Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lv, G.; Wang, J.; Lian, S.; Wang, H.; Wu, R. Genomic Composition of Bovine Leukemia Virus. Encyclopedia. Available online: https://encyclopedia.pub/entry/54175 (accessed on 07 February 2026).
Lv G, Wang J, Lian S, Wang H, Wu R. Genomic Composition of Bovine Leukemia Virus. Encyclopedia. Available at: https://encyclopedia.pub/entry/54175. Accessed February 07, 2026.
Lv, Guanxin, Jianfa Wang, Shuai Lian, Hai Wang, Rui Wu. "Genomic Composition of Bovine Leukemia Virus" Encyclopedia, https://encyclopedia.pub/entry/54175 (accessed February 07, 2026).
Lv, G., Wang, J., Lian, S., Wang, H., & Wu, R. (2024, January 22). Genomic Composition of Bovine Leukemia Virus. In Encyclopedia. https://encyclopedia.pub/entry/54175
Lv, Guanxin, et al. "Genomic Composition of Bovine Leukemia Virus." Encyclopedia. Web. 22 January, 2024.
Copy Citation
Bovine leukemia virus (BLV) is the causative agent of enzootic bovine leukosis (EBL), which is the most significant neoplastic disease in cattle. EBL is often overlooked in daily breeding processes due to the absence of obvious clinical symptoms.
bovine leukemia virus (BLV)
genomic structure
epidemiology
transmission
1. Introduction
Bovine leukemia virus (BLV) is a retrovirus known to cause enzootic bovine leucosis (EBL) [1][2][3][4]. EBL has been successfully eradicated in most European countries [5]. However, recent investigations have shown an increase in infection rates in several countries, including Argentina, Brazil, Canada, Japan, and the United States [6][7][8][9]. The majority of BLV-infected cattle remain asymptomatic, harboring the virus in a latent state of infection, and are thus classified as carriers. As the disease progresses, around one-third of cattle infected with BLV exhibit a non-malignant proliferation of B lymphocytes, a condition referred to as persistent lymphocytosis (PL). After a long period of latent infection lasting 3 to 8 years, less than 5% of these cattle will eventually develop malignant B lymphocyte lymphoma [10][11].
Previous research suggested that lymphosarcoma caused by BLV primarily affected the economics of cattle farming. However, recent research has shown that BLV infection affects the immune system of cattle, even during the latency period, leading to varying degrees of immune suppression and direct economic losses. These losses include reduced milk production, increased susceptibility to infectious diseases (such as mastitis, skin infections, and hoof diseases), and decreased reproductive efficiency [12][13][14][15]. In the United States, where the BLV infection rate exceeds 40%, the annual losses due to reduced milk production have surpassed USD 500 million. Additionally, certain countries and regions have implemented strict import and export regulations for animals infected with BLV, resulting in various indirect economic losses [5][13][15][16][17][18]. As a result, the World Organisation for Animal Health (WOAH) has classified Enzootic Bovine Leukosis (EBL) as a disease that significantly impacts international trade [19].
In addition to its impact on cattle herds, studies have shown that unsterilized or uncooked dairy and beef products contain proviral nucleic acid fragments of BLV, which raises concerns about public health and safety [20]. While it has not been confirmed that BLV can cause human infections, the positive detection rate of BLV in samples from breast cancer patients is significantly higher compared to healthy breast control groups [21][22][23][24][25][26]. Moreover, there is a correlation between the consumption of beef, milk, and dairy products and the incidence of breast cancer in different countries and regions. For example, countries with lower consumption rates like India, Japan, South Korea, and China also exhibit lower incidences of breast cancer. Conversely, countries with higher consumption rates such as the United States, the United Kingdom, Australia, and Germany have a higher incidence of breast cancer [27][28]. However, some studies have not detected proviral DNA of BLV in breast cancer tissue samples [29][30][31].
