You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | SAYED SARTAJ SOHRAB | -- | 3836 | 2024-01-09 12:57:43 | | | |
| 2 | Rita Xu | Meta information modification | 3836 | 2024-01-10 03:18:56 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tabll, A.A.; Sohrab, S.S.; Ali, A.A.; Petrovic, A.; Steiner Srdarevic, S.; Siber, S.; Glasnovic, M.; Smolic, R.; Smolic, M. Hepatitis C Virus Vaccine. Encyclopedia. Available online: https://encyclopedia.pub/entry/53609 (accessed on 17 December 2025).
Tabll AA, Sohrab SS, Ali AA, Petrovic A, Steiner Srdarevic S, Siber S, et al. Hepatitis C Virus Vaccine. Encyclopedia. Available at: https://encyclopedia.pub/entry/53609. Accessed December 17, 2025.
Tabll, Ashraf A., Sayed S. Sohrab, Ahmed A. Ali, Ana Petrovic, Sabina Steiner Srdarevic, Stjepan Siber, Marija Glasnovic, Robert Smolic, Martina Smolic. "Hepatitis C Virus Vaccine" Encyclopedia, https://encyclopedia.pub/entry/53609 (accessed December 17, 2025).
Tabll, A.A., Sohrab, S.S., Ali, A.A., Petrovic, A., Steiner Srdarevic, S., Siber, S., Glasnovic, M., Smolic, R., & Smolic, M. (2024, January 09). Hepatitis C Virus Vaccine. In Encyclopedia. https://encyclopedia.pub/entry/53609
Tabll, Ashraf A., et al. "Hepatitis C Virus Vaccine." Encyclopedia. Web. 09 January, 2024.
Copy Citation
Developing a safe and effective vaccine against the hepatitis C virus (HCV) remains a top priority for global health. Despite recent advances in antiviral therapies, the high cost and limited accessibility of these treatments impede their widespread application, particularly in resource-limited settings. Therefore, the development of the HCV vaccine remains a necessity.
hepatitis C virus
vaccine
SARS-CoV-2
COVID-19
1. Global and Egypt Prevalence of HCV
HCV is a global health problem, with an estimated 58 million people living with the disease, which accounts for about 1% of the world’s population [1]. In 2015, globally, there were 237 cases of HCV per 100,000 people and an estimated 1.75 million new infections of HCV. HCV has been classified into 8 genotypes and 93 subtypes (https://ictv.global/sg_wiki/flaviviridae/hepacivirus (accessed on 29 December 2023) [2]. HCV genotypes 1 (44%), 3 (25%), and 4 (25%) are the most common genotypes in the world. In 20–30 years, about 10–20% of people who have had HCV for a long time are likely to develop problems such as cirrhosis, end-stage liver disease, or hepatocellular carcinoma [3][4]. Only 20% of hepatitis C-infected patients are actually diagnosed, and 15% of those have been treated. To meet the WHO 2030 elimination goals, everyone should get affordable point-of-care diagnostics and pan-genotypic direct-acting antiviral therapy, as reported [1]. Egypt used to have the highest HCV infection rate. Two probability-based studies conducted in 2008 and 2015 suggested a decline in HCV antibody prevalence from 14.7% to 10%, respectively [3][5][6][7]. The most recent prevalence data were gathered from Egypt’s national initiative for HCV screening and treatment. From 2018 to 2019, Waked I [8] found an overall prevalence rate of 4.6% of HCV antibody-positive individuals and 3.5% of individuals with HCV viremia within the screened cohort (consisting of about 50 million Egyptians). This lowering trend in anti-HCV prevalence among Egyptian general populations may be linked to advancements in blood screening and infection control methods over the last two decades, as well as the national treatment campaign and its effectiveness as a preventive tool [9]. However, recent findings by Khattab et al. [10] revealed the persistence of HCV RNA in some HCV patients who achieved a sustained virologic response (SVR) after DAA treatment.
2. Structural and Non-Structural Protein Antigens as Targets for HCV Vaccine
The HCV is a spherical particle and consists of a single-stranded positive-sense RNA molecule enclosed within a capsid composed of multiple subunits of the core protein. The RNA genome and core protein assemble to form the nucleocapsid. Surrounding the nucleocapsid is an envelope composed of two viral glycoproteins, E1 and E2, embedded in a lipid bilayer derived from host membranes [11]. The HCV genome size is approximately 9.6 kb and contains a single open reading frame (ORF) encoding a single polyprotein, which is processed by cellular and viral proteases into structural (Core, E1, and E2) and non-structural proteins (P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (Figure 1). Given the current status of disease burden and available therapeutics, a prophylactic vaccine against HCV is urgently required. Globally, various therapeutic strategies are being used for HCV infections, such as DAAs, host-targeting agents (HTAs), micro-RNAs, nanomedicine, and immunotherapeutic approaches. In parallel, HCV vaccines’ candidates are being designed, developed, and experimentally evaluated using different strategies and platforms, including bioinformatics approaches targeting different regions of the viral genome. The continuous advancement of global research has provided significant and valuable insights into the structural and non-structural proteins of HCV to identify the main targets and epitopes for effective HCV vaccine design and development. While the Core, E1, and E2 proteins have been predominantly utilized as vaccine targets compared to the non-structural proteins, ongoing efforts aim to explore the full potential of these proteins in HCV vaccine design [12][13][14][15][16][17][18][19][20][21][22][23][24][25].
