Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Rajeshwar Sinha | -- | 1855 | 2024-01-16 10:41:34 | | | |
| 2 | Rita Xu | Meta information modification | 1855 | 2024-01-16 10:54:18 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Jha, S.; Singh, V.K.; Singh, A.P.; Gupta, A.; Rana, P.; Sinha, R.P. Cyanobacterial Phycobiliproteins. Encyclopedia. Available online: https://encyclopedia.pub/entry/53886 (accessed on 08 February 2026).
Jha S, Singh VK, Singh AP, Gupta A, Rana P, Sinha RP. Cyanobacterial Phycobiliproteins. Encyclopedia. Available at: https://encyclopedia.pub/entry/53886. Accessed February 08, 2026.
Jha, Sapana, Varsha K. Singh, Ashish P. Singh, Amit Gupta, Palak Rana, Rajeshwar P. Sinha. "Cyanobacterial Phycobiliproteins" Encyclopedia, https://encyclopedia.pub/entry/53886 (accessed February 08, 2026).
Jha, S., Singh, V.K., Singh, A.P., Gupta, A., Rana, P., & Sinha, R.P. (2024, January 16). Cyanobacterial Phycobiliproteins. In Encyclopedia. https://encyclopedia.pub/entry/53886
Jha, Sapana, et al. "Cyanobacterial Phycobiliproteins." Encyclopedia. Web. 16 January, 2024.
Copy Citation
Phycobiliproteins (PBPs) are accessory light-harvesting pigment complexes found in cyanobacteria, red algae, and certain types of cryptophytes. The unique spectral features (strong absorbance and fluorescence), proteinaceous nature, and some imperative properties such as the anti-oxidative, hepato-protective, anti-inflammatory, and anti-aging activity of PBPs allow their use in biomedical industries.
bioavailability
cyanobacteria
phycobiliprotein
1. Introduction
The photosynthetic apparatus of cyanobacteria is composed of three primary light-harvesting systems: two photosystems common to other photosynthetic organisms and a phycobilisome (PBS). Phycobilisomes (PBSs) are large antenna complexes found in cyanobacteria that are crucial for photosynthesis. They capture light energy in the range of 450–650 nm, transfer it within its structure in a unidirectional way, and deliver it to the chlorophyll molecule of photosystems [1]. Although the PBSs of cyanobacteria are mainly composed of phycobiliproteins (PBPs) and linker proteins, their composition may vary from individual organism to different species. PBPs are in charge of absorbing and transmitting light energy, whereas linkers enable proper PBS assembly and control energy transmission [2]. Cyanobacterial PBPs are large, water-soluble supramolecular protein aggregates that are important accessory pigments during photosynthesis. Based on their spectrum features, PBPs can be generically classified into three classes: phycoerythrin (PE), phycocyanin (PC), and allophycocyanin (APC). PBPs are commercially produced from Spirulina platensis, Anabaena sp., and Galdieria sulphuraria and have been extracted and purified from Spirulina sp., Synechococcus sp., Oscillatoria sp., etc. [3]. The structure, amino acid composition, abundance, and types of PBPs of cyanobacteria and algae vary depending on the species and the habitat in which they grow. The spectral properties of PBPs result largely from the environment of the chromophore conferred by the apoprotein rather than the structural properties of the chromophore itself. PBPs in cyanobacteria account for 50% of total cellular protein [4].
Each PBS has two distinct substructures: the rods and the core [5][6]. The core is made up of APC. Several rods made up of PE and PC extend from the core in an outward orientation, which further functions as the light-harvesting antennae. Certain cyanobacteria species, including Spirulina platensis, have rods that solely contain PC and protein linkers that are nonetheless connected to the PBS core [7]. APC and linker proteins combine to create the core. This core transfers the energy captured by the rods to the chlorophyll present in the thylakoid membrane [8].
A growing interest in the study of microorganisms like cyanobacteria has resulted from the search for bioactive compounds in recent decades. This is because these organisms may have commercial applications in a variety of fields, including nutrition, animal and human health, wastewater treatment, energy production, and the chemical and pharmaceutical industries [9]. More specifically, cyanobacterial PBPs have widespread biotechnological applications due to their potent biological and pharmaceutical properties. PBPs, especially PC, have commercial and industrial applications due to their primary functions, including nutraceutical and therapeutic values (due to their pharmacology and biological activities, including anticarcinogenic, antioxidative, and anti-inflammatory activities, as well as a protective effect against various conditions), over-the-top fluorescent properties (high quantum yield, high Stokes shift, and an essential insensitivity to quenching), and natural colorants [10]. PBPs are useful fluorescent tags that find wide-ranging uses in immunoassay, reactive oxygen species (ROS) detection, flow cytometry, and fluorescence-activated cell sorting. These uses take advantage of PBP’s special spectroscopic and physical characteristics. They have a broad range of biomedical applications, such as in food coloring, biotechnology, medicines, cosmetics, and nutraceuticals [11].