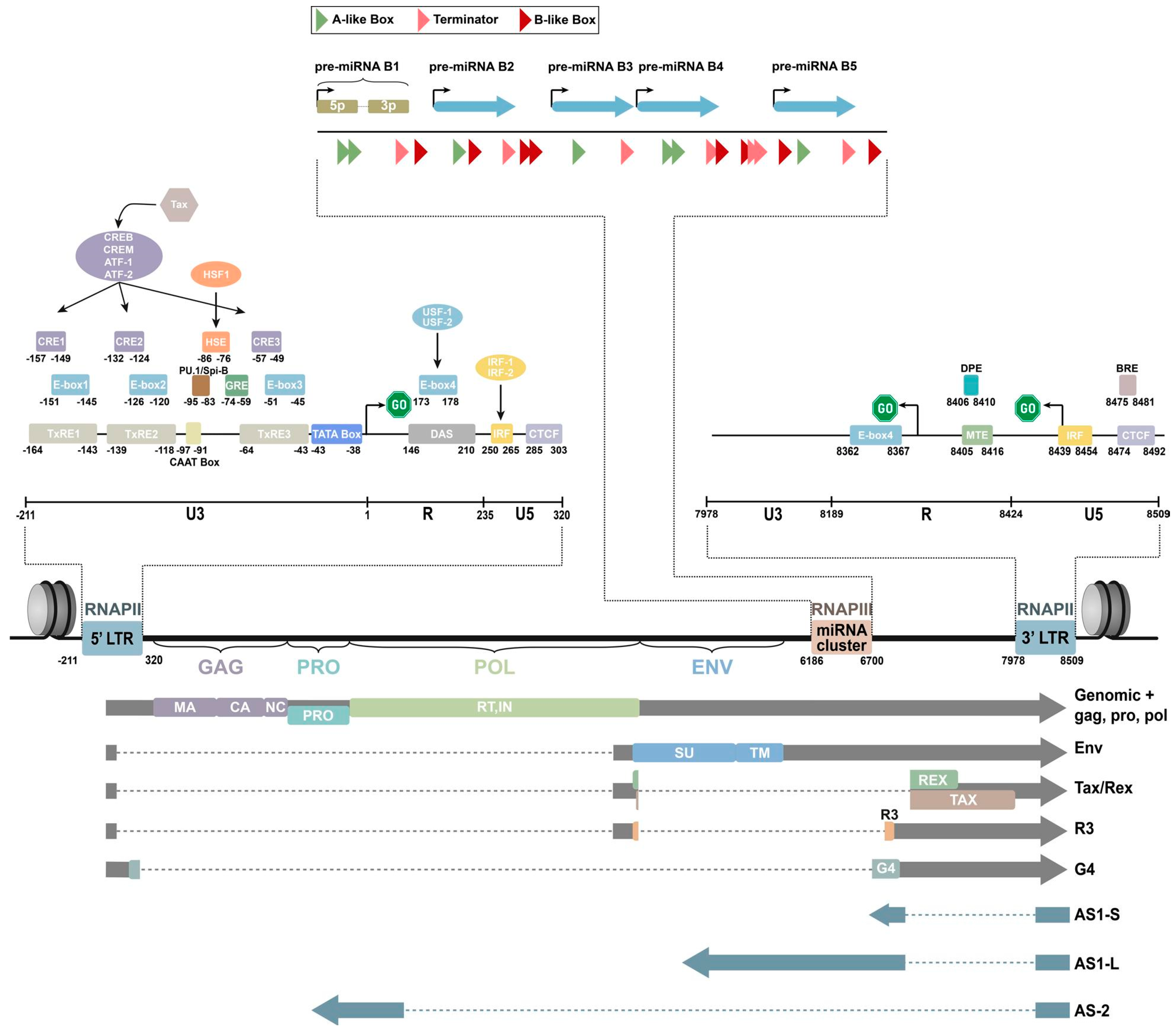
2. Genomic Composition of BLV
BLV is a single-stranded RNA-RT virus that exhibits a spherical or rod-shaped morphology, measuring approximately 60 to 125 nanometers in diameter. It is enveloped by a double-layered membrane structure and its nucleocapsid demonstrates icosahedral symmetry [32][33]. It contains a diploid single-stranded RNA genome. The viral genome of BLV consists of a diploid single-stranded RNA molecule spanning a total of 8714 nucleotides. This genome encompasses structural protein genes, enzyme coding genes, the pX region, and two identical, long, terminal repeat sequences positioned at both ends of the genome (Figure 1) [34][35].

Figure 1. Schematic representation of the BLV genome [35].