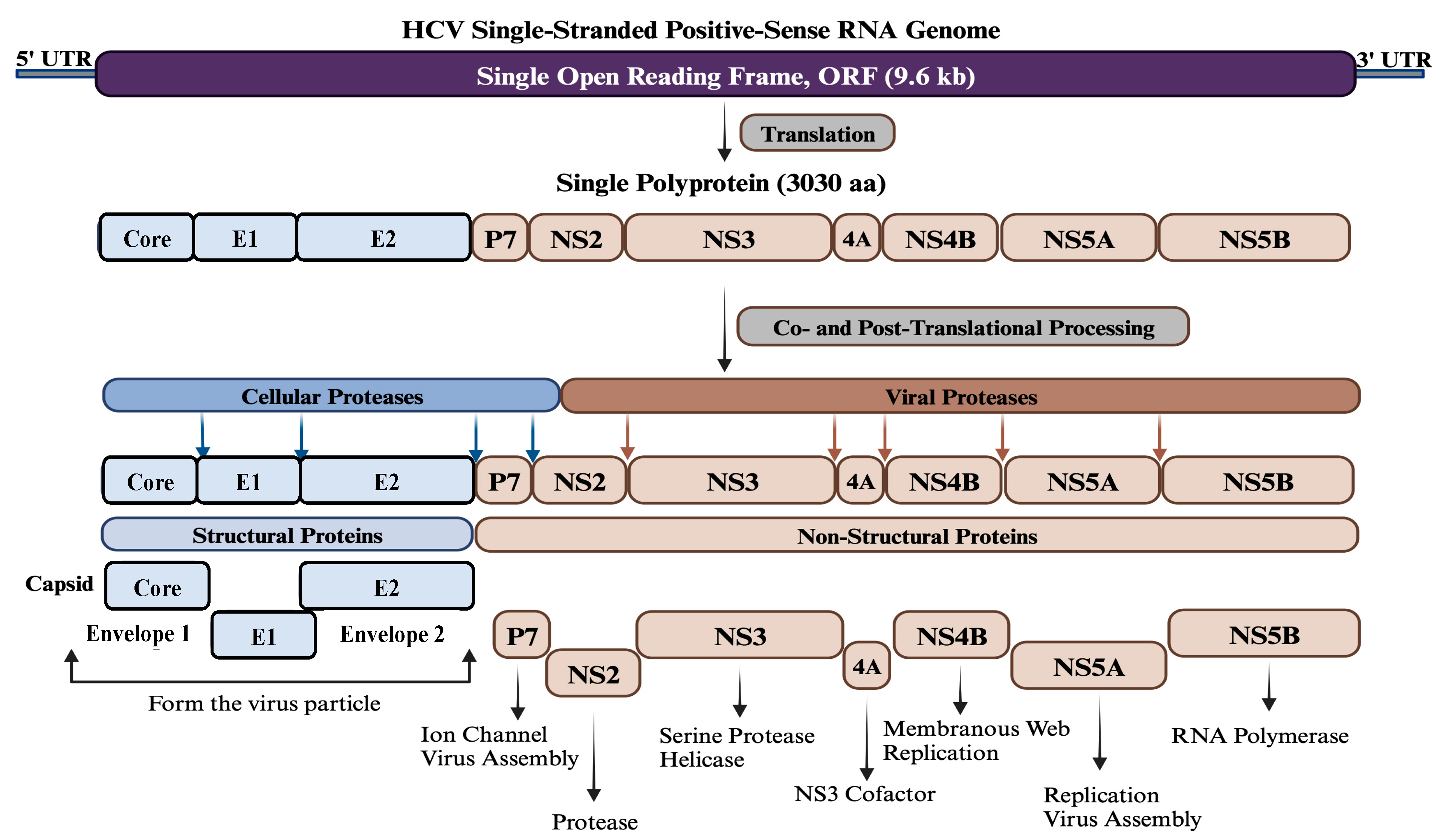
Figure 1. Genome organization of HCV. HCV contains a single-stranded positive-sense RNA genome of about 9.6 kb in size. The genome has a single open reading frame (ORF) flanked by two untranslated regions (UTR) at both ends. The ORF encodes a single polyprotein of approximately 3030 amino acids, which is processed co- and post-translationally by various cellular and viral proteases into three structural proteins (Core, E1, and E2) at the N-terminus and seven non-structural proteins (P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) at the C-terminus. Structural proteins oligomerize and self-assemble to form the viral particle, while non-structural proteins are involved in viral assembly and genome replication.
3. HCV Vaccines Targets and Technologies
Various strategies are being employed in the development of HCV vaccines to trigger immune responses. These strategies include the use of neutralizing antibodies (nAbs) that target exclusively the envelope E1/E2 proteins, the induction of T cell responses against the core protein or non-structural proteins (either individually or in combinations), the utilization of peptides (truncated isoforms or fragmented ones) and the development of multiepitope proteins using different technologies such as RNA interference and nanotechnology. Notably, the envelope glycoproteins (E1/E2) have been extensively utilized in HCV vaccine development. These envelope proteins are well-characterized proteins located on the surface of HCV virion and are known to be highly glycosylated transmembrane proteins (5 glycans on E1 and up to 11 on E2). The generation of neutralizing antibodies (nAbs) directed against E1, E2, and other viral proteins can confer clearance and protection against HCV infection [19][23][25][26][27].
3.1. Structural Proteins
The Envelope Glycoprotein 1 (E1)
The E1 protein has been extensively studied by various research groups and is identified as one of the most important targets for developing nAbs against HCV. While the size of the E1 protein is small, it exhibits better conservation compared to E2 and has been less characterized in terms of its crystal structure. Notably, E1 protein contains antigenic sites in the fragmented residues 192–271 and 314–324 on mature virion. Based on the recently characterized structure of the E1 protein, it has been reported that the E1 protein significantly contributes to the fusion step by using the putative fusion peptide (FP) and transmembrane domain (TMD) (Figure 2). The heterodimer structure of the full-length E1 protein has generated novel elements for the development of an HCV vaccine. The envelop glycoproteins E1 (aa192–383) are located at the N-terminus of the envelope-spanning region of the polyprotein (aa192–746). So far, only two immunogenic regions have been identified for neutralizing antibodies (nAbs) at the N terminus (aa192–207) and C terminus (aa313–328) of the E1 protein [14][22][28].
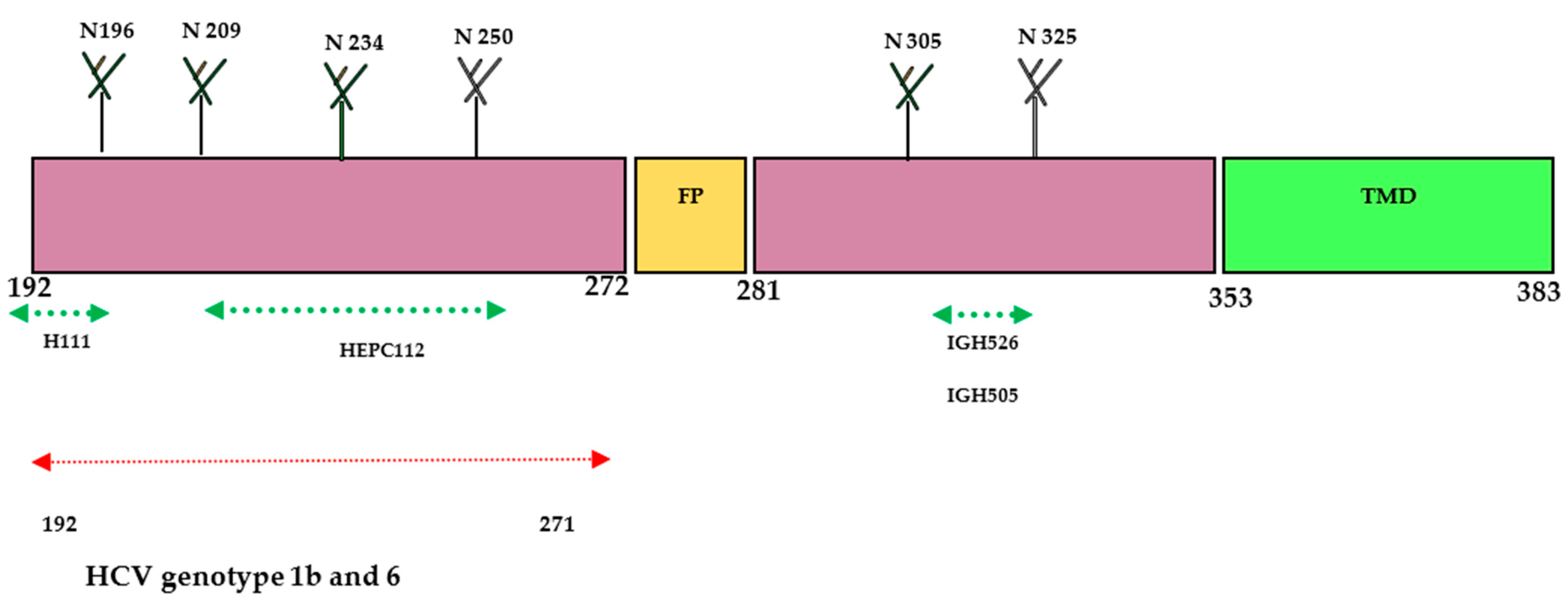
Figure 2. Schematic diagram of HCV- E1 envelope protein. E1 contains N-terminal domain (NTD, 192–383). The size of the E1 protein is 190 aa and is divided into ectodomain (160aa) and transmembrane domain (TMD) (30aa). Four N-glycosylation sites (N196, N209, N234, and N305) are conserved in all genotypes; N250 is found in genotypes 1b and 6, but N325 is absent when a proline residue is present at Asn-X-Ser/Thr. The crystal structure of the N-terminal domain of E1 considered individually was determined (PDB:4UOI), as well as the region 314–324 (PDB: 4N0Y) by co-crystallization with the human antibody IGH526. Antibody binding sites: aa 192–202 for the human monoclonal antibody (mAb) H-111, aa 215–299 for the human mAb HEPC112, and aa 313–324 for the human mAbs IGH505 and IGH526.