2. Spectroscopic and Structural Characteristics of Cyanobacterial Phycobiliproteins
Based on their absorption, PBPs have been divided into three major categories: APC (λmax = 650–660 nm), PC (λmax = 610–625), and PE (λmax = 490–570) [12]. The most diverse chromophores among PBP are found in PE, which has a vibrant pink color. PE can be found in the PBS rods’ distal region. At 542 nm, C-PE exhibits a single absorption maximum. At a maximum emission value of 575 nm, the fluorescence emission of the various types of PE does not demonstrate significant variations [13]. To date, Phycoerythrocyanin (PEC) is exclusively reported in the photosynthetic membrane of Mastigocladus laminosus. It is a PBP with a purple-blue tint. One of the primary features of PEC is that during growth, its production is highly dependent on the kind and intensity of light. A significant increase in abundance was seen under low light conditions with green light [14]. The proximal portion of PBS rods contains the bright blue protein known as PC. There is just a single absorption maximum at 615 nm (Figure 1) [15]. APC, which is only present in the PBS core, is a bright turquoise color with a maximum absorption of 650 nm [8]. A highly effective energy transfer in the PBS is made possible by these spectroscopic features of PBP absorption and emission.
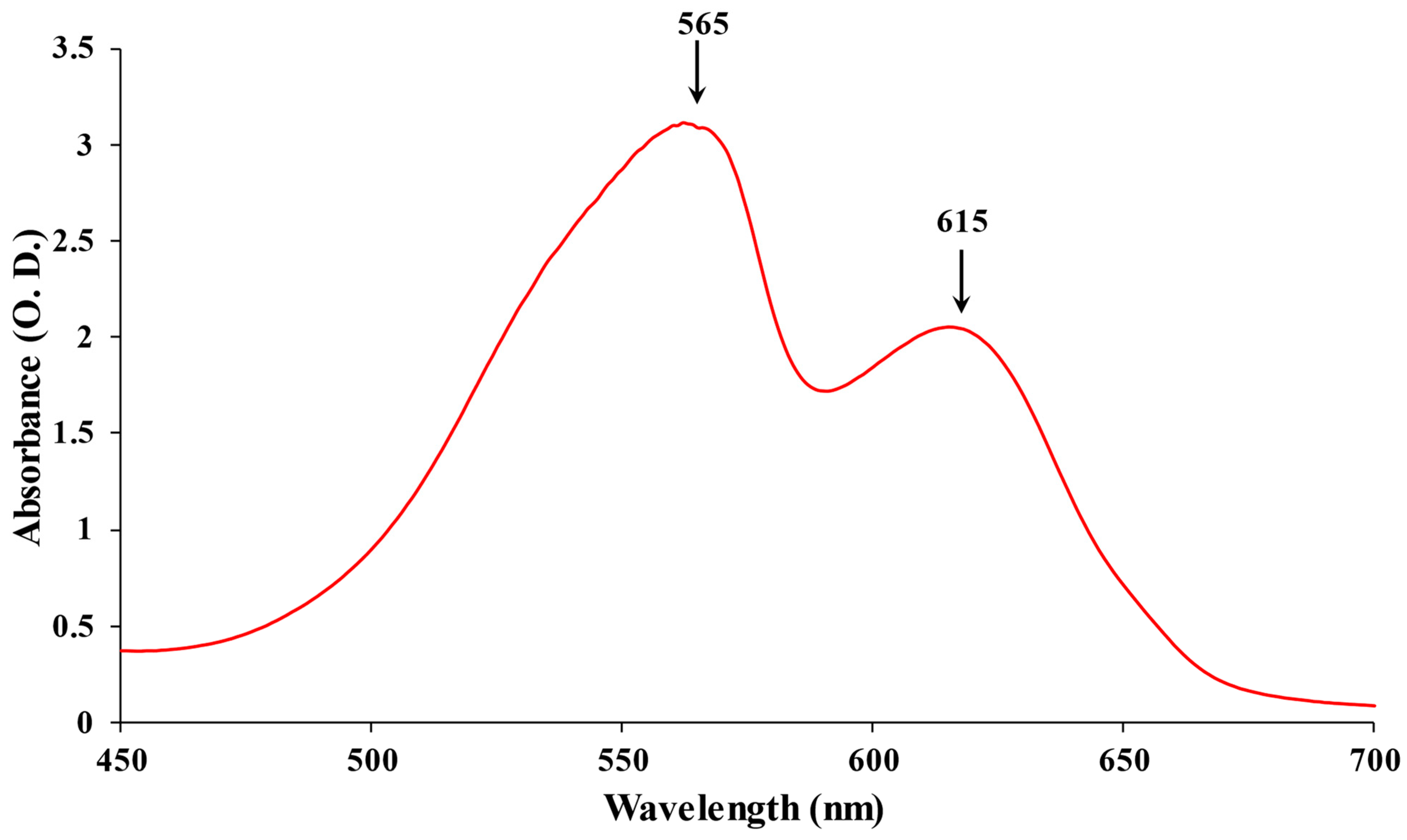
Figure 1. The absorption spectrum of PBPs: PE maximum at 565 nm and PC maximum at 615 nm.