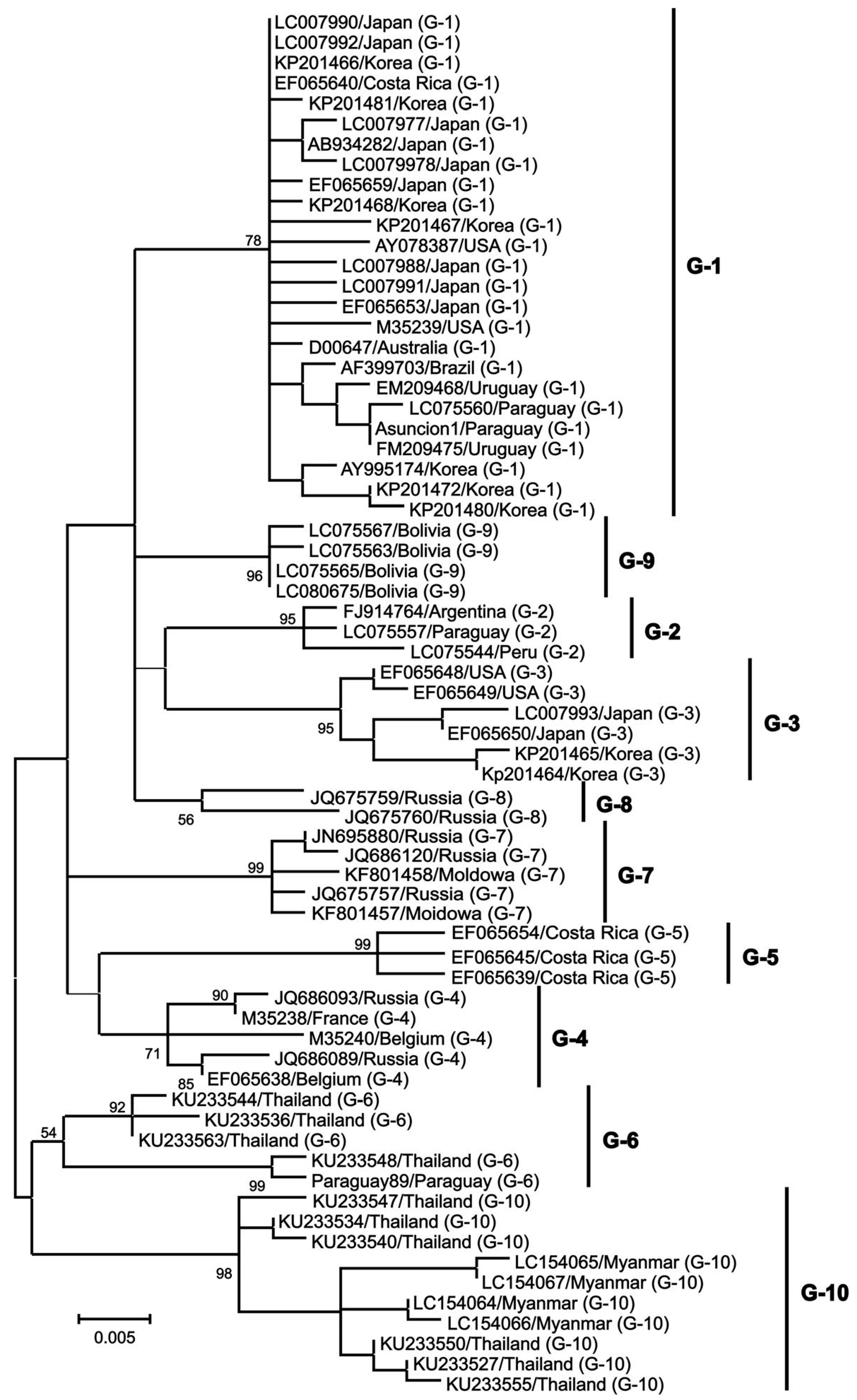
Structural proteins and enzyme coding genes, such as gag, pro, pol, and env, are crucial in the virus lifecycle, infectivity, and the production of infectious viral particles [36][37][38]. The gag gene is highly conserved and responsible for encoding three major non-glycosylated proteins: p12 nucleocapsid protein (which binds with viral genomic RNA), p15 matrix protein (which binds with viral genomic RNA and interacts with the lipid bilayer of the viral membrane), and p24 nucleocapsid protein (which serves as the main target of the host immune response) [37]. The pro gene encodes the viral proteinase p14, which is involved in the post-translation maturation of the virus [36]. The pol gene encodes reverse transcriptase and integrase, which play a role in the reverse transcription process and integration of proviral BLV DNA into the host genome [39]. The env gene encodes two glycosylated proteins: envelope protein gp51 and transmembrane protein gp30, which facilitate virus–cell fusion through interaction with cell membrane receptors [40][41]. The pX region, located between the env gene and 3’LTR, encodes other auxiliary regulatory or non-structural genes, including regulatory proteins Tax and Rex, as well as auxiliary proteins R3 and G4 [42][43][44][45][46][47][48]. Regulatory proteins are important in virus transcription regulation, inducing the malignant transformation of tumors and virus infection output, while R3 and G4 auxiliary proteins help maintain a high viral load [49]. The env gene also shows genetic polymorphism, which is beneficial for phylogenetic studies and classifying BLV isolates [40]. The nucleotide sequence and amino acid composition of the env gene are valuable genomic markers for studying the distribution of BLV and identifying different genotypes based on geographical origin [50]. Researchers have identified 10 BLV genotypes through sequencing and phylogenetic analysis of partial and full-length gp51 env genes. Genotype 1 is the most prevalent and commonly found in the United States, Japan, and South Korea [51][52][53]. South America has genotypes 1, 2, 3, 4, 5, 6, and 9 (with genotype 9 being particularly prevalent in Bolivia). Russia and Eastern Europe commonly have genotypes 4, 7, and 8 [54]. Genotype 10 is prevalent in China, Vietnam, Thailand, and Myanmar [51][55]. Genotype 4 is found in various countries in North and South America, Africa, and Asia [56][57]. Genotype 6 has been found in South American countries such as Argentina, Brazil, Peru, Paraguay, and Bolivia, as well as in Asian countries such as the Philippines, Thailand, and India [56][58][59][60]. Additionally, genotype 1 has been reported in Egypt [61]. Monitoring the emergence of new viral mutations in veterinary medicine and animal husbandry is important since each new genotype may exhibit unique characteristics in its interaction with the host organism and leukemogenesis is influenced by specific genotypes (Figure 2).

