Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tilman Lamparter | -- | 3165 | 2024-01-09 11:02:58 | | | |
| 2 | Peter Tang | Meta information modification | 3165 | 2024-01-10 03:45:41 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kaeser, G.; Krauß, N.; Roughan, C.; Sauthof, L.; Scheerer, P.; Lamparter, T. Phytochrome-Interacting Proteins. Encyclopedia. Available online: https://encyclopedia.pub/entry/53600 (accessed on 28 February 2026).
Kaeser G, Krauß N, Roughan C, Sauthof L, Scheerer P, Lamparter T. Phytochrome-Interacting Proteins. Encyclopedia. Available at: https://encyclopedia.pub/entry/53600. Accessed February 28, 2026.
Kaeser, Gero, Norbert Krauß, Clare Roughan, Luisa Sauthof, Patrick Scheerer, Tilman Lamparter. "Phytochrome-Interacting Proteins" Encyclopedia, https://encyclopedia.pub/entry/53600 (accessed February 28, 2026).
Kaeser, G., Krauß, N., Roughan, C., Sauthof, L., Scheerer, P., & Lamparter, T. (2024, January 09). Phytochrome-Interacting Proteins. In Encyclopedia. https://encyclopedia.pub/entry/53600
Kaeser, Gero, et al. "Phytochrome-Interacting Proteins." Encyclopedia. Web. 09 January, 2024.
Copy Citation
Phytochromes are photoreceptors of plants, fungi, slime molds bacteria and heterokonts. These biliproteins sense red and far-red light and undergo light-induced changes between the two spectral forms, Pr and Pfr. Photoconversion triggered by light induces conformational changes in the bilin chromophore around the ring C-D-connecting methine bridge and is followed by conformational changes in the protein. For plant phytochromes, multiple phytochrome interacting proteins that mediate signal transduction, nuclear translocation or protein degradation have been identified. Few interacting proteins are known as bacterial or fungal phytochromes.
PIF3
PKS2
Cry
plant
bacterial
fungal
interaction
1. Introduction
Phytochromes are photoreceptor proteins that regulate multiple light effects in plants, bacteria, fungi and heterokonts [1][2][3][4]. The protein binds a bilin chromophore covalently to a cysteine residue. As in any photoreceptor, the chromophore absorbs light and this triggers protein conformational changes that initiate a signal transduction cascade. Phytochromes have a special feature, photoreversibility, which describes the switching between two spectrally different forms, Pr and Pfr, by light. This conversion is triggered by an isomerization of the chromophore. Most members of the large group of phytochromes are synthesized in the Pr form, while Pfr can only be formed by light. To achieve conversion from Pfr to Pr, there are two possibilities, either via photoconversion or via dark reversion. Few phytochromes are comparable to this group but have a stable Pfr, i.e., the conversion from Pr to Pfr is only possible by light. A third group, the so-called bathy phytochromes, have a Pfr dark form [5][6]. They are also synthesized in the Pr form, but by dark conversion, Pr converts to Pfr. Pr can then only be formed by light. Three different bilins, phytochromobilin, phycocyanobilin and biliverdin, can be selected by phytochromes (Figure 1). The choice depends on the species, i.e., which chromophore is synthesized, and on the position of the chromophore binding cysteine [7]. There are also few examples of other chromophores with still different spectral properties [8], and the group of cyanobacteriochromes [9], biliproteins that probably evolved out of phytochromes and are absorbed in various different spectral ranges.
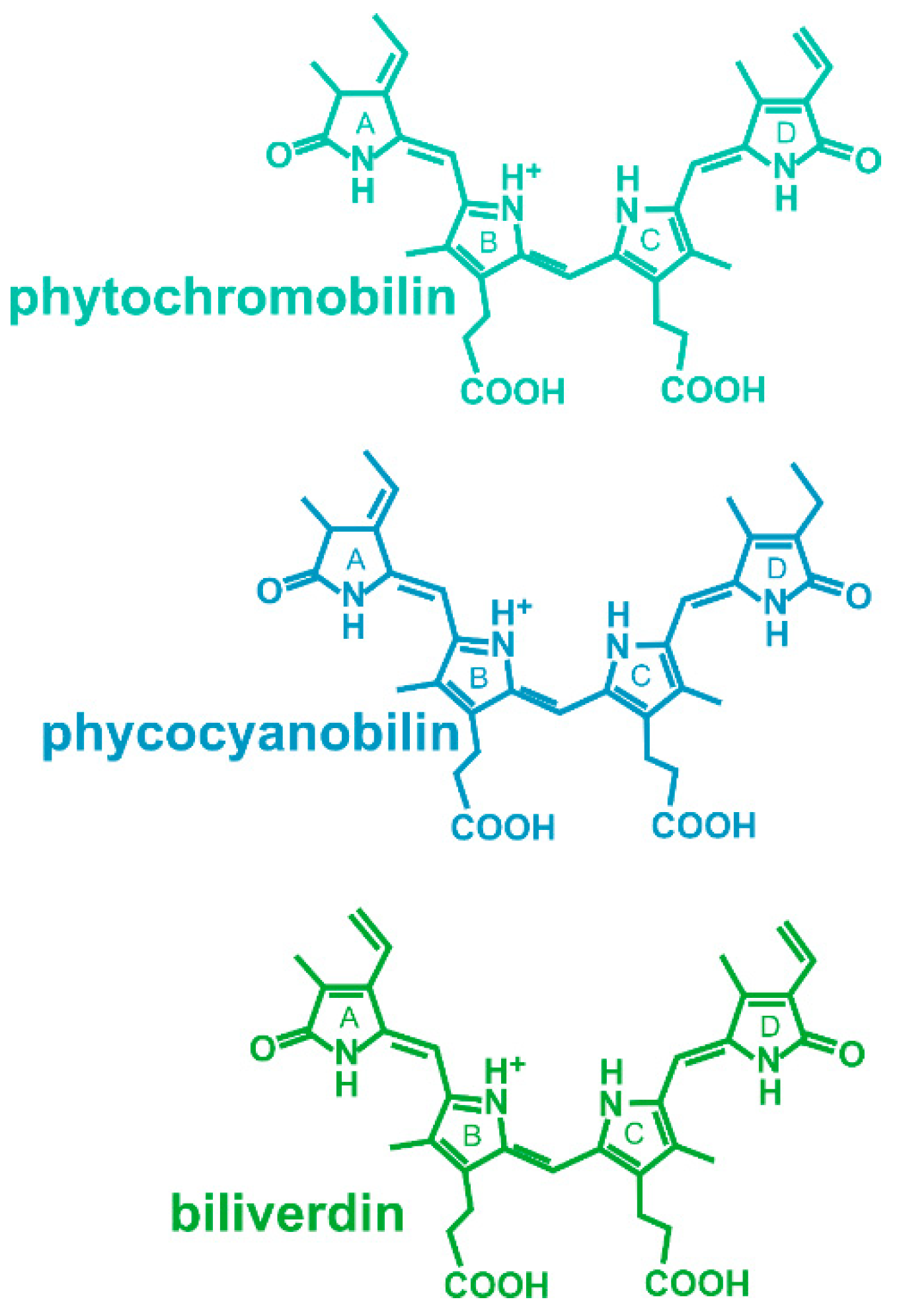
Figure 1. Chromophores of phytochromes. Phytochromobilin is the chromophore of plant phytochromes, phycocyanobilin is the chromophore of some cyanobacterial and green algal phytochromes and of cyanobacteriochromes, and biliverdin is the chromophore of bacterial and fungal phytochromes.
