular carcinoma: A deep learning model validated across hospitals. Cancer Imaging 2021, 21, 56.
- Sun, S.W.; Xu, X.; Liu, Q.P.; Chen, J.N.; Zhu, F.P.; Liu, X.S.; Zhang, Y.D.; Wang, J. LiSNet: An artificial intelligence -based tool for liver imaging staging of hepatocellular carcinoma aggressiveness. Med. Phys. 2022, 49, 6903–6913.
- Chong, H.H.; Yang, L.; Sheng, R.F.; Yu, Y.L.; Wu, D.J.; Rao, S.X.; Yang, C.; Zeng, M.S. Multi-scale and multi-parametric radiomics of gadoxetate disodium-enhanced MRI predicts microvascular invasion and outcome in patients with solitary hepatocellular carcinoma ≤ 5 cm. Eur. Radiol. 2021, 31, 4824–4838.
- Lv, C.; He, N.; Yang, J.J.; Xiao, J.J.; Zhang, Y.; Du, J.; Zuo, S.; Li, H.Y.; Gu, H. Prediction of 3-year recurrence rate of hepatocellular carcinoma after resection based on contrast-enhanced CT: A single-centre study. Br. J. Radiol. 2023, 96, 20220702.
- Yan, M.; Zhang, X.; Zhang, B.; Geng, Z.; Xie, C.; Yang, W.; Zhang, S.; Qi, Z.; Lin, T.; Ke, Q.; et al. Deep learning nomogram based on Gd-EOB-DTPA MRI for predicting early recurrence in hepatocellular carcinoma after hepatectomy. Eur. Radiol. 2023, 33, 4949–4961.
- Ji, G.W.; Zhu, F.P.; Xu, Q.; Wang, K.; Wu, M.Y.; Tang, W.W.; Li, X.C.; Wang, X.H. Radiomic Features at Contrast-enhanced CT Predict Recurrence in Early Stage Hepatocellular Carcinoma: A Multi-Institutional Study. Radiology 2020, 294, 568–579.
- Qu, W.F.; Tian, M.X.; Qiu, J.T.; Guo, Y.C.; Tao, C.Y.; Liu, W.R.; Tang, Z.; Qian, K.; Wang, Z.X.; Li, X.Y.; et al. Exploring pathological signatures for predicting the recurrence of early-stage hepatocellular carcinoma based on deep learning. Front. Oncol. 2022, 12, 968202.
- Saillard, C.; Schmauch, B.; Laifa, O.; Moarii, M.; Toldo, S.; Zaslavskiy, M.; Pronier, E.; Laurent, A.; Amaddeo, G.; Regnault, H.; et al. Predicting Survival after Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology 2020, 72, 2000–2013.
- Kather, J.N.; Heij, L.R.; Grabsch, H.I.; Loeffler, C.; Echle, A.; Muti, H.S.; Krause, J.; Niehues, J.M.; Sommer, K.A.J.; Bankhead, P.; et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 2020, 1, 789–799.
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056.
- Yamashita, R.; Long, J.; Saleem, A.; Rubin, D.L.; Shen, J. Deep learning predicts postsurgical recurrence of hepatocellular carcinoma from digital histopathologic images. Sci. Rep. 2021, 11, 2047.
- Chen, M.; Zhang, B.; Topatana, W.; Cao, J.; Zhu, H.; Juengpanich, S.; Mao, Q.; Yu, H.; Cai, X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis. Oncol. 2020, 4, 14.
- Zeng, Q.; Klein, C.; Caruso, S.; Maille, P.; Laleh, N.G.; Sommacale, D.; Laurent, A.; Amaddeo, G.; Gentien, D.; Rapinat, A.; et al. Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology. J. Hepatol. 2022, 77, 116–127.
- Wu, C.F.; Wu, Y.J.; Liang, P.C.; Wu, C.H.; Peng, S.F.; Chiu, H.W. Disease-free survival assessment by artificial neural networks for hepatocellular carcinoma patients after radiofrequency ablation. J. Formos. Med. Assoc. 2017, 116, 765–773.
- Liu, D.; Liu, F.; Xie, X.; Su, L.; Liu, M.; Xie, X.; Kuang, M.; Huang, G.; Wang, Y.; Zhou, H.; et al. Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound. Eur. Radiol. 2020, 30, 2365–2376.