Figure 2. A maximum likelihood phylogenetic tree was constructed using partial BLV env sequences from various geographical locations worldwide, from [62].
Research indicates that 10 different microRNA (miRNA) can be transcribed under the control of RNA polymerase III in the genomic region spanning from the env gene to the R3 coding area [63]. RNA sequencing analysis has revealed that these miRNAs are highly expressed in primary sheep and cattle cells containing pre-leukemic and malignant B cells, accounting for 40% of the total cellular miRNA [64]. Additionally, elevated levels of BLV miRNA were found in the serum of infected animals, suggesting a potential paracrine role. These findings indicate the important role of BLV miRNAs in the retroviral cycle and/or disease progression, challenging the traditional belief that RNA viruses do not encode miRNAs. They also demonstrate that BLV miRNAs regulate various biological processes, including apoptosis, immunity, signal transduction, and tumorigenesis [65]. Notably, Blv-miR-B4-3p, which shares a seed sequence with the host miRNA miR-29a, is known for downregulating the tumor suppressor genes HMG-box transcription factor 1 (HBP1) and peroxidasin homologs (PXDN) in B-cell tumors [63][65][66]. In vitro studies have confirmed that blv-miR-B4-3p can directly downregulate these genes, although this has not been experimentally validated in cattle infected with BLV. Recently, Petersen et al. used RT-qPCR to analyze hbp1 and pxdn expression in cattle naturally infected with BLV and noted a significant downregulation of pxdn compared to uninfected animals [67]. However, a comprehensive analysis of targets for each mature BLV miRNA is still needed to fully understand the role of BLV in tumorigenesis.
References
- Burny, A.; Bex, F.; Chantrenne, H.; Cleuter, Y.; Dekegel, D.; Ghysdael, J.; Kettmann, R.; Leclercq, M.; Leunen, J.; Mammerickx, M.; et al. Bovine leukemia virus involvement in enzootic bovine leukosis. Adv. Cancer Res. 1978, 28, 251–311.
- Burny, A.; Bruck, C.; Cleuter, Y.; Couez, D.; Deschamps, J.; Gregoire, D.; Ghysdael, J.; Kettmann, R.; Mammerickx, M.; Marbaix, G.; et al. Bovine leukaemia virus and enzootic bovine leukosis. Onderstepoort J. Vet. Res. 1985, 52, 133–144.
- Kettmann, R.; Portetelle, D.; Mammerickx, M.; Cleuter, Y.; Dekegel, D.; Galoux, M.; Ghysdael, J.; Burny, A.; Chantrenne, H. Bovine leukemia virus: An exogenous RNA oncogenic virus. Proc. Natl. Acad. Sci. USA 1976, 73, 1014–1018.
- Reed, V.I. Enzootic bovine leukosis. Can. Vet. J.-Rev. Vet. Can. 1981, 22, 95–102.
- Panel, E.A. Scientific opinion on enzootic bovine leukosis. EFSA J. 2015, 13, 4188.
- Nekouei, O.; Vanleeuwen, J.; Sanchez, J.; Kelton, D.; Tiwari, A.; Keefe, G. Herd-level risk factors for infection with bovine leukemia virus in Canadian dairy herds. Prev. Vet. Med. 2015, 119, 105–113.