Phytochromes are multidomain proteins. The canonical phytochromes have a PAS (period, Arndt, single-minded)-GAF (cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA)-PHY (phytochrome) tridomain in the N-terminus of the protein (Figure 2). The GAF domain gives rise to the cyanobacteriochromes, in which GAF is combined with different other domains. Cyanobacterial proteins, in which GAF and PHY are present but a PAS domain is missing, are termed knotless phytochromes [8]. The focus here is on the canonical phytochromes. In the C-terminus, most phytochromes have a histidine kinase or a histidine kinase-like domain. Plant phytochromes are comparable with the typical bacterial phytochromes but carry two additional PAS domains between the PHY domain and the histidine kinase-like module (Figure 2). However, plant phytochromes lose their histidine kinase activity.
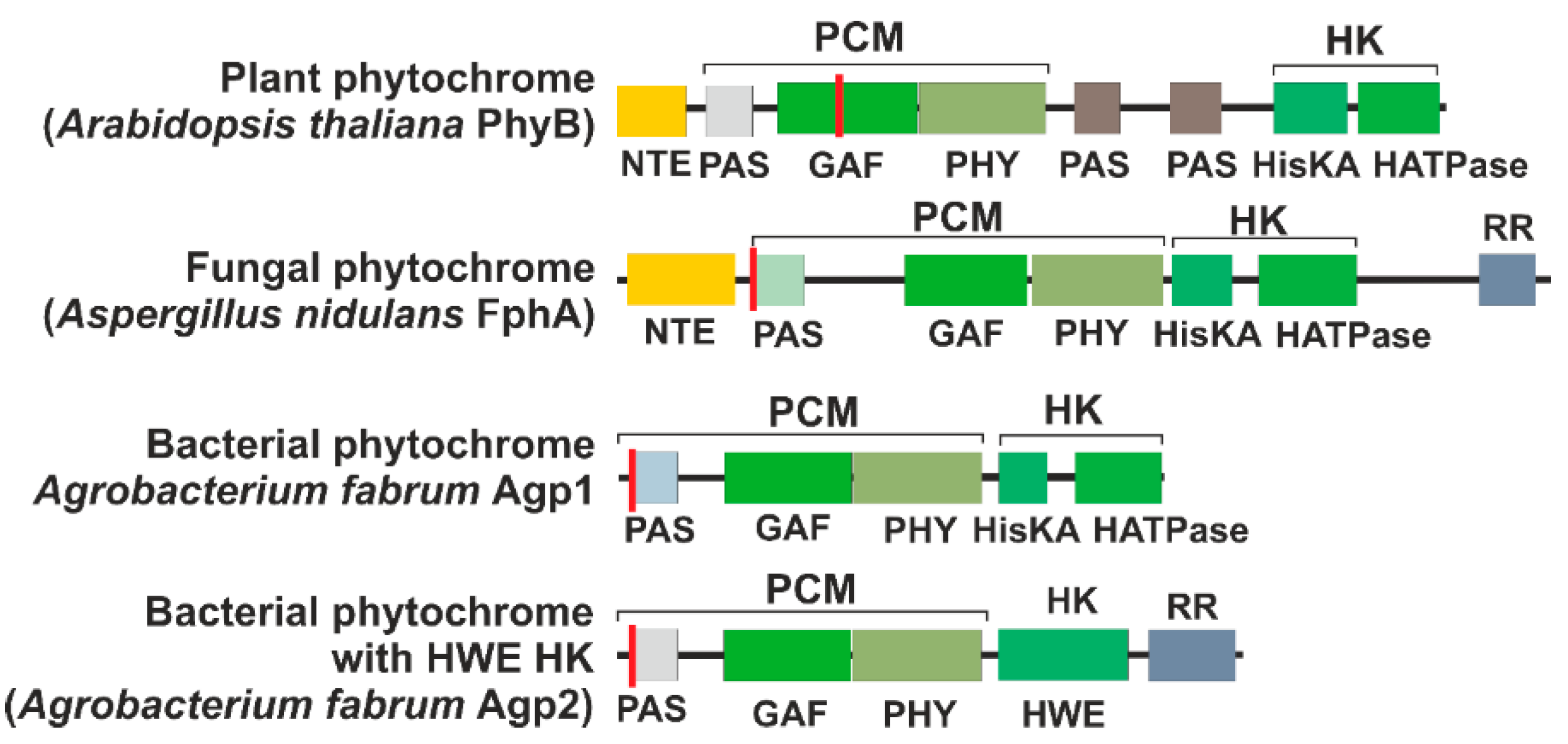
Figure 2. Domain arrangement of representative phytochromes. The red line indicates the position of the chromophore-binding cysteine. The abbreviations are NTE, N-terminal extension; PCM, photosensory core module; HK, histidine kinase (like) module; PAS, PAS domain [10]; GAF, GAF domain [11]; PHY, PHY domain [12]; HisKA, substrate and dimerization domain of histidine kinase; HATPase, ATPase domain of histidine kinase; HWE, HWE type of histidine kinase; RR, response regulator.