The E1 protein participates in the viral entry and fusion step during HCV infection through its interaction with cell membrane receptors known as claudin (CLDN)-1, CLDN-6, and CD36 [22][29]. In 2014, a research group successfully crystallized the N-terminal domain of the E1 protein from genotype 1 (H77 strain) under low-pH conditions [28]. Interestingly, the crystallized protein exhibited a smaller size and resembled phosphatidylcholine transfer protein rather than the fusion proteins of flavivirus. However, another research group reported conflicting findings, suggesting that the N-terminal domain of the E1 protein differed from the structure determined by the earlier group [28][30]. Their research confirmed that E2 protein is needed for the proper folding of E1 protein. Further investigations revealed that the E1 protein can form a trimeric structure through the GxxxG motif situated at its transmembrane domain, which facilitates the interaction between the E1 and E2 proteins [30]. A human monoclonal antibody (mAb) H-111 was generated against Gt 1b HCV, targeting the linear epitope within the amino acid range of 192–202. Although a high titer was observed, it displayed weak neutralizing activity against HCV pseudo particles. Additionally, two other mAbs (IGH505 and IGH526) were identified to target the region between aa 313 and 328. These antibodies demonstrated strong neutralization activity against HCVpp of genotypes 1a and 2a in cell culture. Another study conducted by a different research group showed that a broadly neutralizing antibody called HEPC112, developed against antigenic site (AS) 112 spanning amino acids 215–299 of the E1 protein, was able to neutralize seven strains of HCV [22][31]. These findings highlight the structural and immunological complexity of the E1 protein and underscore the potential of specific antibodies to target various epitopes within this protein. Further research and characterization of the E1 protein hold promise for advancing the understanding of HCV infection and facilitating the design of effective preventive strategies against this viral disease.
3.2. The Envelope Glycoprotein 2 (E2)
The E2 protein has emerged as a prominent target for the development of HCV-specific neutralizing antibodies (nAbs) compared with other proteins, [13][15][22]. The full-length structure of the E2 ectodomain was successfully resolved by several research groups, providing significant structural insights for the development of the HCV vaccine. The E2 protein consists of hypervariable regions (HVR1 and HVR2), an intergenotypic variable region (igVR), β-sandwich (aa492–566) flanked by a front (aa424–459) and a back layer (aa597–645), and a STEM and transmembrane domain (TMD) (Figure 3). The E2 protein contains three variable regions that facilitate viral escape from immune responses. The E2 (aa384–746) is located at the C-terminus of the envelope region of the HCV polyprotein (aa192–746).
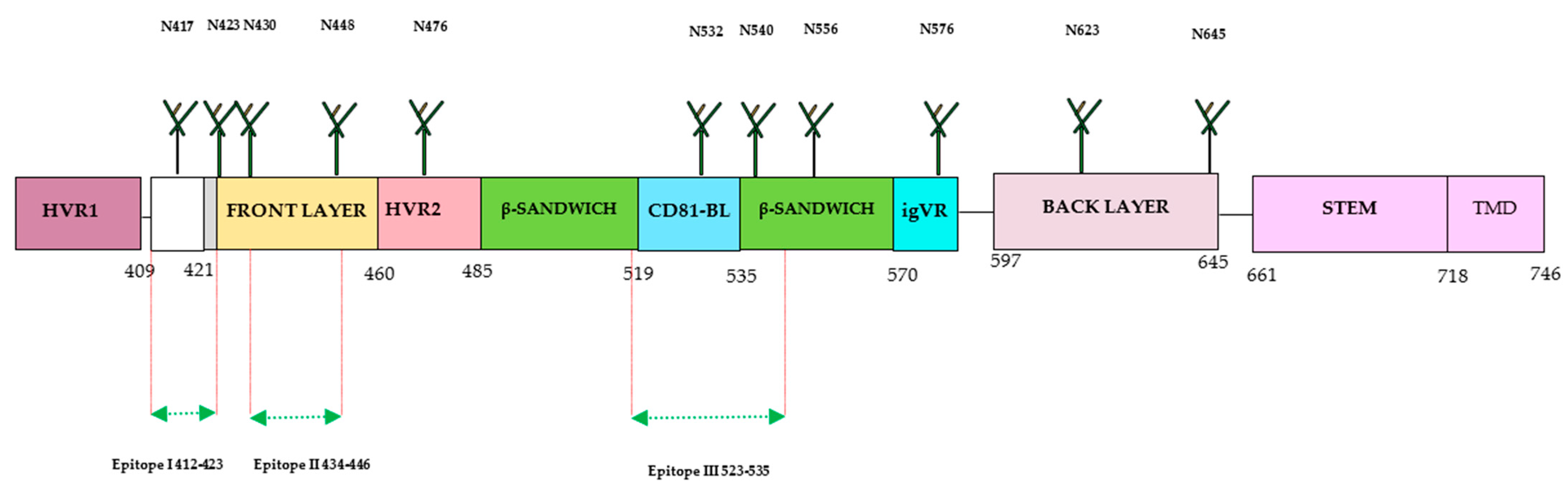
Figure 3. Structure of HCV E2 protein. The E2 consists of 360 aa, which is divided into 30 aa for the transmembrane domain (TMD), and consists of 3 variable regions (HVR1, HVR2, and igVR), including front layer, a back layer, CD81-binding loop (CD81bl), as well as STEM region, TMD regions start at 718 and end at 746, and 11 N-glycosylation sites. The epitope 1 (aa412–423) can adopt three conformations: beta hairpin, semi-open, and open. The epitope II (aa434–446) was co-crystallized with human monoclonal antibodies (mAbs) (H-84-27) and HC-84-1. The crystal structure of epitope III (523–535) was also obtained by co-crystallization with the mouse mAb DAO5 and targeted by the mAbs 1:7 and A8.