- Murakami, K.; Kobayashi, S.; Konishi, M.; Kameyama, K.; Tsutsui, T. Nationwide survey of bovine leukemia virus infection among dairy and beef breeding cattle in Japan from 2009–2011. J. Vet. Med. Sci. 2013, 75, 1123–1126.
- Ma, B.; Gong, Q.; Sheng, C.; Liu, Y.; Ge, G.; Li, D.; Diao, N.; Shi, K.; Li, J.; Sun, Z.; et al. Prevalence of bovine leukemia in 1983–2019 in China: A systematic review and meta-analysis. Microb. Pathog. 2021, 150, 104681.
- Ladronka, R.M.; Ainsworth, S.; Wilkins, M.J.; Norby, B.; Byrem, T.M.; Bartlett, P.C. Prevalence of bovine leukemia virus antibodies in US dairy cattle. Vet. Med. Int. 2018, 2018, 5831278.
- Tsutsui, T.; Kobayashi, S.; Hayama, Y.; Yamamoto, T. Fraction of bovine leukemia virus-infected dairy cattle developing enzootic bovine leukosis. Prev. Vet. Med. 2016, 124, 96–101.
- Gillet, N.; Florins, A.; Boxus, M.; Burteau, C.; Nigro, A.; Vandermeers, F.; Balon, H.; Bouzar, A.B.; Defoiche, J.; Burny, A.; et al. Mechanisms of leukemogenesis induced by bovine leukemia virus: Prospects for novel anti-retroviral therapies in human. Retrovirology 2007, 4, 18.
- Norby, B.; Bartlett, P.C.; Byrem, T.M.; Erskine, R.J. Effect of infection with bovine leukemia virus on milk production in Michigan dairy cows. J. Dairy Sci. 2016, 99, 2043–2052.
- Erskine, R.J.; Bartlett, P.C.; Byrem, T.M.; Render, C.L.; Febvay, C.; Houseman, J.T. Association between bovine leukemia virus, production, and population age in Michigan dairy herds. J. Dairy Sci. 2012, 95, 727–734.
- Bartlett, P.C.; Norby, B.; Byrem, T.M.; Parmelee, A.; Ledergerber, J.T.; Erskine, R.J. Bovine leukemia virus and cow longevity in Michigan dairy herds. J. Dairy Sci. 2013, 96, 1591–1597.
- Frie, M.C.; Coussens, P.M. Bovine leukemia virus: A major silent threat to proper immune responses in cattle. Vet. Immunol. Immunopathol. 2015, 163, 103–114.
- Ruggiero, V.J.; Norby, B.; Benitez, O.J.; Hutchinson, H.; Sporer, K.; Droscha, C.; Swenson, C.L.; Bartlett, P.C. Controlling bovine leukemia virus in dairy herds by identifying and removing cows with the highest proviral load and lymphocyte counts. J. Dairy Sci. 2019, 102, 9165–9175.
- Nuotio, L.; Rusanen, H.; Sihvonen, L.; Neuvonen, E. Eradication of enzootic bovine leukosis from Finland. Prev. Vet. Med. 2003, 59, 43–49.
- Acaite, J.; Tamosiunas, V.; Lukauskas, K.; Milius, J.; Pieskus, J. The eradication experience of enzootic bovine leukosis from Lithuania. Prev. Vet. Med. 2007, 82, 83–89.
- Enzootic Bovine Leukosis, Chapter 3.4.9. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.04.09_EBL.pdf (accessed on 28 December 2023).
- Olaya-Galan, N.N.; Corredor-Figueroa, A.P.; Guzman-Garzon, T.C.; Rios-Hernandez, K.S.; Salas-Cardenas, S.P.; Patarroyo, M.A.; Gutierrez, M.F. Bovine leukaemia virus DNA in fresh milk and raw beef for human consumption. Epidemiol. Infect. 2017, 145, 3125–3130.
- Khatami, A.; Pormohammad, A.; Farzi, R.; Saadati, H.; Mehrabi, M.; Kiani, S.J.; Ghorbani, S. Bovine Leukemia virus (BLV) and risk of breast cancer: A systematic review and meta-analysis of case-control studies. Infect. Agents Cancer 2020, 15, 48.
- Khan, Z.; Abubakar, M.; Arshed, M.J.; Aslam, R.; Sattar, S.; Shah, N.A.; Javed, S.; Tariq, A.; Bostan, N.; Manzoor, S. Molecular investigation of possible relationships concerning bovine leukemia virus and breast cancer. Sci. Rep. 2022, 12, 4161.