All phytochromes analyzed so far have a dimer arrangement with usually two identical subunits, although heterodimer formation is described for few plant phytochromes [13]. As far as recent structure determinations are concerned, subunits of bacterial phytochromes are arranged in a parallel manner (Figure 3). Surprisingly, plant phytochrome structures have parallel histidine kinase-like regions, but the photosensory core modules (PCM modules) are arranged in an antiparallel manner, and the overall dimer forms an unexpected, complex and asymmetric arrangement (Figure 3).
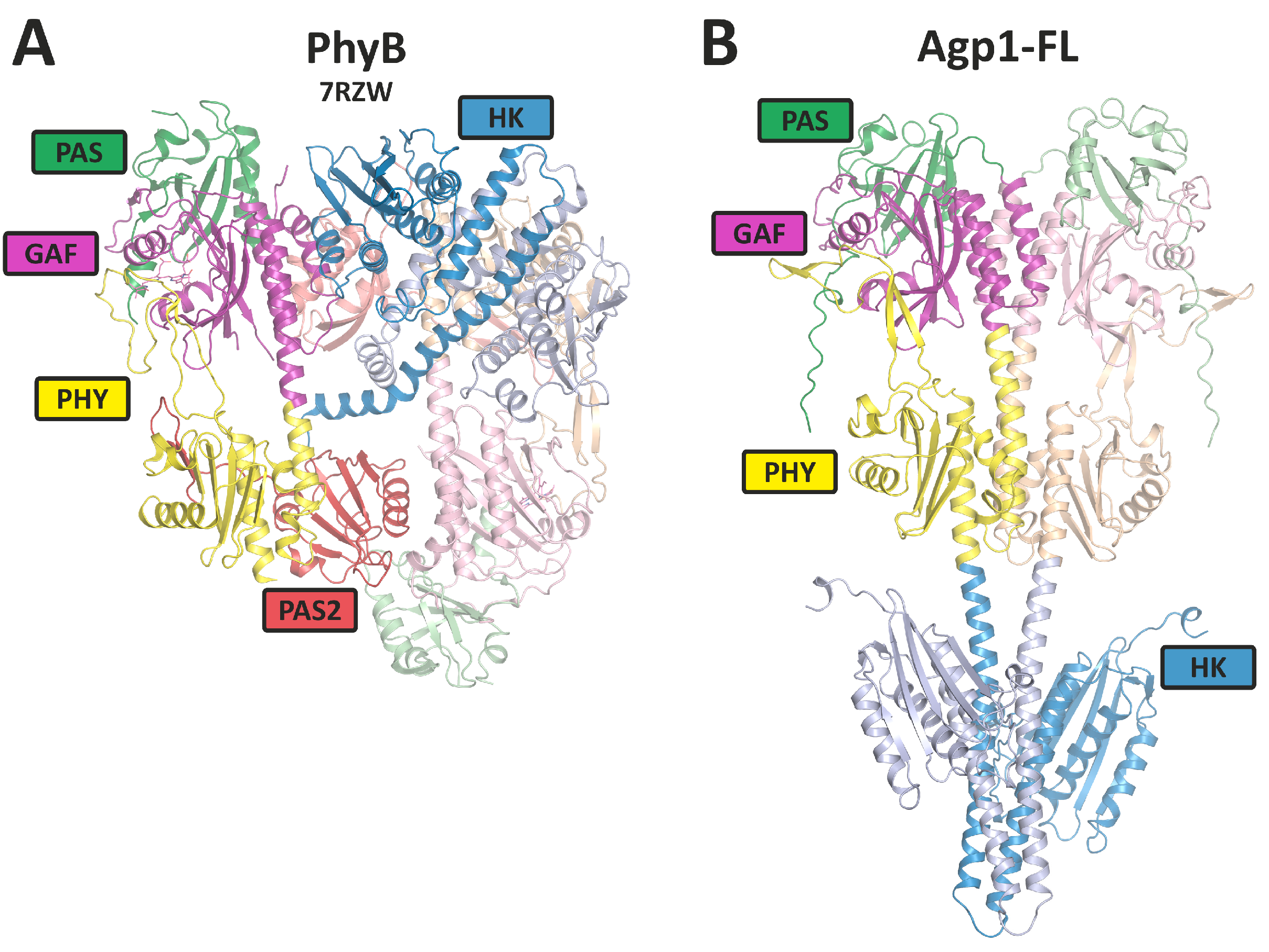
Figure 3. Overall rearrangement of the 3D structures of plant PhyB dimer and a phytochrome Agp1 dimer as representatives for plant and bacterial phytochromes. (A) The PhyB model is from cryo-EM studies (PDB [14] entry 7RZW); (B) the full-length Agp1 (Agp1-FL) model is based on Alphafold 2.3.1 [15] with 2 identical sequences (Uniprot-ID: Q7CY45) as entry (see also [16] for PCM structures). The domains are indicated by different colors. HK stands for histidine kinases (like).
Phytochromes were discovered as the second group of photoreceptors after the opsins. Phytochrome discovery is based on action spectroscopy of plant effects such as flower induction [17], seed germination [18] or de-etiolation [19], followed by spectral detection which is based on the Pr-Pfr photoreversibility [20]. In fact, phytochromes control a large number of developmental effects in plants and can therefore be regarded as the most relevant plant photoreceptors [21]. The later discoveries of phytochromes in bacteria, fungi, slime molds and heterokonts were based on genome sequences, and consequently, the known light effects as controlled by phytochrome are rare in these groups: the organisms are much more diverse, and it is expected that the phytochrome-controlled effects are also diverse, i.e., different from one bacterium to another. Because light effects are hard to recognize in bacteria, they may remain hidden until genes are modified and responses are targeted by researchers. In cyanobacteria, the cyanobacteriochromes control the effects of chromatic adaptation [22] or phototaxis [23], whereas the only effect known for a canonical phytochrome is an effect on biofilm formation, which is probably not light dependent [24].