The entry of HCV virion into hepatocytes is mediated by the interaction of the E1/E2 heterodimer with host cell receptors, including cluster of differentiation 81 (CD81), scavenger receptor class B type I (SRB1), claudin 1 (CLDN1), and occludin (OCLN) [32]. The membrane fusion takes place in endosomes at low pH conditions that trigger conformational changes in the E1/E2 heterodimer located at residues 272–285, leading to the release of the genetic material to the cytoplasm. The first step of viral entry is facilitated by the interaction of the E2 HVR1 domain with the SRB1 receptor. This interaction induces conformational changes and exposes the E2 core region and CD81 binding loop. Clathrin-mediated endocytosis is achieved by the interaction of E2 and CD81 (Figure 4). The neutralization of HCV is usually mediated by the action of nAbs developed against the E2 protein. Many antibodies have been identified and developed against the linear and discontinuous regions of the E2 protein, which are known as Ars 1–5, Epitopes I-III, or domains A-E [13]. The crystal structure of the E2 ectodomain in complex with fragments of two different antibodies has been elucidated by two research groups, and the derived information has significantly advanced the understanding of protein structure. The ectodomain structure consists of a central immunoglobulin-like β-sandwich that is stable through conserved disulfide bonds. The N-terminal region consists of a β-strand and a short α-helix, while the C-terminal region consists of antiparallel β-sheets and short α-helices [33][34].
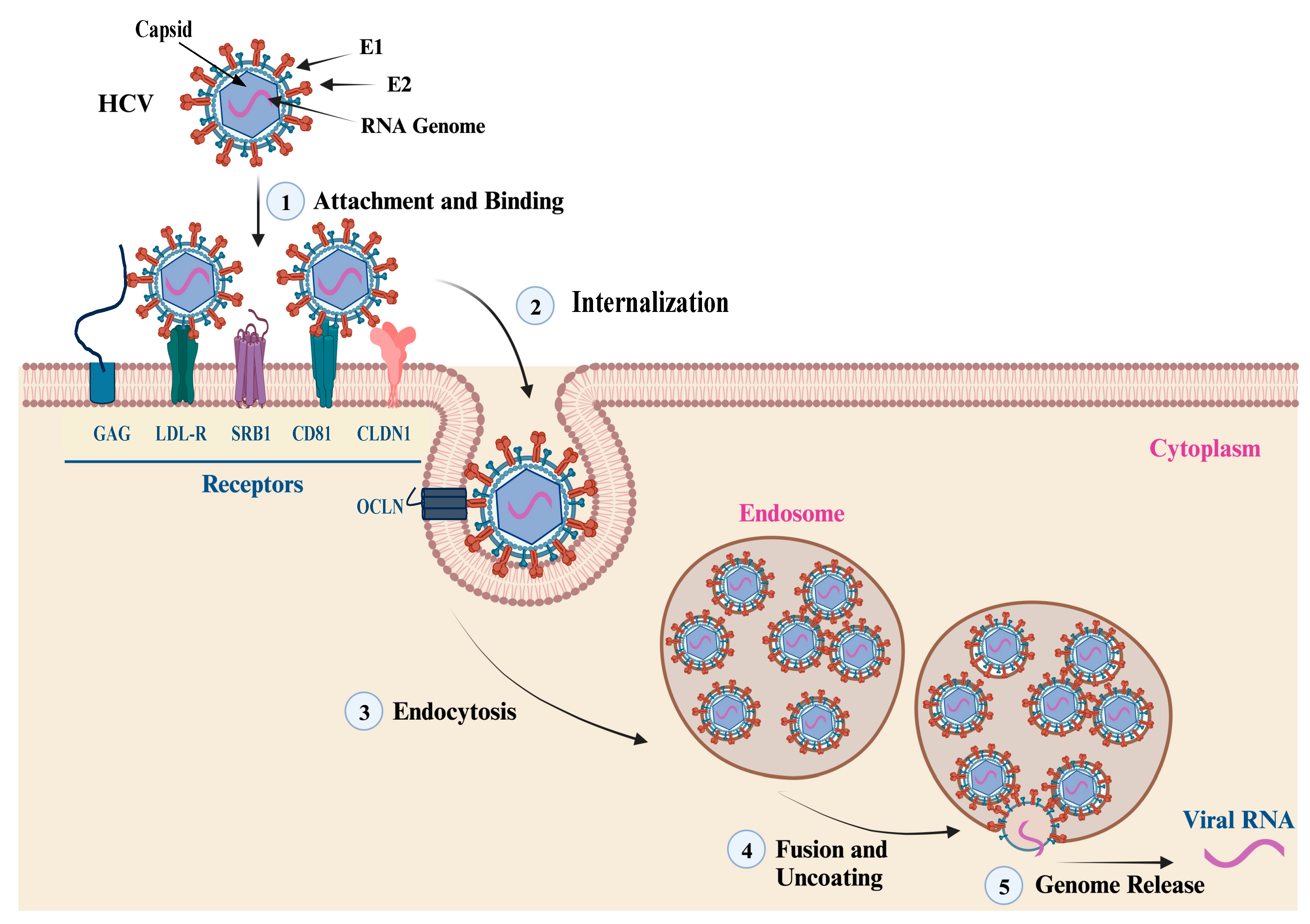
Figure 4. HCV Attachment and Cell Entry. This figure summarizes the main steps of HCV particle cell entry and genome release. (1, 2) The attachment, binding, and internalization are facilitated by the interaction of the viral envelope glycoproteins (E1 and E2) with cell membrane receptors, co-receptors, and host factors. The main cell receptors and factors that mediate the virus attachment and cell entry include glycosaminoglycan (GAG), low-density lipoprotein (LDL-R), scavenger receptor class B type I (SR-BI), cluster of differentiation 81 (CD81), claudin-1 (CLDN1), and occludin (OCLN). (3) Endocytosis: the interaction of viral envelop proteins with the cell receptors and co-receptors triggers the internalization of the virus into an endosome via endocytosis. (4, 5) Fusion, uncoating, and genome release: once inside the endosome, the virus undergoes fusion with the endosome membrane, leading to the uncoating of the nucleocapsid and the release of the viral genetic material into the cytoplasm.
The advancement in understanding the structure of the E2 protein has contributed to the identification of antigenic sites and domains crucial for the development of an HCV vaccine able to induce nAbs. The E2 ectodomain from various HCV genotypes has been characterized through complexing with different antibodies, including broadly neutralizing antibodies (bnAbs). The HCV E2 protein is a highly variable protein and widely used for developing bnAbs. Three hypervariable regions have been identified in the E2 protein: HVR1, aa384–409, HVR2, aa460–485 and the igVR, aa570–580. The HVR1 domain has an immunodominant epitope situated at the N-terminal. During virus infection, mutations in this region can lead to the escape of HCV variants from HCV-specific nAbs. HVR1 exhibits higher variability among the different HCV genotypes and subtypes. Other regions of E2 have less variability or are conserved across many genotypes, especially in areas important for the binding of cellular receptors (CD81) during HCV infection. This region has three conserved residues and encompasses three epitopes recognized by bnAb. These epitopes are referred to as Epitope I (aa412–423) situated at the N-terminal, Epitope II (aa428–446) located at the front layer, and Epitope III (aa518–542) situated at the binding loop of CD81. The effectiveness of antibodies against these epitopes has yielded varying results with different outcomes. The E2 protein exhibits three distinct conformations: the hairpin, open, and semi-open conformations. Monoclonal antibodies (mAb) have been developed against the hairpin conformation, including mouse mAbAP33 and mAb24 human mAb HCV1 [35]. The HCV1 and AP33 mAbs have shown broadly neutralizing activities in mice and chimpanzees. The contact residues of both E1 and E2 were shown to be necessary for antibody-mediated virus neutralization [13][20]. The full-length structure of the E1/E2 heterodimer Gt 1a was further investigated by another group. The group has discovered two additional regions in the E2 protein: the base (aa645–700) and the stem (aa 701–717), which are connected with the transmembrane domain by nine disulfide bonds [30]. Recently, Kumar et al. have characterized the interaction of E2 and CD81 receptors by molecular docking model and found that the region with residues 418–422 in E2 undergoes a displacement. Additionally, they also identified that AS412 in E2 is essential for the proper interaction with CD81 [31][35]. Another study has been conducted recently on the vaccine candidate using recombinant HCVcc with a deletion in HVR1. The study observed increased accessibility of nAbs (AR3A and AR4A) with reduced immunogenicity, suggesting the complex role of this region in immunogenicity [36][37]. Thus, during the design and development of the HCV vaccine using immunogens from different regions, it is crucial to consider the immunogenic parts and epitopes of broadly neutralizing antibodies (bNAbs) for higher elicitation of immunogenic responses [22].