- Adekanmbi, F.; Mcneely, I.; Omeler, S.; Kalalah, A.; Poudel, A.; Merner, N.; Wang, C. Absence of bovine leukemia virus in the buffy coats of breast cancer cases from Alabama, USA. Microb. Pathog. 2021, 161, 105238.
- Canova, R.; Weber, M.N.; Budaszewski, R.F.; Da, S.M.; Schwingel, D.; Canal, C.W.; Kreutz, L.C. Bovine leukemia viral DNA found on human breast tissue is genetically related to the cattle virus. One Health 2021, 13, 100252.
- Buehring, G.C.; Shen, H.M.; Jensen, H.M.; Choi, K.Y.; Sun, D.; Nuovo, G. Bovine leukemia virus DNA in human breast tissue. Emerg. Infect. Dis. 2014, 20, 772–782.
- Olaya-Galan, N.N.; Salas-Cardenas, S.P.; Rodriguez-Sarmiento, J.L.; Ibanez-Pinilla, M.; Monroy, R.; Corredor-Figueroa, A.P.; Rubiano, W.; de la Pena, J.; Shen, H.; Buehring, G.C.; et al. Risk factor for breast cancer development under exposure to bovine leukemia virus in Colombian women: A case-control study. PLoS ONE 2021, 16, e257492.
- Afzal, S.; Fiaz, K.; Noor, A.; Sindhu, A.S.; Hanif, A.; Bibi, A.; Asad, M.; Nawaz, S.; Zafar, S.; Ayub, S.; et al. Interrelated Oncogenic Viruses and Breast Cancer. Front. Mol. Biosci. 2022, 9, 781111.
- Lawson, J.S.; Salmons, B.; Glenn, W.K. Oncogenic Viruses and Breast Cancer: Mouse Mammary Tumor Virus (MMTV), Bovine Leukemia Virus (BLV), Human Papilloma Virus (HPV), and Epstein-Barr Virus (EBV). Front. Oncol. 2018, 8, 1.
- Zhang, R.; Jiang, J.; Sun, W.; Zhang, J.; Huang, K.; Gu, X.; Yang, Y.; Xu, X.; Shi, Y.; Wang, C. Lack of association between bovine leukemia virus and breast cancer in Chinese patients. Breast Cancer Res. 2016, 18, 101.
- Gillet, N.A.; Willems, L. Whole genome sequencing of 51 breast cancers reveals that tumors are devoid of bovine leukemia virus DNA. Retrovirology 2016, 13, 75.
- Bender, A.P.; Robison, L.L.; Kashmiri, S.V.; Mcclain, K.L.; Woods, W.G.; Smithson, W.A.; Heyn, R.; Finlay, J.; Schuman, L.M.; Renier, C.; et al. No involvement of bovine leukemia virus in childhood acute lymphoblastic leukemia and non-Hodgkin’s lymphoma. Cancer Res. 1988, 48, 2919–2922.
- Zhao, X.; Buehring, G.C. Natural genetic variations in bovine leukemia virus envelope gene: Possible effects of selection and escape. Virology 2007, 366, 150–165.
- Boris-Lawrie, K.; Altanerova, V.; Altaner, C.; Kucerova, L.; Temin, H.M. In vivo study of genetically simplified bovine leukemia virus derivatives that lack tax and rex. J. Virol. 1997, 71, 1514–1520.
- Aida, Y.; Murakami, H.; Takahashi, M.; Takeshima, S.N. Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Front. Microbiol. 2013, 4, 328.
- Plant, E.; Bellefroid, M.; Van Lint, C. A complex network of transcription factors and epigenetic regulators involved in bovine leukemia virus transcriptional regulation. Retrovirology 2023, 20, 11.
- Llames, L.; Goyache, J.; Domenech, A.; Montana, A.V.; Suarez, G.; Gomez-Lucia, E. Cellular distribution of bovine leukemia virus proteins gp51SU, Pr72(env), and Pr66(gag-pro) in persistently infected cells. Virus Res. 2001, 79, 47–57.
- Katoh, I.; Kyushiki, H.; Sakamoto, Y.; Ikawa, Y.; Yoshinaka, Y. Bovine leukemia virus matrix-associated protein MA(p15): Further processing and formation of a specific complex with the dimer of the 5′-terminal genomic RNA fragment. J. Virol. 1991, 65, 6845–6855.