2. Interaction Partners of Plant Phytochromes
Most phytochrome interaction proteins have been found for the plant Arabidopsis thaliana. This model plant has five phytochromes. Interacting proteins are listed in the Biogrid database [25]. The entries for PhyA are, for example, found here: https://thebiogrid.org/22724/table/arabidopsis-thaliana/PhyA.html. The selection below covers most of the database entries. In most cases, PhyA and/or PhyB are the interaction partners, and the experiments are usually carried out with the model plant Arabidopsis thaliana.
3. Homodimer and Heterodimer Formation
Phytochromes are dimeric proteins. The interaction between monomers may be regarded as a very stable protein interaction of phytochromes. Dimerization has been long documented and analyzed [26][27][28]. Recently, it has been discovered that the arrangement of subunits in PhyA and PhyB is significantly different from what had been expected. The arrangement is asymmetric, and the subunits are aligned parallel to each other in the C-terminal part, the histidine kinase-like region, and antiparallel to each other in the N-terminal PCM. This asymmetric arrangement poses new questions about the interaction with other proteins. Is it a specific interaction with one subunit, is it an interaction with either subunit of both sides, or are both subunits required?
Typically, both subunits of the dimer have identical sequences. However, since there are different phytochromes in a species, there is the possibility that subunits of different phytochromes form a dimer, although a mixed formation would not necessarily turn out from standard biochemical assays such as Western blotting. Indeed, co-precipitation of Myc-tagged phytochromes showed that heterodimers can be formed between all type II phytochromes of Arabidopsis (PhyB, PhyC, PhyD and PhyE), whereas PhyA seems to be excluded from mixed dimer formation [13].
4. Interaction with PIF3 and Other PIF Proteins
The interaction between phytochromes and phytochrome interacting factor 3 (PIF3) was the first interaction for a phytochrome with another protein to be discovered. This is probably the best-studied interaction of a phytochrome with another protein, as seen by the literature statistics: in Web of Science (a paid-access platform for reference and citation data from academic journals), there are 420 articles and reviews on PIF3 [29][30][31][32]. PIF3 was found by a yeast two-hybrid screening using an Arabidopsis cDNA library as prey and PhyB as bait [33]. Considering the evidence indicating that the “signaling” C-terminal domain of the phytochromes might be directly involved in the interaction with signaling partners [34], Ni et al. used the non-photoactive C-terminal PAS-PAS-HK (see Figure 2) of PhyB as bait in the yeast two-hybrid system.
To investigate the functional relevance to phytochrome signaling, transgenic Arabidopsis seedlings expressing both sense and antisense PIF3 constructs were examined, evaluating their impact on photoresponsiveness under constant red light or constant far-red light conditions, which are indicative of PhyB and PhyA activity, respectively. Transgenic Arabidopsis seedlings, which had reduced levels of PIF3 through antisense techniques, showed significantly decreased responsiveness to light signals detected by either PhyB or PhyA. Thus, PIF3 is functionally involved in both the PhyA and PhyB signaling pathways within plant cells, which aligns with its ability to bind to both photoreceptors [35].
Following the discovery of the interaction between PIF3 with the C-terminal domain of PhyB, an in vitro pull-down assay was performed to verify the interaction with the full-length, photoactive PhyB holoprotein. In their study, Ni et al. [36] used PIF3 immobilized on beads as bait and full-length phytochrome molecules carrying a chromophore as prey. The phytochrome molecules were synthesized in vitro [36]. The results demonstrated that PhyB binds to PIF3 only upon light-induced conversion to its biologically active Pfr form. When PhyB reverted to its inactive Pr form, PIF3 dissociated from the phytochrome. This indicates that PhyB signaling relies on the specific recognition of PIF3 by the active Pfr form of the photoreceptor. While initial experiments indicated an interaction between PIF3 and the C-terminal region of the phytochrome, it became evident that a phytochrome lacking a C-terminus can initiate light-induced signal transduction, and in fact, it can do so even more efficiently than the full-length protein [37]. The C-terminus might function in a modulatory way or could initiate pathways that differ from the standard signaling pathways, providing an explanation for PIF3’s interaction with the C-terminus. Indeed, the Ni et al. experiments (1999) [36] showed that there is a Pfr-dependent interaction of PIF3 with the N-terminal PAS-GAF-PHY domain of the phytochrome.
In a comprehensive mutagenesis-based “yeast reverse-hybrid screen”, Kikis et al. identified four amino acids of PhyB that are necessary for PIF3 binding [38]. The same PhyB mutants were also reduced in their light responses in Arabidopsis. All four amino acids clustered in the knot region of PhyB, which is formed by PAS and GAF domains in the N-terminus of the protein. These experiments gave deep insight into the spatial interactions between PhyB and PIF3. Because the C-terminus of PhyB is too distant from the knot region, it seems unlikely that PIF3 interacts with the C-terminus and the N-terminal knot at the same time.
In 2006, Al Sady et al. [39] developed antibodies targeting the low-abundance native PIF3 protein. They also created various transgenic Arabidopsis lines that expressed different forms of PIF3 fusion proteins labeled with fluorescent markers and epitope tags. Through Western blot analysis, they confirmed and expanded the data of Bauer et al. [40], who demonstrated that the immunochemically measurable native PIF3 protein decreases below detectable levels in wild-type Arabidopsis seedlings within 260 min of exposure to continuous red light. These studies showed that the interaction with Phy induces the degradation of PIF3.
PIF3 is a member of the basic helix-loop-helix (bHLH) family and localized in the nucleus [33][36]. It functions as a negative regulator of photomorphogenesis by repressing the expression of genes involved, like those associated with chlorophyll biosynthesis and light-induced growth processes [38][41]. Under dark conditions or in the presence of inactive phytochromes, PIF3 is stable and able to bind to specific DNA sequences in the promoters of target genes, thereby repressing their expression. In the light, plant phytochrome in its active Pfr conformation is quickly translocated into the nucleus. These active phytochromes can interact with PIF3 and induce its degradation by phosphorylation [42] and sumoylation [43] or prevent its binding to DNA by competing with the DNA binding. The degradation or inhibition of PIF3 by active phytochromes allows for the activation of photomorphogenesis-related genes and promotes plant growth and development [39][44][45].