3.3. Core Proteins
Various vaccine candidates have been designed against HCV by using different strategies and multiple combinations of proteins (structural and non-structural proteins). They have been evaluated for the stimulation of immunogenic responses, but only a few reports have explored the use of Core protein as an antigen for HCV vaccine development [17][38][39][40]. Some recent publications have reported that the vaccine based on a combination of core E1/E2 may induce a better immunogenic response than the virus-like particle assembly and chimeric protein of NS3 epitopes from three HCV genotypes [17][26][41]. Additionally, some HCV vaccines have been developed in transgenic plants. HCV Core protein was expressed in transgenic canola seed, and the extract from these seeds was mixed with an oil adjuvant for mouse vaccination. The vaccination has induced significant humoral (IgG) and Th1-biased immunogenic responses. The cytokine staining has shown that the immunization of mice with the total seed extract induced both CD4+ and CD8+ T cells to release IFN-γ [42]. Another recent study has shown that plasmid DNA carrying the HCV Core sequence was transfected into mesenchymal stem cells (MCS) before the injection of cells into Balb/c mice. The immunized mice demonstrated a superior humoral immunogenic response and produced core-specific antibodies. The transfected MCS-treated mice also exhibited a significant increase in splenocyte proliferation compared to the control group l [43].
3.4. Non-Structural Proteins
Some published papers from different research groups have discussed the use of non-structural proteins for HCV vaccine development, either alone or in combination. Among the HCV proteins, the NS3 and Core proteins have shown high conservation and contain many T cell determinants. Various technologies have been employed to study combinations of these proteins, either alone or in multi-tope formulations, aiming to induce Th1-biased immunogenic responses and clear HCV infection. However, the inclusion of an adjuvant in the vaccine formulation has also been shown to be necessary to elicit robust immune responses [19][44]. The heat shock protein (HSP) gp96 has widely been used as a natural adjuvant in viral vaccine formulations to induce innate and adaptive immune responses. In a recent study, the HCV Core-NS3 and Core-NS3-NT (gp96) proteins were expressed in cos-7 cells, and the purified proteins were used for mouse vaccination. A significantly high level of total IgG was observed in vaccinated mice as compared to the control group at weeks 3 and 11. The IFN-production level was high at weeks 3 and 11, but drastic variation was observed for the IL-4 level in the vaccinated and control groups at week 3. Based on the results obtained from different mouse groups, the long-term potency was observed by using the protein/protein + rNT (gp96) vaccine formulation [19]. In another study, it was demonstrated that the immunogenic epitope (1095–1379 aa) of the partial NS3 gene significantly induced the total antibodies and IgG2a, Interferon (IFN)-γ, and Interleukin (IL)-4 [45]. In NS4A and NS3 mice, immunization was performed, which resulted in higher expression levels of NS3 and NS3-specific antibodies and an IgG2a/IgG1 ratio (420 vs. 3) [46]. A DNA vaccine encoding HCV-3a NS3/NS4A was employed for the immunization of C57BL/6 mice, and a significant cell-mediated response was observed [46].
3.5. Use of Multi-Epitopes
Previous studies have reported that multi-epitope vaccines can elicit varying levels of immune responses. A plasmid DNA containing a combination of peptides and CD8 + T cell epitopes from the Core (132–142), NS3 (1073–1081), and NS5B (2727–2735), along with a ThCD4 + epitope from NS3 (1248–1262) and combined with a B-cell epitope from E2 (412–426) can trigger a higher immune response against HCV. The level of IFN-γ was significantly higher in peptide vaccine adjuvanted with Montanide ISA 720 after three doses of vaccinations as compared to two doses of plasmid and single doses of peptide vaccine. The triple doses of peptide vaccine induced a higher level of IFN-γ/IL-4 ratio as compared to other tested combinations [47]. Another study has indicated that the vaccination of BALB/c mice with a dose of 800 ng to 16 µg of multiple antigenic peptides made by combining conserved epitopes of HCV E1, E2, NS4B, NS5A, and NS5B has produced elevated immunogenic responses and caused an induction of IFN-γ-producing CD4+/CD8+ T-lymphocytes in the immunized mice [15][48].
3.6. Contributions of Monoclonal Antibodies to HCV Vaccine Development
Monoclonal antibodies (mAbs) play an important role in the development of HCV vaccines by aiding in understanding the viral proteins and immune responses during the viral infection. Their significant role in the development of the HCV vaccine can be attributed to various mechanisms, such as: (1) Identification of neutralizing antibodies: Monoclonal antibodies can be derived from individuals who have successfully eliminated the viral infection, enabling researchers to discern and analyze antibodies with the capacity to neutralize the virus. Neutralizing antibodies are valuable targets for the development of vaccines due to their ability to interfere with HCV-infecting liver cells [49]. (2) Epitope mapping: Monoclonal antibodies can locate precise epitopes on viral proteins that neutralizing antibodies specifically recognize. Researchers can identify critical areas of the virus that the immune system can attack by gaining insight regarding monoclonal antibody interactions with viral proteins. The acquisition of this information is crucial to developing vaccine candidates capable of inducing a comparable immune response [50][51]. (3) Vaccine design and evaluation: The efficacy of HCV vaccine candidates can be assessed through the utilization of monoclonal antibodies. Researchers can evaluate the capacity of vaccine-induced antibodies to neutralize various strains or variants of HCV. This procedure aids in determining the potency of the immune response the vaccine elicits, thereby guiding the selection of the most promising vaccine candidates for further development [16]. (4) Passive immunization involves the direct administration of purified antibodies, specifically monoclonal antibodies, to at-risk patients. This strategy offers prompt protection against the virus and can be employed in circumstances where a vaccine has not yet been developed or an insufficient immune reaction to vaccination is exhibited [52][53]. In conclusion, monoclonal antibodies have made a significant contribution to the progress of HCV vaccine development by helping in the identification of viral targets, facilitating the design, testing the vaccines, and opening up new ways to prevent and treat HCV infection.