- Müller, L.; Kloeckner, R.; Mähringer-Kunz, A.; Stoehr, F.; Düber, C.; Arnhold, G.; Gairing, S.J.; Foerster, F.; Weinmann, A.; Galle, P.R.; et al. Fully automated AI-based splenic segmentation for predicting survival and estimating the risk of hepatic decompensation in TACE patients with HCC. Eur. Radiol. 2022, 32, 6302–6313.
- Wang, J.; Zhang, X.; Lv, P.; Wang, H.; Cheng, Y. Automatic Liver Segmentation Using EfficientNet and Attention-Based Residual U-Net in CT. J. Digit. Imaging 2022, 35, 1479–1493.
- Tan, M.; Wu, F.; Kong, D.; Mao, X. Automatic liver segmentation using 3D convolutional neural networks with a hybrid loss function. Med. Phys. 2021, 48, 1707–1719.
- Özcan, F.; Uçan, O.N.; Karaçam, S.; Tunçman, D. Fully Automatic Liver and Tumor Segmentation from CT Image Using an AIM-Unet. Bioengineering 2023, 10, 215.
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Şahin, Y.; Özkan, S.; Baydar, B.; Yüksel, U.; Kılıkçıer, Ç.; Olut, Ş.; Bozdağı Akar, G.; et al. Comparison of semi-automatic and deep learning-based automatic methods for liver segmentation in living liver transplant donors. Diagn. Interv. Radiol. 2020, 26, 11–21.
- Meng, L.; Tian, Y.; Bu, S. Liver tumor segmentation based on 3D convolutional neural network with dual scale. J. Appl. Clin. Med. Phys. 2020, 21, 144–157.
- Zheng, R.; Wang, Q.; Lv, S.; Li, C.; Wang, C.; Chen, W.; Wang, H. Automatic Liver Tumor Segmentation on Dynamic Contrast Enhanced MRI Using 4D Information: Deep Learning Model Based on 3D Convolution and Convolutional LSTM. IEEE Trans. Med. Imaging 2022, 41, 2965–2976.
- Wang, J.; Zheng, T.; Liao, Y.; Geng, S.; Li, J.; Zhang, Z.; Shang, D.; Liu, C.; Yu, P.; Huang, Y.; et al. Machine learning prediction model for post- hepatectomy liver failure in hepatocellular carcinoma: A multicenter study. Front. Oncol. 2022, 12, 986867.
- Laino, M.E.; Fiz, F.; Morandini, P.; Costa, G.; Maffia, F.; Giuffrida, M.; Pecorella, I.; Gionso, M.; Wheeler, D.R.; Cambiaghi, M.; et al. A virtual biopsy of liver parenchyma to predict the outcome of liver resection. Updates Surg. 2023, 75, 1519–1531.
- Liu, Y.; Wang, Q.; Du, B.; Wang, X.; Xue, Q.; Gao, W. A meta-analysis of the three-dimensional reconstruction visualization technology for hepatectomy. Asian J. Surg. 2023, 46, 669–676.
- Takamoto, T.; Ban, D.; Nara, S.; Mizui, T.; Nagashima, D.; Esaki, M.; Shimada, K. Automated Three-Dimensional Liver Reconstruction with Artificial Intelligence for Virtual Hepatectomy. J. Gastrointest. Surg. 2022, 26, 2119–2127.
- Barash, Y.; Klang, E.; Lux, A.; Konen, E.; Horesh, N.; Pery, R.; Zilka, N.; Eshkenazy, R.; Nachmany, I.; Pencovich, N. Artificial intelligence for identification of focal lesions in intraoperative liver ultrasonography. Langenbecks Arch. Surg. 2022, 407, 3553–3560.
- Takamoto, T.; Nara, S.; Ban, D.; Mizui, T.; Murase, Y.; Esaki, M.; Shimada, K. Enhanced Recognition Confidence of Millimeter-Sized Intrahepatic Targets by Real-Time Virtual Sonography. J. Ultrasound Med. 2023, 42, 1789–1797.
- Golse, N.; Petit, A.; Lewin, M.; Vibert, E.; Cotin, S. Augmented Reality during Open Liver Surgery Using a Markerless Non-rigid Registration System. J. Gastrointest. Surg. 2021, 25, 662–671.
More
Information
Subjects:
Surgery
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
577
Revisions:
2 times
(View History)
Update Date:
25 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