In Arabidopsis, there are six other proteins that are homologous to PIF3, namely, PIF1, PIF4 [46], PIF5 [47], PIF6 [48], PIF7 [49] and PIF8 [46]. Homologous PIFs have also been examined in other plants [50]. All members seem to bind phytochrome specifically in the Pfr form, but the DNA binding positions differ. The variability in PIFs is thus one way to allow for multiple variations in light control via phytochrome.
The signal transduction cascade for PIF3 performs as follows: Upon irradiation of the plant and photoconversion from Pr to Pfr, the phytochrome transfers from the cytosol to the nucleus. Here, it interacts with PIF3 in a Pfr specific way. This leads to the phosphorylation of PIF3 through a kinase which also interacts with the phytochrome and PIF3. Upon sumoylation, PIF3 is targeted for degradation. Because, in general, PIF3 acts as a negative transcriptional regulator, the degradation of this protein leads to an induction of expression in the light (see model in Figure 4). Due to its early discovery, PIF3 is the best-analyzed member of the PIF family but probably also the most relevant with respect to light regulation of plant responses.
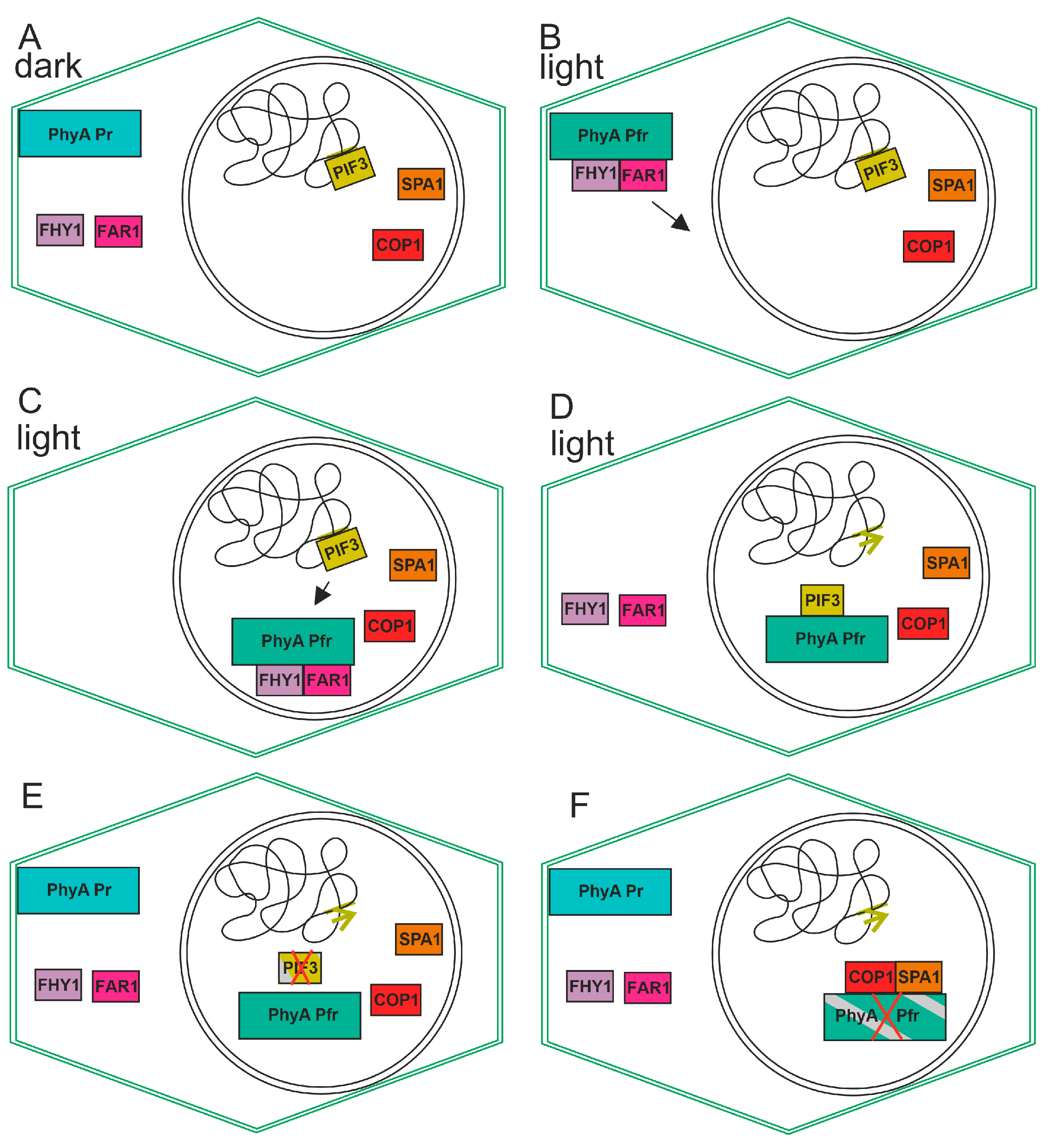
Figure 4. Cartoon for the action of plant phytochrome A. The cell borders are defined by the outer lines, while the nucleus is represented by the inner circle. (A) In darkness, phytochrome resides in the cytosol. PIF3 interacts with the DNA as negative transcription factor and inhibits transcription of certain genes. (B) In the light, phytochrome converts to Pfr, and this form interacts with FAR1 and FHY1. The complex moves into the nucleus (arrow) (C) This triggers transfer to the nucleus. PIF3 moves towards PhyA (D) FAR1 and FHY1 move back to the cytosol and phytochrome interacts with PIF3, thereby triggering its degradation possibly by phosphorylation and sumoylation. The green arrow indicates that specific transcription can start due to removal of PIF3 from the promotor. (E) PIF3 degrades, and new phytochrome in the Pr form appears in the cytosol. (F) PhyA interacts with SPA1 and with COP1, and PhyA is degraded.
5. Interaction with Cryptochrome (Cry)
Cryptochromes are widely distributed flavoprotein blue light photoreceptors that are homologous to the photolyases, which are enzymes that repair DNA damage. Plants typically contain two to three cryptochromes and two other flavoprotein photoreceptors, the phototropins. Already, in the early days of phytochrome research [51][52], it was found that many phytochrome effects can also be induced by blue light. Because the bilin chromophore of the phytochrome absorbs not only red light but to a lesser extent also blue light, both light qualities could equally induce responses via the phytochrome. However, separate blue light photoreceptors could as also cause the blue light effects. The gene of cryptochrome was discovered in 1994 [53] and the gene of phototropin was discovered in 1997 [54].