4. General Advancement and Considerations in HCV Vaccine
The current status of vaccine development for HCV involves various initiatives and considerations that are being addressed [13][54]. The researchers want to specifically target and address comprehensive immune responses, with a particular focus on the production of antibodies that can efficiently kill the virus [55][56]. However, it is important for future research to explore more extensive immune responses, such as cellular immunity, to achieve long-lasting immunity and successfully eliminate the virus [57]. The use of novel vaccination platforms, such as mRNA and viral vector-based technologies, has promising promise for the development of a vaccine against HCV [58][59]. The effectiveness of these platforms in managing other viral infections has been well documented, indicating their potential suitability for the production of HCV vaccines. HCV has considerable genetic heterogeneity, posing a substantial challenge that calls for the investigation of customized vaccine approaches targeting certain HCV genotypes or subtypes prevalent in specific geographic regions [60][61]. Moreover, the incorporation of HCV vaccinations alongside other vaccines, such as those specifically targeting the hepatitis B virus, presents a promising approach toward a comprehensive preventive strategy against both hepatitis C and hepatitis B infections [62][63]. This technique has significant potential for patients who are at an elevated risk of getting both disorders concurrently. The present initiatives are focused on advancing the development of effective HCV vaccines and addressing the complexities associated with this infectious disease [64][65]. Future research and collaboration in the development of a vaccine for HCV should prioritize many crucial proposals [66]. Before using antibody-based strategies, it is important to take a close look at and learn more about larger immune responses, such as cellular immunity [14][67]. This has the potential to result in enhanced and enduring protection against HCV. Also, more research needs to be done on new platforms for vaccination, like mRNA and viral vector-based technologies, so that their full potential can be used and they can be changed to help make an HCV vaccine [67][68]. The acceleration of translating these technologies into viable vaccines necessitates collaborative endeavors among researchers, industry stakeholders, and regulatory agencies [69][70][71]. Furthermore, it is imperative to explore individualized vaccine strategies that are specifically designed to target distinct HCV genotypes or subtypes that are widespread in various geographical regions [72][73][74].
References
- Roudot-Thoraval, F. Epidemiology of hepatitis C virus infection. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101596.
- Smith, D.B.; Bukh, J.; Kuiken, C.; Muerhoff, A.S.; Rice, C.M.; Stapleton, J.T.; Simmonds, P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology 2014, 59, 318–327.
- El-Zanaty, F.; Way, A. Egypt Demographic and Health Survey 2008; Ministry of Health: Cairo, Egypt, 2009.
- Hassanin, A.; Kamel, S.; Waked, I.; Fort, M. Egypt’s Ambitious Strategy to Eliminate Hepatitis C Virus: A Case Study. Glob. Health Sci. Pract. 2021, 9, 187–200.
- Hatzakis, A.; Lazarus, J.V.; Cholongitas, E.; Baptista-Leite, R.; Boucher, C.; Busoi, C.S.; Deuffic-Burban, S.; Chhatwal, J.; Esmat, G.; Hutchinson, S.; et al. Securing sustainable funding for viral hepatitis elimination plans. Liver Int. 2020, 40, 260–270.
- Elrashdy, F.; Hagag, S.; Mohamed, R.; Alem, S.A.; Meshaal, S.; Cordie, A.; Elsharkawy, A.; Esmat, G. Incidence of hepatitis C virus infection among people living with HIV: An Egyptian cohort study. South. Afr. J. HIV Med. 2022, 23, 1–5.
- Omran, D.; Alboraie, M.; Zayed, R.A.; Wifi, M.N.; Naguib, M.; Eltabbakh, M.; Abdellah, M.; Sherief, A.F.; Maklad, S.; Eldemellawy, H.H.; et al. Towards hepatitis C virus elimination: Egyptian experience, achievements and limitations. World J. Gastroenterol. 2018, 24, 4330–4340.
- Waked, I. Case study of hepatitis C virus control in Egypt: Impact of access program. Antivir. Ther. 2022, 27, 13596535211067592.
- Ayoub, H.H.; Abu-Raddad, L.J. Impact of treatment on hepatitis C virus transmission and incidence in Egypt: A case for treatment as prevention. J. Viral Hepat. 2017, 24, 486–495.
- Khattab, M.A.; Zakaria, Y.; Sadek, E.; Abd El Fatah, A.S.; Fouad, M.; Khattab, M.; Moness, H.M.; Adel, N.M.; Ahmed, E. Detection of hepatitis C virus (HCV) RNA in the peripheral blood mononuclear cells of HCV-infected patients following sustained virologic response. Clin. Exp. Med. 2023, 23, 131–140.
- Kaito, M.; Watanabe, S.; Tanaka, H.; Fujita, N.; Konishi, M.; Iwasa, M.; Kobayashi, Y.; Gabazza, E.C.; Adachi, Y.; Tsukiyama-Kohara, K.; et al. Morphological identification of hepatitis C virus E1 and E2 envelope glycoproteins on the virion surface using immunogold electron microscopy. Int. J. Mol. Med. 2006, 18, 673–678.
- Ikram, A.; Zaheer, T.; Awan, F.M.; Obaid, A.; Naz, A.; Hanif, R.; Paracha, R.Z.; Ali, A.; Naveed, A.K.; Janjua, H.A. Exploring NS3/4A, NS5A and NS5B proteins to design conserved subunit multi-epitope vaccine against HCV utilizing immunoinformatics approaches. Sci. Rep. 2018, 8, 16107.
- Duncan, J.D.; Urbanowicz, R.A.; Tarr, A.W.; Ball, J.K. Hepatitis C Virus Vaccine: Challenges and Prospects. Vaccines 2020, 8, 90.
- Sepulveda-Crespo, D.; Resino, S.; Martinez, I. Hepatitis C virus vaccine design: Focus on the humoral immune response. J. Biomed. Sci. 2020, 27, 78.
- Shayeghpour, A.; Kianfar, R.; Hosseini, P.; Ajorloo, M.; Aghajanian, S.; Hedayat Yaghoobi, M.; Hashempour, T.; Mozhgani, S.H. Hepatitis C virus DNA vaccines: A systematic review. Virol. J. 2021, 18, 248.
- Hartlage, A.S.; Kapoor, A. Hepatitis C Virus Vaccine Research: Time to Put Up or Shut Up. Viruses 2021, 13, 1596.
- Guest, J.D.; Pierce, B.G. Structure-Based and Rational Design of a Hepatitis C Virus Vaccine. Viruses 2021, 13, 837.
- Yechezkel, I.; Law, M.; Tzarum, N. From Structural Studies to HCV Vaccine Design. Viruses 2021, 13, 833.
- Hajikhezri, Z.; Roohvand, F.; Maleki, M.; Shahmahmoodi, S.; Amirzargar, A.A.; Keshavarz, A.; Seyed, N.; Farahmand, M.; Samimi-Rad, K. HCV Core/NS3 Protein Immunization with “N-Terminal Heat Shock gp96 Protein (rNT (gp96))” Induced Strong and Sustained Th1-Type Cytokines in Immunized Mice. Vaccines 2021, 9, 215.
- Behmard, E.; Abdulabbas, H.T.; Abdalkareem Jasim, S.; Najafipour, S.; Ghasemian, A.; Farjadfar, A.; Barzegari, E.; Kouhpayeh, A.; Abdolmaleki, P. Design of a novel multi-epitope vaccine candidate against hepatitis C virus using structural and nonstructural proteins: An immunoinformatics approach. PLoS ONE 2022, 17, e0272582.
- Zhao, Q.; He, K.; Zhang, X.; Xu, M.; Zhang, X.; Li, H. Production and immunogenicity of different prophylactic vaccines for hepatitis C virus (Review). Exp. Ther. Med. 2022, 24, 474.