2.1. Subunits and Heterodimers (αβ)
The basic component of PBPs is its subunits, specifically α and β subunits, which stabilize each other to form an αβ heterodimer. The self-assembly of all PBPs appears when two subunits, α and β, dock together [16][17]. This α and β heterodimer is known as the fundamental component of PBPs’ monomeric structure. It subsequently comes together to form trimeric discs (αβ)3. Monomer subunits are primarily distinguished from native trimeric units by dissociated subunits, which are usually less intense in color. In various PBPs, native trimeric and hexameric units have different molecular weights. When it comes to capturing solar light energy, the hexamer assembly is more functional and stable. On the other hand, the trimeric form of the majority of PCs has demonstrated an intriguing rod structure assembly without a requirement for linker polypeptides [18]. Four trimers of APC biliproteins come together to form core cylinders when they are assembled. A full functional structure is then formed by the subsequent assembly of two to five core cylinders into the core substructure with the help of linker polypeptides, as in Figure 2.
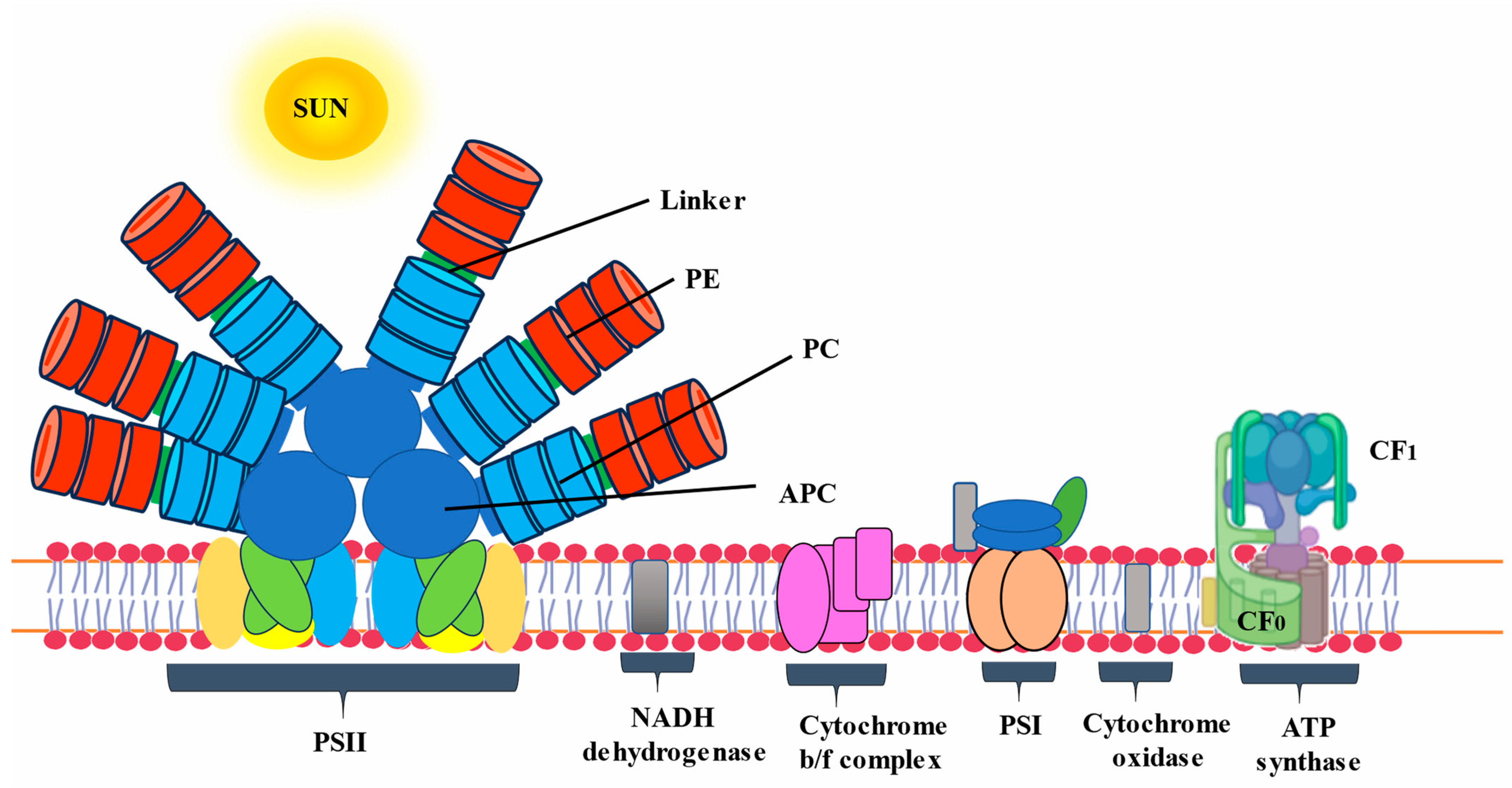
Figure 2. Schematic diagram of phycobilisome located on the thylakoid membrane.