- Mamoun, R.Z.; Morisson, M.; Rebeyrotte, N.; Busetta, B.; Couez, D.; Kettmann, R.; Hospital, M.; Guillemain, B. Sequence variability of bovine leukemia virus env gene and its relevance to the structure and antigenicity of the glycoproteins. J. Virol. 1990, 64, 4180–4188.
- Lairmore, M.D. Animal models of bovine leukemia virus and human T-lymphotrophic virus type-1: Insights in transmission and pathogenesis. Annu. Rev. Anim. Biosci. 2014, 2, 189–208.
- Rola-Luszczak, M.; Sakhawat, A.; Pluta, A.; Rylo, A.; Bomba, A.; Bibi, N.; Kuzmak, J. Molecular Characterization of the env Gene of Bovine Leukemia Virus in Cattle from Pakistan with NGS-Based Evidence of Virus Heterogeneity. Pathogens 2021, 10, 910.
- de Brogniez, A.; Bouzar, A.B.; Jacques, J.R.; Cosse, J.P.; Gillet, N.; Callebaut, I.; Reichert, M.; Willems, L. Mutation of a Single Envelope N-Linked Glycosylation Site Enhances the Pathogenicity of Bovine Leukemia Virus. J. Virol. 2015, 89, 8945–8956.
- Gillet, N.A.; Gutierrez, G.; Rodriguez, S.M.; de Brogniez, A.; Renotte, N.; Alvarez, I.; Trono, K.; Willems, L. Massive depletion of bovine leukemia virus proviral clones located in genomic transcriptionally active sites during primary infection. PLoS Pathog. 2013, 9, e1003687.
- Lefebvre, L.; Ciminale, V.; Vanderplasschen, A.; D’Agostino, D.; Burny, A.; Willems, L.; Kettmann, R. Subcellular localization of the bovine leukemia virus R3 and G4 accessory proteins. J. Virol. 2002, 76, 7843–7854.
- Willems, L.; Kerkhofs, P.; Dequiedt, F.; Portetelle, D.; Mammerickx, M.; Burny, A.; Kettmann, R. Attenuation of bovine leukemia virus by deletion of R3 and G4 open reading frames. Proc. Natl. Acad. Sci. USA 1994, 91, 11532–11536.
- Reichert, M.; Cantor, G.H.; Willems, L.; Kettmann, R. Protective effects of a live attenuated bovine leukaemia virus vaccine with deletion in the R3 and G4 genes. J. Gen. Virol. 2000, 81, 965–969.
- Zyrianova, I.M.; Kovalchuk, S.N. Bovine leukemia virus tax gene/Tax protein polymorphism and its relation to Enzootic Bovine Leukosis. Virulence 2020, 11, 80–87.
- Willems, L.; Grimonpont, C.; Kerkhofs, P.; Capiau, C.; Gheysen, D.; Conrath, K.; Roussef, R.; Mamoun, R.; Portetelle, D.; Burny, A.; et al. Phosphorylation of bovine leukemia virus Tax protein is required for in vitro transformation but not for transactivation. Oncogene 1998, 16, 2165–2176.
- Pyeon, D.; Splitter, G.A. Regulation of bovine leukemia virus tax and pol mRNA levels by interleukin-2 and -10. J. Virol. 1999, 73, 8427–8434.
- Kerkhofs, P.; Heremans, H.; Burny, A.; Kettmann, R.; Willems, L. In vitro and in vivo oncogenic potential of bovine leukemia virus G4 protein. J. Virol. 1998, 72, 2554–2559.
- Licursi, M.; Inoshima, Y.; Wu, D.; Yokoyama, T.; Gonzalez, E.T.; Sentsui, H. Provirus variants of bovine leukemia virus in naturally infected cattle from Argentina and Japan. Vet. Microbiol. 2003, 96, 17–23.
- Lee, E.; Kim, E.J.; Joung, H.K.; Kim, B.H.; Song, J.Y.; Cho, I.S.; Lee, K.K.; Shin, Y.K. Sequencing and phylogenetic analysis of the gp51 gene from Korean bovine leukemia virus isolates. Virol. J. 2015, 12, 64.
- Corredor-Figueroa, A.P.; Salas, S.; Olaya-Galan, N.N.; Quintero, J.S.; Fajardo, A.; Sonora, M.; Moreno, P.; Cristina, J.; Sanchez, A.; Tobon, J.; et al. Prevalence and molecular epidemiology of bovine leukemia virus in Colombian cattle. Infect. Genet. Evol. 2020, 80, 104171.