- Gomez-Escobar, E.; Roingeard, P.; Beaumont, E. Current Hepatitis C Vaccine Candidates Based on the Induction of Neutralizing Antibodies. Viruses 2023, 15, 1151.
- Bernal, L.A.; Soti, V. Hepatitis C Virus: Insights Into Its History, Treatment, Challenges, and Future Directions. Cureus 2023, 15, e43924.
- Toth, E.A.; Andrianov, A.K.; Fuerst, T.R. Prospects for developing an Hepatitis C virus E1E2-based nanoparticle vaccine. Rev. Med. Virol. 2023, 33, e2474.
- Adugna, A. Therapeutic strategies and promising vaccine for hepatitis C virus infection. Immun. Inflamm. Dis. 2023, 11, e977.
- Olivera, S.; Perez, A.; Falcon, V.; Urquiza, D.; Pichardo, D.; Martinez-Donato, G. Protective cellular immune response against hepatitis C virus elicited by chimeric protein formulations in BALB/c mice. Arch. Virol. 2020, 165, 593–607.
- Donnison, T.; von Delft, A.; Brown, A.; Swadling, L.; Hutchings, C.; Hanke, T.; Chinnakannan, S.; Barnes, E. Viral vectored hepatitis C virus vaccines generate pan-genotypic T cell responses to conserved subdominant epitopes. Vaccine 2020, 38, 5036–5048.
- El Omari, K.; Iourin, O.; Kadlec, J.; Sutton, G.; Harlos, K.; Grimes, J.M.; Stuart, D.I. Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat. Commun. 2014, 5, 4874.
- Douam, F.; Thi, V.L.D.; Maurin, G.; Fresquet, J.; Mompelat, D.; Zeisel, M.B.; Baumert, T.F.; Cosset, F.L.; Lavillette, D. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology 2014, 59, 776–788.
- Torrents de la Peña, A.; Sliepen, K.; Eshun-Wilson, L.; Newby, M.L.; Allen, J.D.; Zon, I.; Koekkoek, S.; Chumbe, A.; Crispin, M.; Schinkel, J.; et al. Structure of the hepatitis C virus E1E2 glycoprotein complex. Science 2022, 378, 263–269.
- Colbert, M.D.; Flyak, A.I.; Ogega, C.O.; Kinchen, V.J.; Massaccesi, G.; Hernandez, M.; Davidson, E.; Doranz, B.J.; Cox, A.L.; Crowe, J.E., Jr.; et al. Broadly Neutralizing Antibodies Targeting New Sites of Vulnerability in Hepatitis C Virus E1E2. J. Virol. 2019, 93, 10–1128.
- Kumar, A.; Rohe, T.C.; Elrod, E.J.; Khan, A.G.; Dearborn, A.D.; Kissinger, R.; Grakoui, A.; Marcotrigiano, J. Regions of hepatitis C virus E2 required for membrane association. Nat. Commun. 2023, 14, 433.
- Kong, L.; Giang, E.; Nieusma, T.; Kadam, R.U.; Cogburn, K.E.; Hua, Y.; Dai, X.; Stanfield, R.L.; Burton, D.R.; Ward, A.B.; et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science 2013, 342, 1090–1094.
- Khan, A.G.; Whidby, J.; Miller, M.T.; Scarborough, H.; Zatorski, A.V.; Cygan, A.; Price, A.A.; Yost, S.A.; Bohannon, C.D.; Jacob, J.; et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 2014, 509, 381–384.
- Alhammad, Y.; Gu, J.; Boo, I.; Harrison, D.; McCaffrey, K.; Vietheer, P.T.; Edwards, S.; Quinn, C.; Coulibaly, F.; Poumbourios, P.; et al. Monoclonal Antibodies Directed toward the Hepatitis C Virus Glycoprotein E2 Detect Antigenic Differences Modulated by the N-Terminal Hypervariable Region 1 (HVR1), HVR2, and Intergenotypic Variable Region. J. Virol. 2015, 89, 12245–12261.
- Alzua, G.P.; Pihl, A.F.; Offersgaard, A.; Duarte Hernandez, C.R.; Duan, Z.; Feng, S.; Fahnøe, U.; Sølund, C.; Weis, N.; Law, M.; et al. Inactivated genotype 1a, 2a and 3a HCV vaccine candidates induced broadly neutralising antibodies in mice. Gut 2023, 72, 560–572.
- Pihl, A.F.; Feng, S.; Offersgaard, A.; Alzua, G.P.; Augestad, E.H.; Mathiesen, C.K.; Jensen, T.B.; Krarup, H.; Law, M.; Prentoe, J.; et al. Inactivated whole hepatitis C virus vaccine employing a licensed adjuvant elicits cross-genotype neutralizing antibodies in mice. J. Hepatol. 2022, 76, 1051–1061.
- Roohvand, F.; Aghasadeghi, M.R.; Sadat, S.M.; Budkowska, A.; Khabiri, A.R. HCV core protein immunization with Montanide/CpG elicits strong Th1/Th2 and long-lived CTL responses. Biochem. Biophys. Res. Commun. 2007, 354, 641–649.
- Drane, D.; Maraskovsky, E.; Gibson, R.; Mitchell, S.; Barnden, M.; Moskwa, A.; Shaw, D.; Gervase, B.; Coates, S.; Houghton, M.; et al. Priming of CD4+ and CD8+ T cell responses using a HCV core ISCOMATRIX vaccine: A phase I study in healthy volunteers. Hum. Vaccines 2009, 5, 151–157.
- Gawlik, K.; Gallay, P.A. HCV core protein and virus assembly: What we know without structures. Immunol. Res. 2014, 60, 1–10.
- Christiansen, D.; Earnest-Silveira, L.; Grubor-Bauk, B.; Wijesundara, D.K.; Boo, I.; Ramsland, P.A.; Vincan, E.; Drummer, H.E.; Gowans, E.J.; Torresi, J. Pre-clinical evaluation of a quadrivalent HCV VLP vaccine in pigs following microneedle delivery. Sci. Rep. 2019, 9, 9251.
- Mohammadzadeh, S.; Roohvand, F.; Ehsani, P.; Salmanian, A.H.; Ajdary, S. Canola oilseed- and Escherichia coli- derived hepatitis C virus (HCV) core proteins adjuvanted with oil bodies, induced robust Th1-oriented immune responses in immunized mice. J. Pathol. Microbiol. Immunol. APMIS 2020, 128, 593–602.
- Masoudi, M.R.; Rafati, A. Immunogenicity against hepatitis C virus with mesenchymal stem cells of inbreed BALB/c mice sub cloned with HCVcp protein gene. Transpl. Immunol. 2022, 74, 101651.
- Bastola, R.; Noh, G.; Keum, T.; Bashyal, S.; Seo, J.E.; Choi, J.; Oh, Y.; Cho, Y.; Lee, S. Vaccine adjuvants: Smart components to boost the immune system. Arch. Pharmacal Res. 2017, 40, 1238–1248.
- Pouriayevali, M.H.; Bamdad, T.; Aghasadeghi, M.R.; Sadat, S.M.; Sabahi, F. Construction and Immunogenicity Analysis of Hepatitis C Virus (HCV) Truncated Non-Structural Protein 3 (NS3) Plasmid Vaccine. Jundishapur J. Microbiol. 2016, 9, e33909.