Ultimately, a complete PBS is formed by the association of both core cylinders and rods (Figure 3). When it comes to high-resolution views of proteins, X-ray crystallography has emerged as an extremely valuable instrument. Initially, the entire structure of PBS was explored at low resolution through electron microscopy. PBS crystal structures have also significantly improved our potential to analyze the tertiary, or hexameric, structure of proteins as well as their stability and functionalities in a range of environmental circumstances [19].
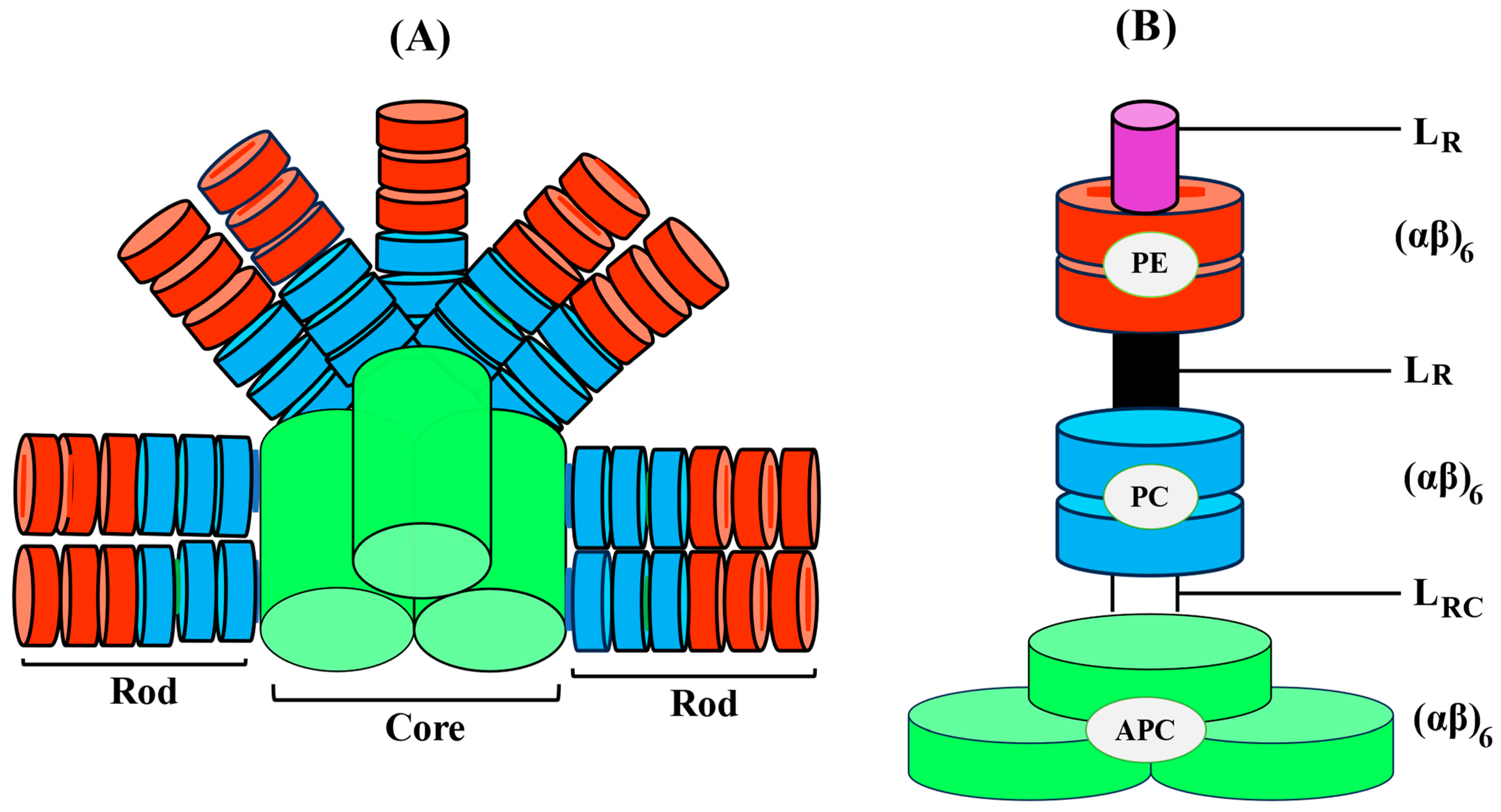
Figure 3. Diagrammatic illustration of the three-dimensional intact PBS with the PC/PE rod organized in a parallel orientation around the APC core (A), Dimension of PBP and associated linker polypeptides and, (B) schematic arrangement of cylindrical core (APC).