An interaction between Cry1 and plant phytochrome A was first demonstrated by the phosphorylation of Cry1 by purified PhyA, and by yeast two-hybrid techniques [55]. The interaction seems independent of the photochromic state (Pr or Pfr) of the phytochrome, because phosphorylation of Cry by the phytochrome was independent of the respective form of the latter (but see [56]).
While the interaction between Phy and PIF3 results in a clear action, i.e., the degradation of PIF3, the result of the phytochrome/cryptochrome interaction is open. However, studies on the coaction of both photoreceptors have continued [56][57].
6. Interaction with ARR4
Bacterial and fungal phytochromes often have histidine kinase domains and function as light-regulated histidine kinases that phosphorylate cognate response regulator proteins. Plant phytochromes have a module at the C-terminus which has evolved from histidine kinases but has no phosphorylating function and the regions do not dimerize in the way characteristic for histidine kinases [58]. Nevertheless, the attention has been drawn to Arabidopsis homologs for the E. coli CheY response regulator. Mutant studies have already identified response regulator sequences in cytokinin signal transduction [59]. One of these, ARR4 (Arabidopsis response regulator 4), was found to be involved in light signal transduction. Subsequent in vitro studies demonstrated that ARR4 interacts with the N-terminus of PhyB, but not of PhyA [60]. ARR4 knockout mutants show reduced sensitivity to red light. One action of ARR4 is that it modulates dark reversion of PhyB.
7. Interaction with Phototropin
While interactions with cryptochrome and PIFs probably take place in the nucleus of a plant cell, an interaction between the phytochrome and phototropin at the plasma membrane has been shown for the moss Physcomitrium patens [61]. The tip cell of moss filaments grows phototropically toward the light. This response is controlled by the phytochrome (and possibly phototropin). A gradient of activated photoreceptor can only be formed by unilateral light if the photoreceptors are immobilized, most likely at the plasma membrane. The interaction between the phytochrome and phototropin at the plasma membrane could fulfil both functions.
8. Interaction with COP1
Constitutive photomorphogenesis 1 (COP1) is a ubiquitin ligase that is involved in protein degradation. Dark grown COP1 and DET (de-etiolated) mutant seedlings are similar in many aspects to wild-types grown in the light. The interaction of COP1 with the N-terminal of PhyB in vitro has been shown by pull-down assays [62]. COP1 is involved in many photomorphogenic responses, probably by inducing degradation as part of plant proteasomes. The light-induced degradation of plant PhyA was found already very early in phytochrome research. Here, PhyB is thought to be degraded through the COP1 proteasome in the nucleus.
9. Interaction with NDPK2
In a yeast two-hybrid screen using the C-terminal part of PhyA as a bait, the nucleoside diphosphate kinase 2 (NDPK2) was isolated [63]. In vitro characterization showed that the interaction takes place between one PAS domain A of the C-terminus of PhyA and the C-terminal part of NDPK2 [64]. The enzyme catalyzes the phosphotransfer of the gamma phosphate of ATP to other nucleoside diphosphates and thus plays an important role in nucleotide metabolism. NDPK2 was shown to be involved in phytochrome photoregulation, e.g., of starch formation [64][65][66][67].
An open question in NDPK2 function is its localization to the chloroplast [67] via a chloroplast target sequence. Plant phytochromes reside in the cytoplasm, at the plasmalemma or in the nucleus, but have never been reported to be in the chloroplast. The interaction between NDPK2 and the phytochrome is thus either not relevant for in vivo functions or is relevant for the protein on its way to the chloroplast.
10. Interaction with PKS1
PKS1 (PHYTOCHROME KINASE SUBSTRATE 1) is a protein in Arabidopsis thaliana that has been identified in a yeast two-hybrid screen with C-terminal PhyA and PhyB as bait. The interaction in vitro with the full-length GST-tagged PhyA and PhyB has been demonstrated: the interaction appears to be independent from Pr or Pfr [68]. A homolog PKS2 fulfils similar functions [69]. PKS1 is phosphorylated by phytochromes in response to light, and this phosphorylation is thought to activate downstream signaling components. In addition to its role as a substrate of phytochrome-mediated signaling, PKS1 has also been shown to interact with other proteins involved in light signaling pathways, such as the photoreceptors cryptochrome or phytotropin. This suggests that PKS1 may play a broader role in regulating plant responses to light beyond its interactions with phytochromes.
11. Interaction with SPA1
The suppressor of phytochrome A (SPA1) has been shown to interact with phytochromes by using the yeast two-hybrid system and by using FRET-based methods in plant cells [70]. SPA1 is involved in the regulation of photoperiodic flowering and regulates circadian rhythms. It is required for the suppression of photomorphogenesis in dark-grown seedlings and for normal elongation growth of adult plants. As an integral component of the COP1-SPA-E3 ubiquitin–protein ligase complex, it is involved in degradation of PhyA as well as in HY5, HFR1, LAF1 and CO degradation.
References
- Huq, E.; Quail, P.H. Phytochrome signaling. In Handbook of Photosensory Receptors; Wiley: Hoboken, NJ, USA, 2005; pp. 151–170.
- Tu, S.-L.; Lagarias, J.C. The phytochromes. In Handbook of Photosensory Receptors; Wiley: Hoboken, NJ, USA, 2005; pp. 121–149.
- Vierstra, R.D.; Karniol, B. Phytochromes in microorganisms. In Handbook of Photosensory Receptors; Wiley: Hoboken, NJ, USA, 2005; pp. 171–195.
- Rockwell, N.C.; Duanmu, D.; Martin, S.S.; Bachy, C.; Price, D.C.; Bhattacharya, D.; Worden, A.Z.; Lagarias, J.C. Eukaryotic algal phytochromes span the visible spectrum. Proc. Natl. Acad. Sci. USA 2024, 111, 3871–3876.
