Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Xueyan Wang | -- | 1299 | 2024-01-10 12:27:02 | | | |
| 2 | Camila Xu | Meta information modification | 1299 | 2024-01-11 02:08:56 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lin, M.; Wang, X. CRISPR/Cas9-Mediated Gene Editing System. Encyclopedia. Available online: https://encyclopedia.pub/entry/53682 (accessed on 02 March 2026).
Lin M, Wang X. CRISPR/Cas9-Mediated Gene Editing System. Encyclopedia. Available at: https://encyclopedia.pub/entry/53682. Accessed March 02, 2026.
Lin, Meng, Xueyan Wang. "CRISPR/Cas9-Mediated Gene Editing System" Encyclopedia, https://encyclopedia.pub/entry/53682 (accessed March 02, 2026).
Lin, M., & Wang, X. (2024, January 10). CRISPR/Cas9-Mediated Gene Editing System. In Encyclopedia. https://encyclopedia.pub/entry/53682
Lin, Meng and Xueyan Wang. "CRISPR/Cas9-Mediated Gene Editing System." Encyclopedia. Web. 10 January, 2024.
Copy Citation
The natural CRISPR-Cas9 system are composed of Cas9, crRNA, and tracrRNA. The artificial CRISPR/Cas9 system usually consists of two components: the Cas9 endonuclease and the sgRNA, which form the ribonucleoprotein complex via base pairing to mediate the gene editing.
CRISPR/Cas9
natural biopolymer
gene delivery
tumor therapy
1. Mechanism of CRISPR/Cas9
CRISPR/Cas9 stemmed from the adaptive immune systems of most archaea and many bacteria and has been widely utilized in genome editing [1]. The natural CRISPR-Cas9 system are composed of Cas9, crRNA, and tracrRNA. The crRNA contains a sequence for target DNA recognition and a sequence for tracrRNA binding. The tracrRNA can bind with the tracrRNA via complementary pairing. In mammalian genome editing with CRISPR-Cas9, crRNA, and tracrRNA have been engineered into sgRNA. Therefore, the artificial CRISPR/Cas9 system usually consists of two components: the Cas9 endonuclease and the sgRNA, which form the ribonucleoprotein complex via base pairing to mediate the gene editing. That is, the Cas9 is directed to the upstream of protospacer adjacent motif (PAM) under the guidance of sgRNA and cleaves the target gene to generate the double-strand breaks (DSBs). DSBs can be repaired by cells via two pathways: non-homologous end joining (NHEJ) and homology-directed repair (HDR) [1][2]. The insertion/deletion (InDel) of edited DNA strands often occurs during NHEJ, leading to the frameshifts and/or the premature of stop codons [3]. Unlike NHEJ, which joins the breaks together, the donor DNA template was inserted at the specific sites during HDR to edit the gene accurately (Figure 1). Although both pathways exist in cells simultaneously, only 25% of genome repair occurs via the HDR pathway, while the remaining 75% of DSBs are repaired by the error-prone NHEJ mechanism [4][5]. Strategies to improve the efficiency of HDR have been developed, such as using NHEJ inhibitors or HDR enhancers [6].