2.2. PBPs Chromophore
Chromophores are open-chain tetrapyrroles covalently linked to cysteine via thioether bonds. PBPs can vary in the number of chromophores they contain. Based on their structural characteristics, the chromophores can be categorized into the following categories: phycocyanobilin (PCB), phycoerythrobilin (PEB), phycoviolobilin (PVB), or phycourobilin (PUB) (Figure 4) [20]. Each biliprotein consists of the α and β subunits that are linked through covalent bonds to the apoprotein’s cysteines through a thioether bond to C-3 on ring A, and occasionally through an extra thioether bond to C-18 on ring D. The chromophore that is developed from heme is joined to the apoproteins of bilin by an ethylidene bond, which adds a thiol group [21]. A single cysteinyl thioester linkage via the vinyl substituents on the pyrrole ring A of the tetrapyrrole, or periodically two cysteine linkages through the vinyl substituents on both the A and D pyrrole rings, are typically responsible for binding chromophores to the polypeptide chain at the conserved position. The assembly of chromophores within the clearly identified geometries of proteins’ cores determines the properties of light-harvesting PBPs in photosynthetic cyanobacteria [22]. The electronic state of chromophores and related apoproteins plays a major role in the establishment of the stable chemical structure of proteins. Consequently, the chromophore serves as a report on the integrity of the protein structure; the protein possesses a stunning dark blue color when it is in its native form, but this color disappears when it is denatured [23]. PBPs have been studied using fluorescence techniques because, when detached from reaction centers, they show strong fluorescence. The fluorescence quantum yield of the tetrapyrrole chromophore in C-PC is roughly 60%, with a maximum fluorescence of 650 nm when it is maintained in a linear alignment by the protein scaffolding [24].
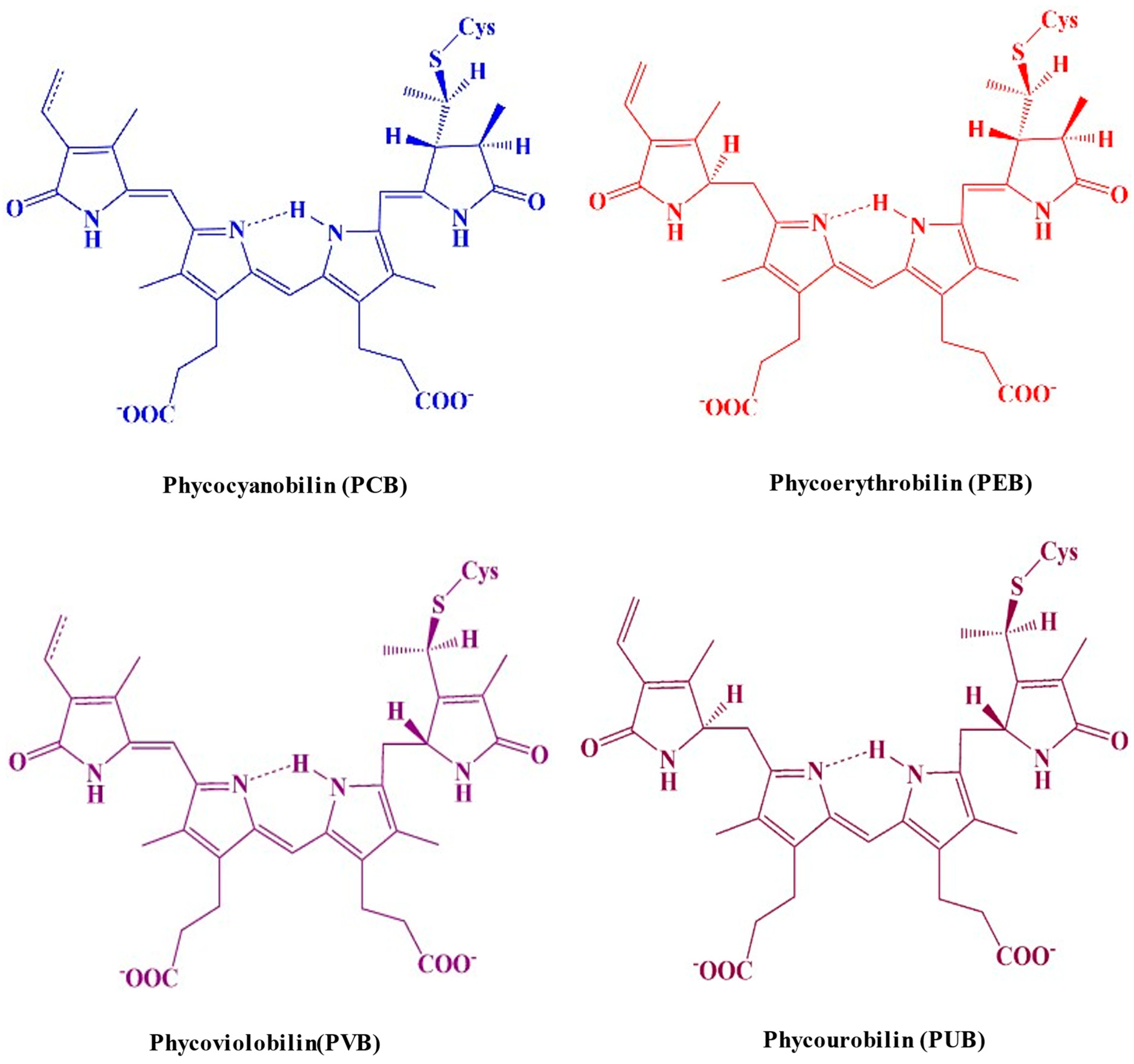
Figure 4. A molecular structure illustrating the distinguished colors of the cysteine-linked chromophores in PBP complexes.