- Giraud, E.; Fardoux, J.; Fourrier, N.; Hannibal, L.; Genty, B.; Bouyer, P.; Dreyfus, B.; Vermeglio, A. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 2002, 417, 202–205.
- Rottwinkel, G.; Oberpichler, I.; Lamparter, T. Bathy phytochromes in rhizobial soil bacteria. J. Bacteriol. 2010, 192, 5124–5133.
- Lamparter, T.; Carrascal, M.; Michael, N.; Martinez, E.; Rottwinkel, G.; Abian, J. The biliverdin chromophore binds covalently to a conserved cysteine residue in the n-terminus of Agrobacterium phytochrome Agp1. Biochemistry 2004, 43, 3659–3669.
- Giraud, E.; Vuillet, L.; Hannibal, L.; Fardoux, J.; Zappa, S.; Adriano, J.M.; Berthomieu, C.; Bouyer, P.; Pignol, D.; Verméglio, A. A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. J. Biol. Chem. 2005, 280, 32389–32397.
- Ishizuka, T.; Shimada, T.; Okajima, K.; Yoshihara, S.; Ochiai, Y.; Katayama, M.; Ikeuchi, M. Characterization of cyanobacteriochrome TePixJ from a thermophilic cyanobacterium Thermosynechococcus elongatus strain bp-1. Plant Cell Physiol. 2006, 47, 1251–1261.
- Ponting, C.P.; Aravind, L. PAS: A multifunctional domain family comes to light. Curr. Biol. 1997, 7, R674–R677.
- Aravind, L.; Ponting, C.P. The GAF domain: An evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 1997, 22, 458–459.
- Oka, Y.; Matsushita, T.; Mochizuki, N.; Suzuki, T.; Tokutomi, S.; Nagatani, A. Functional analysis of a 450-amino acid n-terminal fragment of phytochrome b in Arabidopsis. Plant Cell 2004, 16, 2104–2116.
- Sharrock, R.A.; Clack, T. Heterodimerization of type ii phytochromes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 11500–11505.
- Armstrong, D.R.; Berrisford, J.M.; Conroy, M.J.; Gutmanas, A.; Anyango, S.; Choudhary, P.; Clark, A.R.; Dana, J.M.; Deshpande, M.; Dunlop, R.; et al. Pdbe: Improved findability of macromolecular structure data in the PDB. Nucleic Acids Res. 2020, 48, D335–D343.
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with alphafold. Nature 2021, 596, 583–589.
- Nagano, S.; Scheerer, P.; Zubow, K.; Michael, N.; Inomata, K.; Lamparter, T.; Krauss, N. The crystal structures of the n-terminal photosensory core module of Agrobacterium phytochrome Agp1 as parallel and anti-parallel dimers. J. Biol. Chem. 2016, 291, 20674–20691.
- Parker, M.W.; Hendricks, S.B.; Borthwick, H.A.; Scully, N.J. Action spectrum for the photoperiodic control of floral initiation in biloxi soybean. Science 1945, 102, 152–155.
- Flint, L.H.; McAlister, E.D. Wavelengths of radiation in the visible spectrum promoting the germination of light-sensitive lettuce seed. Smithson. Misc. Coll. 1937, 96, 1–8.
- Virgin, H.I. Action spectrum for elimination of lag phase in chlorophyll formation in previously dark grown leaves of wheat. Physiol. Plant. 1961, 14, 439–452.
- Butler, W.L.; Norris, K.H.; Siegelman, H.W.; Hendricks, S.B. Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc. Natl. Acad. Sci. USA 1959, 45, 1703–1708.
- Kendrick, R.E.; Kronenberg, G.H.M. Photomorphogenesis in Plants, 2nd ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994.
- Kehoe, D.M.; Grossman, A.R. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 1996, 273, 1409–1412.
- Ng, W.O.; Grossman, A.R.; Bhaya, D. Multiple light inputs control phototaxis in synechocystis sp. Strain pcc6803. J. Bacteriol. 2003, 185, 1599–1607.
- Lamparter, T.; Babian, J.; Frohlich, K.; Mielke, M.; Weber, N.; Wunsch, N.; Zais, F.; Schulz, K.; Aschmann, V.; Spohrer, N.; et al. The involvement of type iv pili and the phytochrome Cpha in gliding motility, lateral motility and photophobotaxis of the cyanobacterium Phormidium lacuna. PLoS ONE 2022, 17, e0249509.
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The biogrid interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541.
- Hunt, R.E.; Pratt, L.H. Partial characterization of undegraded oat phytochrome. Biochemistry 1980, 19, 390–394.
- Lagarias, J.C.; Mercurio, F.M. Structure function studies on phytochrome. Identification of light- induced conformational changes in 124-kda avena phytochrome in vitro. J. Biol. Chem. 1985, 260, 2415–2423.
- Edgerton, M.D.; Jones, A.M. Subunit interactions in the carboxy-terminal domain of phytochrome. Biochemistry 1993, 32, 8239–8245.
- Cheng, M.C.; Kathare, P.K.; Paik, I.; Huq, E. Phytochrome signaling networks. Annu. Rev. Plant Biol. 2021, 72, 217–244.
- Castillon, A.; Shen, H.; Huq, E. Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521.
- Duek, P.D.; Fankhauser, C. Bhlh class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54.
- Sharma, A.; Samtani, H.; Sahu, K.; Sharma, A.K.; Khurana, J.P.; Khurana, P. Functions of phytochrome-interacting factors (pifs) in the regulation of plant growth and development: A comprehensive review. Int. J. Biol. Macromol. 2023, 244, 125234.
- Ni, M.; Tepperman, J.M.; Quail, P.H. Pif3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 1998, 95, 657–667.
- Quail, P.H.; Boylan, M.T.; Parks, B.M.; Short, T.W.; Xu, Y.; Wagner, D. Phytochromes: Photosensory perception and signal transduction. Science 1995, 268, 675–680.
- Quail, P.H. Phytochrome-interacting factors. Semin. Cell Dev. Biol. 2000, 11, 457–466.
- Ni, M.; Tepperman, J.M.; Quail, P.H. Binding of phytochrome b to its nuclear signaling partner pif3 is reversibly induced by light. Nature 1999, 400, 781–784.