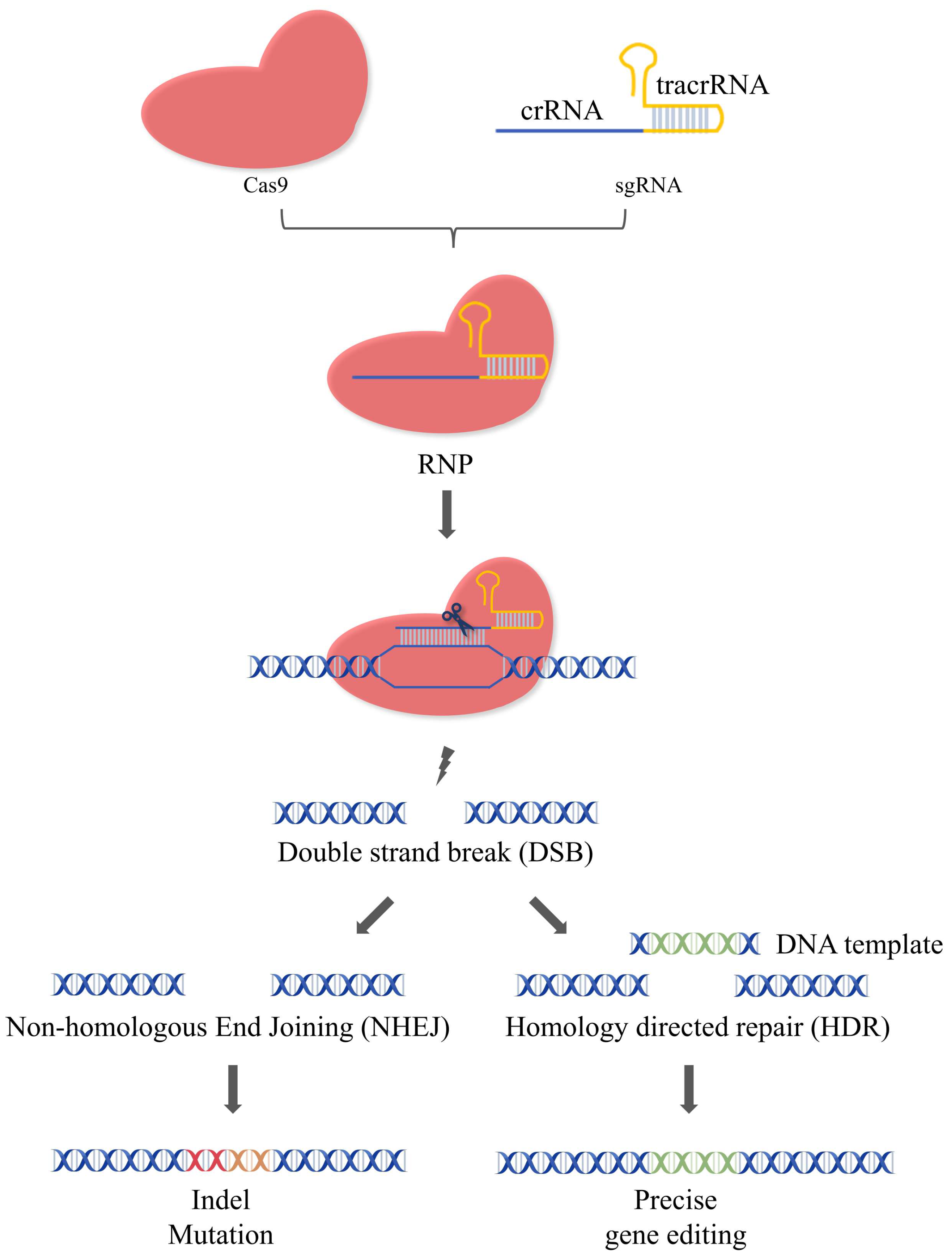
Figure 1. Mechanism of the CRISPR/Cas9 system. The CRISPR/Cas9 system is composed of Cas9 and sgRNA. The Cas9 can specifically cleave the target gene under the guidance of sgRNA to generate DSBs. The cell can repair the DSBs via two pathways: NHEJ or HDR. During NHEJ, indel mutations of edited DNA strands often occur, leading to the frameshifts and/or the formation of premature termination codons. During HDR, the donor DNA template is inserted at the specific sites for precise gene editing.
Different from conventional gene editing tools like ZFNs and TALENs, it is more convenient and easier to personalize the CRISPR/Cas9 complex by only changing the sgRNA sequence [7], rendering it possible to edit multiple independent sites simultaneously. Thus, CRISPR/Cas9 gene editing technology has been broadly utilized to correct mutated genes to treat various diseases, including Duchenne muscular dystrophy (DMD) [8], hereditary tyrosinemia I [9], hypercholesterolemia [10], and cancer [11]. Since the main cause of cancer is the dysregulation of cell growth, knocking out the oncogenes or repairing the tumor-suppressive genes by CRISPR/Cas9 gene engineering tools has shown promising potential in cancer treatment. Up to now, CRISPR/Cas9-based therapy has been utilized to treat multiple tumors, including lung cancer, breast cancer, colon cancer, melanoma, hepatocellular carcinoma, etc. The therapeutic targets for CRISPR/Cas9-mediated cancer treatment include oncogenes (Kras) [12], cell death-related genes (MTH1) [13], epigenetic genes (DNMT1) [14], immune-related genes (CD47) [15], viral oncogenes (E6 or E7) [16], and tumor microenvironment-associated gene targets (VEGFA) [17], etc.