The heterodimeric (αβ) protomer of C-phycocyanin (C-PC) contains three PCB chromophores at cysteines α-84, β-84, and β-155. This residue is also conserved in the β subunit of the C-PC chromophore, where it is classified as Asp39 in the β-155 chromophore and Asp87 in β-84. It is present in both C-APC and C-PE. The chromophore’s electronic state is established by an important amino acid called the Asp87 residue [25]. Position Cys84 on the α subunit contains a single PVB chromophore, while positions Cys82 and Cys153 on the β subunit contain two PVB chromophores. PEB comprises a set of two to three covalently linked tetrapyrrole chromophores integrated with the α (164 amino acid) and β (177 amino acid) subunits of C-PE, respectively [26].
2.3. Linker Polypeptides
The PBP structure involves the non-pigmented linker polypeptides [27]. The polypeptides have molecular masses varying between 8 and 120 kDa. Two possible functions of linker polypeptides in the PBS have been proposed: (1) maintain the PBS structures and establish structural links between the nearest PBPs, and (2) adjust both the properties of absorption and fluorescence to support or directly take part in the energy transfer from the rod to the core and ultimately to the thylakoid membrane of the photosynthetic cells, which contain chlorophyll [28]. The two basic subunits (α and β) of PBPs along with its heterodimer, i.e., αβ are represented in Figure 5.
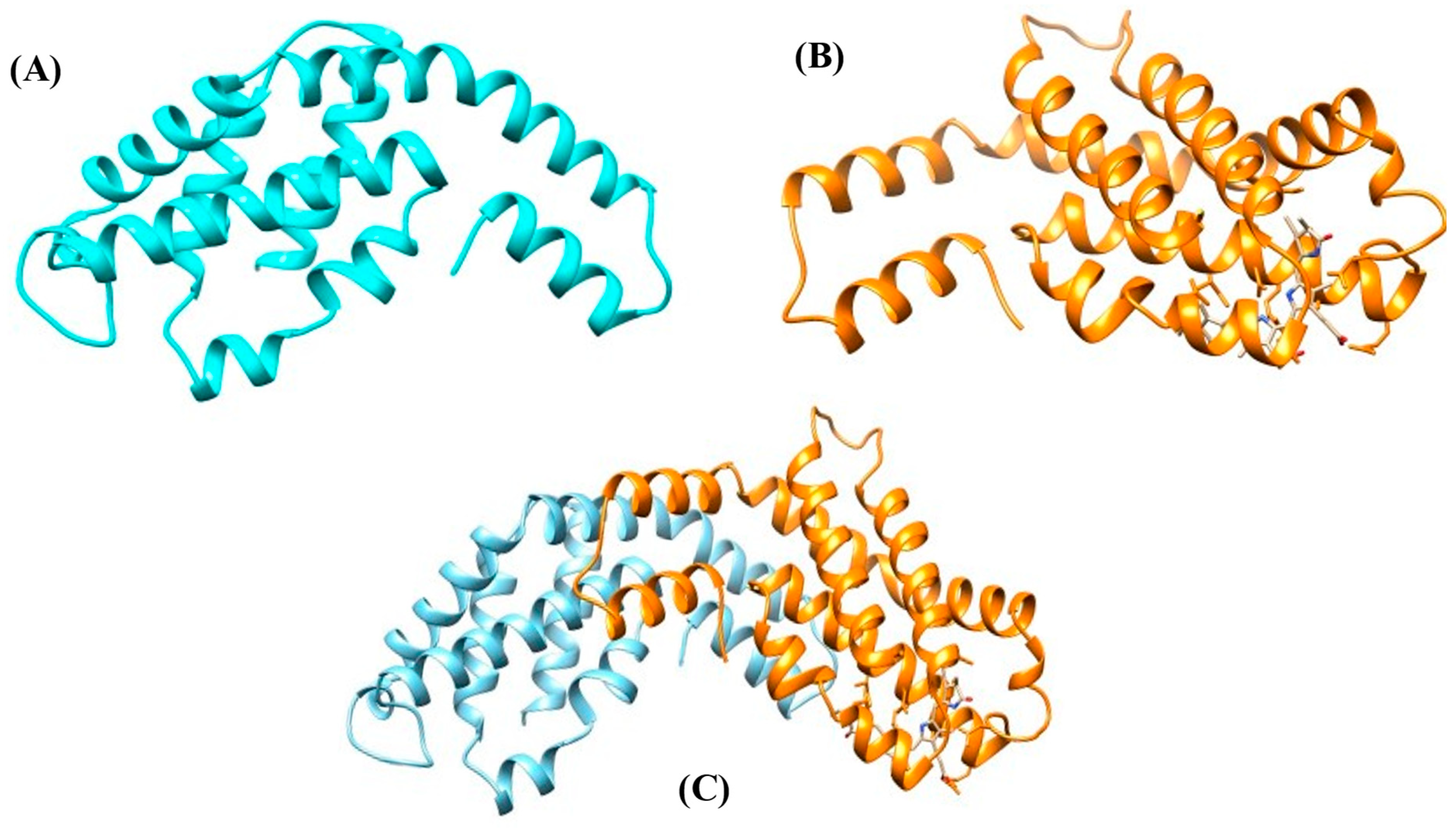
Figure 5. Protein model of PBPs. (A) α subunit, (B) β subunit, and (C) αβ heterodimer.