- Marawan, M.A.; Mekata, H.; Hayashi, T.; Sekiguchi, S.; Kirino, Y.; Horii, Y.; Moustafa, A.M.; Arnaout, F.K.; Galila, E.; Norimine, J. Phylogenetic analysis of env gene of bovine leukemia virus strains spread in Miyazaki prefecture, Japan. J. Vet. Med. Sci. 2017, 79, 912–916.
- Pluta, A.; Rola-Luszczak, M.; Kubis, P.; Balov, S.; Moskalik, R.; Choudhury, B.; Kuzmak, J. Molecular characterization of bovine leukemia virus from Moldovan dairy cattle. Arch. Virol. 2017, 162, 1563–1576.
- Le, D.T.; Yamashita-Kawanishi, N.; Okamoto, M.; Nguyen, S.V.; Nguyen, N.H.; Sugiura, K.; Miura, T.; Haga, T. Detection and genotyping of bovine leukemia virus (BLV) in Vietnamese cattle. J. Vet. Med. Sci. 2020, 82, 1042–1050.
- Polat, M.; Takeshima, S.N.; Hosomichi, K.; Kim, J.; Miyasaka, T.; Yamada, K.; Arainga, M.; Murakami, T.; Matsumoto, Y.; de la Barra, D.V.; et al. A new genotype of bovine leukemia virus in South America identified by NGS-based whole genome sequencing and molecular evolutionary genetic analysis. Retrovirology 2016, 13, 4.
- Hamada, R.; Metwally, S.; Polat, M.; Borjigin, L.; Ali, A.O.; Abdel-Hady, A.; Mohamed, A.; Wada, S.; Aida, Y. Detection and Molecular Characterization of Bovine Leukemia Virus in Egyptian Dairy Cattle. Front. Vet. Sci. 2020, 7, 608.
- Rodriguez, S.M.; Golemba, M.D.; Campos, R.H.; Trono, K.; Jones, L.R. Bovine leukemia virus can be classified into seven genotypes: Evidence for the existence of two novel clades. J. Gen. Virol. 2009, 90, 2788–2797.
- Gautam, S.; Mishra, N.; Kalaiyarasu, S.; Jhade, S.K.; Sood, R. Molecular Characterization of Bovine Leukaemia Virus (BLV) Strains Reveals Existence of Genotype 6 in Cattle in India with evidence of a new subgenotype. Transbound. Emerg. Dis. 2018, 65, 1968–1978.
- Lee, E.; Kim, E.J.; Ratthanophart, J.; Vitoonpong, R.; Kim, B.H.; Cho, I.S.; Song, J.Y.; Lee, K.K.; Shin, Y.K. Molecular epidemiological and serological studies of bovine leukemia virus (BLV) infection in Thailand cattle. Infect. Genet. Evol. 2016, 41, 245–254.
- Selim, A.; Manaa, E.A.; Alanazi, A.D.; Alyousif, M.S. Seroprevalence, Risk Factors and Molecular Identification of Bovine Leukemia Virus in Egyptian Cattle. Animals 2021, 11, 319.
- Polat, M.; Takeshima, S.N.; Aida, Y. Epidemiology and genetic diversity of bovine leukemia virus. Virol. J. 2017, 14, 209.
- Kincaid, R.P.; Burke, J.M.; Sullivan, C.S. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. USA 2012, 109, 3077–3082.
- Rosewick, N.; Momont, M.; Durkin, K.; Takeda, H.; Caiment, F.; Cleuter, Y.; Vernin, C.; Mortreux, F.; Wattel, E.; Burny, A.; et al. Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 2306–2311.
- Gillet, N.A.; Hamaidia, M.; de Brogniez, A.; Gutierrez, G.; Renotte, N.; Reichert, M.; Trono, K.; Willems, L. Bovine Leukemia Virus Small Noncoding RNAs Are Functional Elements That Regulate Replication and Contribute to Oncogenesis In Vivo. PLoS Pathog. 2016, 12, e1005588.
- Santanam, U.; Zanesi, N.; Efanov, A.; Costinean, S.; Palamarchuk, A.; Hagan, J.P.; Volinia, S.; Alder, H.; Rassenti, L.; Kipps, T.; et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc. Natl. Acad. Sci. USA 2010, 107, 12210–12215.
- Petersen, M.I.; Carignano, H.A.; Mongini, C.; Gonzalez, D.D.; Jaworski, J.P. Bovine leukemia virus encoded blv-miR-b4-3p microRNA is associated with reduced expression of anti-oncogenic gene in vivo. PLoS ONE 2023, 18, e281317.
More
Information
Subjects:
Virology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
559
Revisions:
2 times
(View History)
Update Date:
22 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