2. Three Forms of CRISPR/Cas9 Delivery
There are three forms of delivery when using the CRISPR/Cas9 system: DNA, mRNA, and ribonucleoprotein (Figure 2). Each of these delivery forms has advantages and disadvantages in efficiency and accuracy. Regardless of the different delivery forms, the ribonucleoprotein complex formed by Cas9 and sgRNA is the key to gene editing.
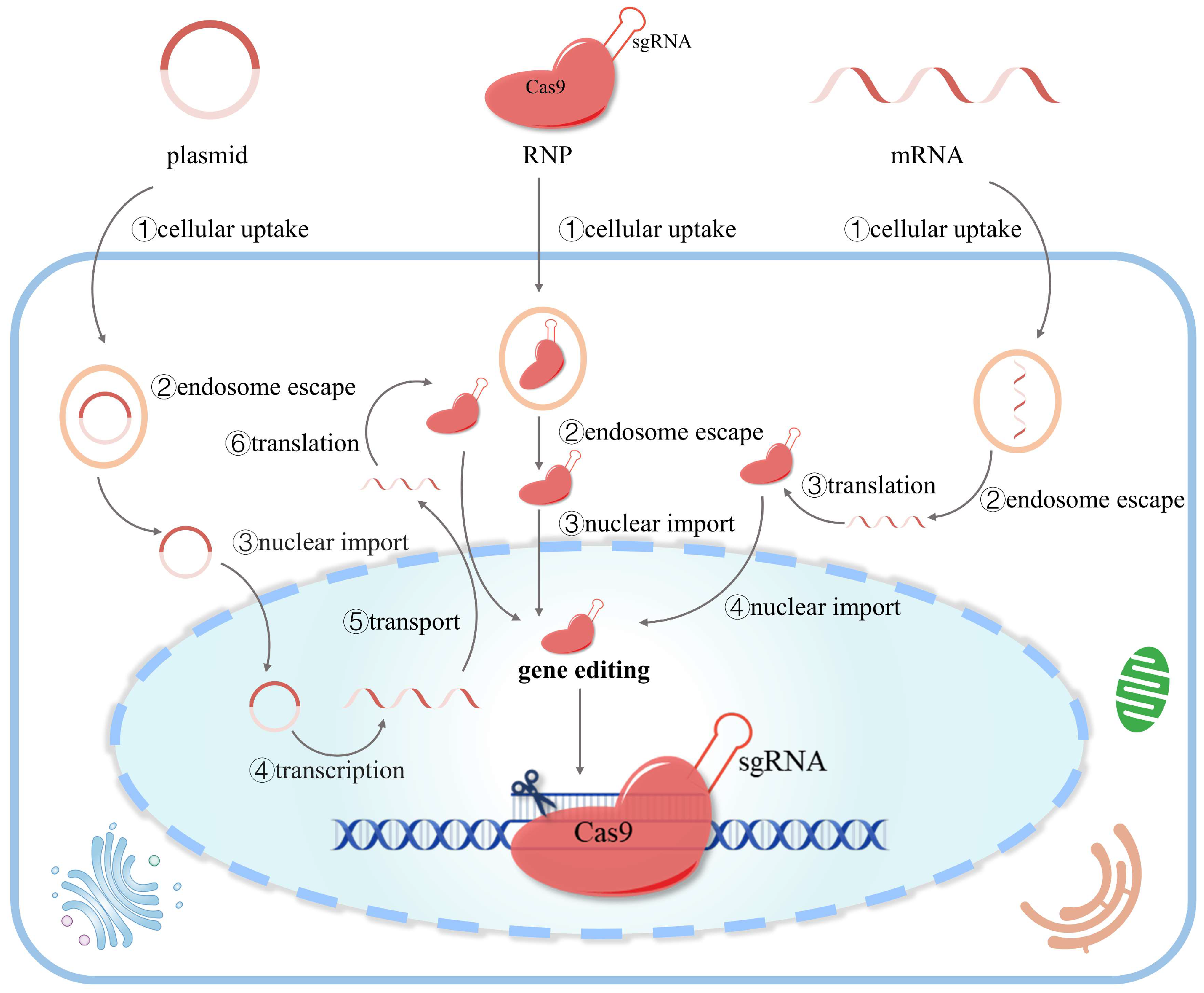
Figure 2. Different forms of CRISPR/Cas9-based gene editing. There are three strategies to edit the genome using CRISPR/Cas9: DNA (plasmid), mRNA, and ribonucleoprotein (RNP). The plasmid has to undergo multiple biological processes (cellular uptake, endosome escape, nuclear import, transcription, transport, and translation) to express the RNP for efficient gene editing. mRNA is translated to RNP in the cytoplasm and then transported to the nucleus for gene editing. RNP can initiate gene editing after entering the nucleus without transcription or translation.