The standard acronym for linker polypeptides is LXY, where X represents the location and Y represents the molecular mass of the linker polypeptide (L) PBS complex. In addition, X can be assigned either to the letter R (rod) or C (core) for the main chain, while RC (rod-core) and CM (core-membrane) are used to represent junctions. In class I and II of R-PE, the γ-subunits and the core-membrane linker (LCM) PCB-ApcE are two of the linker polypeptides that contain covalently attached chromophores, while the majority of them are colorless [29]. Based on their location and functional characteristics, the linker polypeptides are divided into four groups. PBS: Group I consists of LR polypeptides (27–35 kDa) involved in the peripheral rod assembly, together with a small number of 10 kDa rod linker polypeptides (LR10, LR33, and LR35, for example, which connect trimeric or hexameric PC/PE structural components into rod segments); Group II LRC polypeptides, which are between 25 and 27 kDa, take part in establishing the peripheral rods to the core subunits; LC polypeptides (8 kDa) belonging to Group III are essential for joining core components and ensuring their functional properties; and the main terminal energy emitter for PSII is Group IV, LCM (70–120 kDa), with a greater molecular weight of polypeptides that connects PBS to the photosynthetic membrane [30]. Six conserved domains (N-terminus) in rod linker polypeptides have been recognized by structural motif analysis as being crucial for the packing and assembly of rod discs into hexamers (Figure 3). There are connections between PBPs and linker polypeptides because the surfaces of linker polypeptides possess a positive charge, while the majority of globular proteins are probably hydrophobic [28].
References
- Masojídek, J.; Torzillo, G.; Koblížek, M. Photosynthesis in microalgae. In Handbook of Microalgal Culture: Applied Phycology and Biotechnology; Wiley: Hoboken, NJ, USA, 2013; pp. 21–36.
- Pagels, F.; Guedes, A.C.; Amaro, H.M.; Kijjoa, A.; Vasconcelos, V. Phycobiliproteins from cyanobacteria: Chemistry and biotechnological applications. Biotechnol. Adv. 2019, 37, 422–443.
- Kannaujiya, V.K.; Sundaram, S.; Sinha, R.P. Structural and functional significance of phycobiliproteins. In Phycobiliproteins: Recent Developments and Future Applications; Springer: Berlin/Heidelberg, Germany, 2017; pp. 21–44.
- Bryant, D.A.; Guglielmi, G.; de Marsac, N.T.; Castets, A.M.; Cohen-Bazire, G. The structure of cyanobacterial phycobilisomes: A model. Arch. Microbiol. 1979, 123, 113–127.
- Arteni, A.A.; Ajlani, G.; Boekema, E.J. Structural organisation of phycobilisomes from Synechocystis sp. strain PCC 6803 and their interaction with the membrane. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 272–279.
- Figueroa, M.; Martínez-Oyanedel, J.; Matamala, A.R.; Dagnino-Leone, J.; Mella, C.; Fritz, R.; Bunster, M. In silico model of an antenna of a phycobilisome and energy transfer rates determination by theoretical Förster approach. Protein Sci. 2012, 21, 1921–1928.
- Su, H.N.; Xie, B.B.; Chen, X.L.; Wang, J.X.; Zhang, X.Y.; Zhou, B.C.; Zhang, Y.Z. Efficient separation and purification of allophycocyanin from Spirulina (Arthrospira) platensis. J. Appl. Phycol. 2010, 22, 65–70.
- Dagnino-Leone, J.; Figueroa, M.; Uribe, E.; Hinrichs, M.V.; Ortiz-López, D.; Martínez-Oyanedel, J.; Bunster, M. Biosynthesis and characterization of a recombinant eukaryotic allophycocyanin using prokaryotic accessory enzymes. Microbiology 2020, 9, e989.
- Singh, V.K.; Jha, S.; Rana, P.; Mishra, S.; Kumari, N.; Singh, S.C.; Sinha, R.P. Resilience and mitigation strategies of cyanobacteria under ultraviolet radiation stress. Int. J. Mol. Sci. 2023, 24, 12381.
- Manirafasha, E.; Guo, L.; Jing, K. Nutraceutical and pharmaceutical applications of phycobiliproteins. In Pigments from Microalgae Handbook; Springer: Berlin/Heidelberg, Germany, 2020; pp. 575–584.