- Behzadi, M.A.; Alborzi, A.; Kalani, M.; Pouladfar, G.; Dianatpour, M.; Ziyaeyan, M. Immunization with a Recombinant Expression Vector Encoding NS3/NS4A of Hepatitis C Virus Genotype 3a Elicits Cell-Mediated Immune Responses in C57BL/6 Mice. Viral Immunol. 2016, 29, 138–147.
- Pishraft Sabet, L.; Taheri, T.; Memarnejadian, A.; Mokhtari Azad, T.; Asgari, F.; Rahimnia, R.; Alavian, S.M.; Rafati, S.; Samimi Rad, K. Immunogenicity of Multi-Epitope DNA and Peptide Vaccine Candidates Based on Core, E2, NS3 and NS5B HCV Epitopes in BALB/c Mice. Hepat. Mon. 2014, 14, e22215.
- Dawood, R.M.; Moustafa, R.I.; Abdelhafez, T.H.; El-Shenawy, R.; El-Abd, Y.; Bader El Din, N.G.; Dubuisson, J.; El Awady, M.K. A multiepitope peptide vaccine against HCV stimulates neutralizing humoral and persistent cellular responses in mice. BMC Infect. Dis. 2019, 19, 932.
- Tong, Y.; Li, Q.; Li, R.; Xu, Y.; Pan, Y.; Niu, J.; Zhong, J. A Novel Approach To Display Structural Proteins of Hepatitis C Virus Quasispecies in Patients Reveals a Key Role of E2 HVR1 in Viral Evolution. J. Virol. 2020, 94, e00622-20.
- Brasher, N.A.; Adhikari, A.; Lloyd, A.R.; Tedla, N.; Bull, R.A. Hepatitis C Virus Epitope Immunodominance and B Cell Repertoire Diversity. Viruses 2021, 13, 983.
- Quadeer, A.A.; Louie, R.H.Y.; McKay, M.R. Identifying immunologically-vulnerable regions of the HCV E2 glycoprotein and broadly neutralizing antibodies that target them. Nat. Commun. 2019, 10, 2073.
- Tabll, A.; Abbas, A.T.; El-Kafrawy, S.; Wahid, A. Monoclonal antibodies: Principles and applications of immmunodiagnosis and immunotherapy for hepatitis C virus. World J. Hepatol. 2015, 7, 2369–2383.
- Vlatkovic, I.; Ludwig, J.; Boros, G.; Szabó, G.T.; Reichert, J.; Buff, M.; Baiersdörfer, M.; Reinholz, J.; Mahiny, A.J.; Şahin, U.; et al. Ribozyme Assays to Quantify the Capping Efficiency of In Vitro-Transcribed mRNA. Pharmaceutics 2022, 14, 328.
- Feld, J.J.; Bruneau, J.; Dore, G.J.; Ghany, M.G.; Hansen, B.; Sulkowski, M.; Thomas, D.L. Controlled human infection model for hepatitis C virus vaccine development: Trial design considerations. Clin. Infect. Dis. 2023, 77 (Suppl. S3), S262–S269.
- Obaid, A.; Naz, A.; Ikram, A.; Awan, F.M.; Raza, A.; Ahmad, J.; Ali, A. Model of the adaptive immune response system against HCV infection reveals potential immunomodulatory agents for combination therapy. Sci. Rep. 2018, 8, 8874.
- Wang, X.Y.; Wang, B.; Wen, Y.M. From therapeutic antibodies to immune complex vaccines. NPJ Vaccines 2019, 4, 2.
- Kemming, J.; Thimme, R.; Neumann-Haefelin, C. Adaptive Immune Response against Hepatitis C Virus. Int. J. Mol. Sci. 2020, 21, 5644.
- Matić, Z.; Šantak, M. Biotechnology, Current view on novel vaccine technologies to combat human infectious diseases. Appl. Microbiol. Biotechnol. 2022, 106, 25–56.
- Vargová, S. Current Approaches in the Development of Vaccines against Infectious Viral Diseases. Bachelor’s Thesis, Charles University—Faculty of Science, Prague, Czech Republic, 2022.
- Guo, X.; Zhong, J.Y.; Li, J.W. Hepatitis C virus infection and vaccine development. J. Clin. Exp. Hepatol. 2018, 8, 195–204.
- Mohamed, Z. Assessing and Improving the Hepatitis C Virus Cascade of Care for Marginalised Populations. Ph.D. Thesis, Imperial College London, London, UK, 2022.
- Cox, A.L. Challenges and promise of a hepatitis C virus vaccine. Cold Spring Harb. Perspect. Med. 2020, 10, a036947.
- Locarnini, S.; Hatzakis, A.; Chen, D.S.; Lok, A. Strategies to control hepatitis B: Public policy, epidemiology, vaccine and drugs. J. Hepatol. 2015, 62, S76–S86.
- Fries, C.N.; Curvino, E.J.; Chen, J.L.; Permar, S.R.; Fouda, G.G.; Collier, J.H. Advances in nanomaterial vaccine strategies to address infectious diseases impacting global health. Nat. Nanotechnol. 2021, 16, 1–14.
- Duraisamy, G.S.; Bhosale, D.; Lipenská, I.; Huvarova, I.; Růžek, D.; Windisch, M.P.; Miller, A.D. Advanced therapeutics, vaccinations, and precision medicine in the treatment and management of chronic hepatitis b viral infections: Where are we and where are we going? Viruses 2020, 12, 998.
- Wedemeyer, H.; Tergast, T.L.; Lazarus, J.V.; Razavi, H.; Bakoyannis, K.; Baptista-Leite, R.; Bartoli, M.; Bruggmann, P.; Buşoi, C.S.; Buti, M.; et al. Securing wider EU commitment to the elimination of hepatitis C virus. Liver Int. 2023, 43, 276–291.
- Shoukry, N.H. Hepatitis C vaccines, antibodies, and T cells. Liver Int. 2018, 9, 1480.
- Echeverría, N.; Comas, V.; Aldunate, F.; Perbolianachis, P.; Moreno, P.; Cristina, J. In the era of rapid mRNA-based vaccines: Why is there no effective hepatitis C virus vaccine yet? World J. Hepatol. 2021, 13, 1234–1268.
- Travieso, T.; Li, J.; Mahesh, S.; Mello, J.; Blasi, M. The use of viral vectors in vaccine development. NPJ Vaccines 2022, 7, 75.
- Stevens, H.; Debackere, K.; Goldman, M.; Mahoney, R.T.; Stevens, P.; Huys, I. Vaccines: Accelerating Innovation and Access; World Intellectual Property Organization: Geneva, Switzerland, 2017.
- Hayman, B.; Suri, R.K.; Downham, M. Sustainable vaccine manufacturing in Low-and middle-income countries. Vaccine 2022, 40, 7288–7304.
- Halliday, J.; Klenerman, P.; Barnes, E. Vaccination for hepatitis C virus: Closing in on an evasive target. Expert Rev. Vaccines 2011, 10, 659–672.
- Kwok, A.J.; Mentzer, A.; Knight, J.C. Host genetics and infectious disease: New tools, insights and translational opportunities. Nat. Rev. Genet. 2021, 22, 137–153.
- Bukh, J. The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J. Hepatol. 2016, 65, S2–S21.
More
Information
Subjects:
Virology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
563
Revisions:
2 times
(View History)
Update Date:
10 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