2.1. DNA (Plasmid)-Based Delivery
Cas9 and sgRNA cassettes can be packed together in the same plasmid or separately in two plasmids. After transcription and translation, the formed ribonucleoprotein complex enables gene editing at the target locus. Due to its good stability and relatively low cost, DNA-based gene engineering may be easier to scale up and translate into the clinic. However, the large size of Cas9 and the plasmid makes it even harder for efficient delivery. An additional obstacle for DNA-based gene editing is the requirement for multiple biological processes, including transcription and translation, which delay the onset of gene editing and may decrease the editing efficiency. Moreover, the continuous expression of ribonucleoprotein increases the risk of off-target mutagenesis [18].
2.2. mRNA-Based Delivery
Cas9 mRNA can be obtained by in vitro transcription and mediate genome editing after translation in cells. Usually, mRNA-based delivery enables a quicker onset of gene editing than DNA-based delivery, as it does not require nuclear entry for DNA transcription. The transient expression of Cas9 mediated by mRNA also reduces the risk of off-target effects. However, mRNA is less stable than DNA and is prone to degradation, increasing the difficulty of production, storage, and clinical use. In addition, when delivering in the form of mRNA, the right timing of delivery has to be taken into account. For efficient genome editing, Cas9 and sgRNA should be presented at the target site simultaneously. Therefore, delivering the mRNA into the cells before sgRNA is optimal since the mRNA encoding Cas9 has to be translated first. One study showed that injection of Cas9 mRNA followed by an injection of sgRNA 6 h later in mice is helpful to increase the efficiency of gene editing [19].
2.3. Protein-Based Delivery
It is the most straightforward way to deliver the ribonucleoprotein complex formed by Cas9 and sgRNA. It does not require these biological processes, such as transcription and translation, thus enabling the quickest onset of gene editing [20]. However, the large size of Cas9 protein (160 kDa) and gRNA (34 kDa) increases the difficulty of delivery. Moreover, it is difficult and costly to obtain proteins of high purity, and proteins isolated from bacteria may contain toxins, increasing the risk of safety. Additionally, the use of proteins in the body may trigger immune responses.
3. Multiple Barriers of Delivery
High molecular weight and massive charge are the common characteristics of DNA, mRNA, and ribonucleoprotein, and the use of these biomacromolecules all face similar dilemmas in their delivery. Firstly, these cargoes should be packed with suitable materials to facilitate delivery. Then, during circulation, these biomacromolecules should resist the harsh environment in vivo. Especially for mRNA and proteins of poor stability, it is vital to protect them from enzymes to maintain their stability for in vivo application. Also, it is essential to prevent clearance by the mononuclear phagocyte system (MPS) and to maintain long circulation time for efficient genome editing [21]. Moreover, the efficiency and accuracy of CRISPR/Cas9-based gene engineering relies on the accumulation of these cargoes at their target organs, cells, or even organelles. After accumulating at their target organs, they have to cross the extracellular matrix and accurately identify the target cells. However, the CRISPR/Cas9 machinery is unlikely to pass through the cell membranes without the assistance of a carrier due to their high molecular weight and massive charge. In addition to cellular uptake, intracellular barriers, including endosomal escape and nuclear entry, also severely restrict the editing efficacy. For mRNA and proteins whose target sites are in the cytoplasm, the key to delivery is cellular uptake and endosomal escape, while for DNA, whose target sites are in the nucleus, the determining step is nuclear entry. Therefore, it is crucial to design suitable delivery systems on the basis of their own characteristics to maximize the genome editing efficiency.
References
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823.
- Mout, R.; Ray, M.; Lee, Y.W.; Scaletti, F.; Rotello, V.M. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: Progress and challenges. Bioconjugate Chem. 2017, 28, 880–884.
- Liu, C.; Zhang, L.; Liu, H.; Cheng, K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release 2017, 266, 17–26.
- Johnson, R.D.; Jasin, M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000, 19, 3398–3407.
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 2008, 7, 1765–1771.