- Gupta, A.; Singh, A.P.; Singh, V.K.; Singh, P.R.; Jaiswal, J.; Kumari, N.; Sinha, R.P. Natural sun-screening compounds and DNA-repair enzymes: Photoprotection and photoaging. Catalysts 2023, 13, 745.
- Scheer, H.; Zhao, K.H. Biliprotein maturation: The chromophore attachment. Mol. Microbiol. 2008, 68, 263–276.
- Ma, J.; You, X.; Sun, S.; Wang, X.; Qin, S.; Sui, S.F. Structural basis of energy transfer in Porphyridium purpureum phycobilisome. Nature 2020, 579, 146–151.
- Hirose, Y.; Chihong, S.; Watanabe, M.; Yonekawa, C.; Murata, K.; Ikeuchi, M.; Eki, T. Diverse chromatic acclimation processes regulating phycoerythrocyanin and rod-shaped phycobilisome in cyanobacteria. Mol. Plant 2019, 12, 715–725.
- Alvey, R.M.; Biswas, A.; Schluchter, W.M.; Bryant, D.A. Attachment of noncognate chromophores to CpcA of Synechocystis sp. PCC 6803 and Synechococcus sp. PCC 7002 by heterologous expression in Escherichia coli. Biochemistry 2011, 50, 4890–4902.
- McGregor, A.; Klartag, M.; David, L.; Adir, N. Allophycocyanin trimer stability and functionality are primarily due to polar enhanced hydrophobicity of the phycocyanobilin binding pocket. J. Mol. Biol. 2008, 384, 406–421.
- Li, W.; Su, H.N.; Pu, Y.; Chen, J.; Liu, L.N.; Liu, Q.; Qin, S. Phycobiliproteins: Molecular structure, production, applications, and prospects. Biotechnol. Adv. 2019, 37, 340–353.
- Marx, A.; Adir, N. Allophycocyanin and phycocyanin crystal structures reveal facets of phycobilisome assembly. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 311–318.
- Zhang, J.; Ma, J.; Liu, D.; Qin, S.; Sun, S.; Zhao, J.; Sui, S.F. Structure of phycobilisome from the red alga Griffithsia pacifica. Nature 2017, 551, 57–63.
- Sui, S.F. Structure of phycobilisomes. Annu. Rev. Biophys. 2021, 50, 53–72.
- Dagnino-Leone, J.; Figueroa, C.P.; Castañeda, M.L.; Youlton, A.D.; Vallejos-Almirall, A.; Agurto-Muñoz, A.; Agurto-Muñoz, C. Phycobiliproteins: Structural aspects, functional characteristics, and biotechnological perspectives. Comput. Struct. Biotechnol. J. 2022, 20, 1506–1527.
- Stadnichuk, I.N.; Krasilnikov, P.M.; Zlenko, D.V. Cyanobacterial phycobilisomes and phycobiliproteins. Microbiology 2015, 84, 101–111.
- Chen, H.; Qi, H.; Xiong, P. Phycobiliproteins-A family of algae-derived biliproteins: Productions, characterization and pharmaceutical potentials. Mar. Drugs 2022, 20, 450.
- Zehetmayer, P.; Hellerer, T.; Parbel, A.; Scheer, H.; Zumbusch, A. Spectroscopy of single phycoerythrocyanin monomers: Dark state identification and observation of energy transfer heterogeneities. Biophys. J. 2002, 83, 407–415.
- Kikuchi, H. Redshifting and blueshifting of β82 chromophores in the phycocyanin hexamer of Porphyridium purpureum phycobilisomes due to linker proteins. Life 2022, 12, 1833.
- MacColl, R.; Eisele, L.E.; Malak, H.; Endres, R.L.; Williams, E.C.; Bowser, S.S. Studies on R-phycoerythrins from two Antarctic marine red algae and a mesophilic red alga. Polar Biol. 1999, 22, 384–388.
- Stadnichuk, I.N.; Tropin, I.V. Phycobiliproteins: Structure, functions and biotechnological applications. Appl. Biochem. Microbiol. 2017, 53, 1–10.
- Liu, L.N.; Chen, X.L.; Zhang, Y.Z.; Zhou, B.C. Characterization, structure and function of linker polypeptides in phycobilisomes of cyanobacteria and red algae: An overview. Biochim. Biophys. Acta Bioenerg. 2005, 1708, 133–142.
- Stadnichuk, I.N.; Kusnetsov, V.V. Phycobilisomes and phycobiliproteins in the pigment apparatus of oxygenic photosynthetics: From cyanobacteria to tertiary endosymbiosis. Int. J. Mol. Sci. 2023, 24, 2290.
- Nganou, C.; David, L.; Adir, N.; Mkandawire, M. Linker proteins enable ultrafast excitation energy transfer in the phycobilisome antenna system of Thermosynechococcus vulcanus. Photochem. Photobiol. Sci. 2016, 15, 31–44.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
603
Revisions:
2 times
(View History)
Update Date:
16 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