- Aravalli, R.N.; Steer, C.J. CRISPR/Cas9 therapeutics for liver diseases. J. Cell. Biochem. 2018, 119, 4265–4278.
- Yi, L.; Li, J. CRISPR-Cas9 therapeutics in cancer: Promising strategies and present challenges. Biochim. Biophys. Acta 2016, 1866, 197–207.
- Min, Y.L.; Li, H.; Rodriguez-Caycedo, C.; Mireault, A.A.; Huang, J.; Shelton, J.M.; McAnally, J.R.; Amoasii, L.; Mammen, P.P.A.; Bassel-Duby, R.; et al. CRISPR-Cas9 corrects duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 2019, 5, eaav4324.
- Yin, H.; Xue, W.; Chen, S.; Bogorad, R.L.; Benedetti, E.; Grompe, M.; Koteliansky, V.; Sharp, P.A.; Jacks, T.; Anderson, D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014, 32, 551–553.
- Ding, Q.; Strong, A.; Patel, K.M.; Ng, S.L.; Gosis, B.S.; Regan, S.N.; Cowan, C.A.; Rader, D.J.; Musunuru, K. Permanent alteration of pcsk9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 2014, 115, 488–492.
- Zhang, B.-C.; Luo, B.-Y.; Zou, J.-J.; Wu, P.-Y.; Jiang, J.-L.; Le, J.-Q.; Zhao, R.-R.; Chen, L.; Shao, J.-W. Co-delivery of sorafenib and CRISPR/Cas9 based on targeted core–shell hollow mesoporous organosilica nanoparticles for synergistic hcc therapy. ACS Appl. Mater. Interfaces 2020, 12, 57362–57372.
- Kodama, M.; Kodama, T.; Newberg, J.Y.; Katayama, H.; Kobayashi, M.; Hanash, S.M.; Yoshihara, K.; Wei, Z.; Tien, J.C.; Rangel, R.; et al. In vivo loss-of-function screens identify kpnb1 as a new druggable oncogene in epithelial ovarian cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E7301–E7310.
- Li, L.; Song, L.; Liu, X.; Yang, X.; Li, X.; He, T.; Wang, N.; Yang, S.; Yu, C.; Yin, T.; et al. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano 2017, 11, 95–111.
- He, Z.Y.; Zhang, Y.G.; Yang, Y.H.; Ma, C.C.; Wang, P.; Du, W.; Li, L.; Xiang, R.; Song, X.R.; Zhao, X.; et al. In vivo ovarian cancer gene therapy using CRISPR-Cas9. Hum. Gene Ther. 2018, 29, 223–233.
- Lin, M.; Yang, Z.; Yang, Y.; Peng, Y.; Li, J.; Du, Y.; Sun, Q.; Gao, D.; Yuan, Q.; Zhou, Y.; et al. CRISPR-based in situ engineering tumor cells to reprogram macrophages for effective cancer immunotherapy. Nano Today 2022, 42, 101359.
- Kennedy, E.M.; Kornepati, A.V.; Goldstein, M.; Bogerd, H.P.; Poling, B.C.; Whisnant, A.W.; Kastan, M.B.; Cullen, B.R. Inactivation of the human papillomavirus e6 or e7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 2014, 88, 11965–11972.
- Liang, C.; Li, F.; Wang, L.; Zhang, Z.K.; Wang, C.; He, B.; Li, J.; Chen, Z.; Shaikh, A.B.; Liu, J.; et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of vegfa in osteosarcoma. Biomaterials 2017, 147, 68–85.
- Ishida, K.; Gee, P.; Hotta, A. Minimizing off-target mutagenesis risks caused by programmable nucleases. Int. J. Mol. Sci. 2015, 16, 24751–24771.
- Jiang, C.; Mei, M.; Li, B.; Zhu, X.; Zu, W.; Tian, Y.; Wang, Q.; Guo, Y.; Dong, Y.; Tan, X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting hbv DNA and pcsk9 in vivo. Cell Res. 2017, 27, 440–443.
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015, 208, 44–53.
- Tong, S.; Moyo, B.; Lee, C.M.; Leong, K.; Bao, G. Engineered materials for in vivo delivery of genome-editing machinery. Nat. Rev. Mater. 2019, 4, 726–737.
More
Information
Subjects:
Genetics & Heredity
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
665
Revisions:
2 times
(View History)
Update Date:
11 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