- Matsushita, T.; Mochizuki, N.; Nagatani, A. Dimers of the n-terminal domain of phytochrome b are functional in the nucleus. Nature 2003, 424, 571–574.
- Kikis, E.A.; Oka, Y.; Hudson, M.E.; Nagatani, A.; Quail, P.H. Residues clustered in the light-sensing knot of phytochrome b are necessary for conformer-specific binding to signaling partner pif3. PLoS Genet. 2009, 5, e1000352.
- Al Sady, B.; Ni, W.; Kircher, S.; Schäfer, E.; Quail, P.H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 2006, 23, 439–446.
- Bauer, D.; Viczian, A.; Kircher, S.; Nobis, T.; Nitschke, R.; Kunkel, T.; Panigrahi, K.C.; Adam, E.; Fejes, E.; Schäfer, E.; et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 2004, 16, 1433–1445.
- Monte, E.; Tepperman, J.M.; Al Sady, B.; Kaczorowski, K.A.; Alonso, J.M.; Ecker, J.R.; Li, X.; Zhang, Y.; Quail, P.H. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 2004, 101, 16091–16098.
- Phee, B.K.; Kim, J.I.; Shin, D.H.; Yoo, J.; Park, K.J.; Han, Y.J.; Kwon, Y.K.; Cho, M.H.; Jeon, J.S.; Bhoo, S.H.; et al. A novel protein phosphatase indirectly regulates phytochrome-interacting factor 3 via phytochrome. Biochem. J. 2008, 415, 247–255.
- Bernula, P.; Pettkó-Szandtner, A.; Hajdu, A.; Kozma-Bognár, L.; Josse, E.M.; Adám, E.; Nagy, F.; Viczián, A. Sumoylation of phytochrome interacting factor 3 promotes photomorphogenesis in Arabidopsis thaliana. New Phytol. 2021, 229, 2050–2061.
- Ni, W.M.; Xu, S.L.; Tepperman, J.M.; Stanley, D.J.; Maltby, D.A.; Gross, J.D.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 2014, 344, 1160–1164.
- Leivar, P.; Tepperman, J.M.; Monte, E.; Calderon, R.H.; Liu, T.L.; Quail, P.H. Definition of early transcriptional circuitry involved in light-induced reversal of pif-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 2009, 21, 3535–3553.
- Xu, Y.; Zhu, Z.Q. Pif4 and pif4-interacting proteins: At the nexus of plant light, temperature and hormone signal integrations. Int. J. Mol. Sci. 2021, 22, 10304.
- Tavridou, E.; Pireyre, M.; Ulm, R. Degradation of the transcription factors pif4 and pif5 under UV-b promotes UVR8-mediated inhibition of hypocotyl growth in Arabidopsis. Plant J. 2020, 101, 507–517.
- Khanna, R.; Huq, E.; Kikis, E.A.; Al Sady, B.; Lanzatella, C.; Quail, P.H. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 2004, 16, 3033–3044.
- Zhou, Y.; Park, S.H.; Soh, M.Y.; Chua, N.H. Ubiquitin-specific proteases ubp12 and ubp13 promote shade avoidance response by enhancing pif7 stability. Proc. Natl. Acad. Sci. USA 2021, 118, e2103633118.
- Xu, T.F.; Hiltbrunner, A. Phytochrome interacting factors from physcomitrella patens are active in Arabidopsis and complement the pif quadruple mutant. Plant Signal. Behav. 2017, 12, e1388975.
- Wareing, P.F.; Black, M. Similar effects of blue and infra-red radiation on light-sensitive seeds. Nature 1958, 181, 1420–1421.
- Sugai, M.; Furuya, M. Photomorphogenesis in pteris vittata I. Phytochrome-mediated spore germination and blue light interaction. Plant Cell Physiol. 1967, 8, 737–748.
- Ahmad, M.; Cashmore, A.R. Hy4 gene of a. Thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 1993, 366, 162–166.
- Huala, E.; Oeller, P.W.; Liscum, E.; Han, I.S.; Larsen, E.; Briggs, W.R. Arabidopsis nph1: A protein kinase with a putative redox-sensing domain. Science 1997, 278, 2120–2123.
- Ahmad, M.; Jarillo, J.A.; Smirnova, O.; Cashmore, A.R. The Cry1 blue light photoreceptor of Arabidopsis interacts with phytochrome a in vitro. Mol. Cell 1998, 1, 939–948.
- Neff, M.M.; Chory, J. Genetic interactions between phytochrome a, phytochrome b, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998, 118, 27–36.
- de Wit, M.; Keuskamp, D.H.; Bongers, F.J.; Hornitschek, P.; Gommers, C.M.M.; Reinen, E.; Martinez-Ceron, C.; Fankhauser, C.; Pierik, R. Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr. Biol. 2016, 26, 3320–3326.
- Li, H.; Burgie, E.S.; Gannam, Z.T.K.; Li, H.; Vierstra, R.D. Plant phytochrome b is an asymmetric dimer with unique signalling potential. Nature 2022, 604, 127–133.
- Brandstatter, I.; Kieber, J.J. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 1998, 10, 1009–1019.
- Sweere, U.; Eichenberg, K.; Lohrmann, J.; Mira-Rodado, V.; Baurle, I.; Kudla, J.; Nagy, F.; Schäfer, E.; Harter, K. Interaction of the response regulator arr4 with phytochrome b in modulating red light signaling. Science 2001, 294, 1108–1111.
- Jaedicke, K.; Lichtenthaler, A.L.; Meyberg, R.; Zeidler, M.; Hughes, J. A phytochrome-phototropin light signaling complex at the plasma membrane. Proc. Natl. Acad. Sci. USA 2012, 109, 12231–12236.
- Jang, I.C.; Henriques, R.; Seo, H.S.; Nagatani, A.; Chua, N.H. Arabidopsis phytochrome interacting factor proteins promote phytochrome b polyubiquitination by cop1 e3 ligase in the nucleus. Plant Cell 2010, 22, 2370–2383.
- Choi, G.; Yi, H.; Lee, J.; Kwon, Y.-K.; Soh, M.S.; Shin, B.; Luka, Z.; Hahn, T.-R.; Song, P.-S. Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 1999, 401, 610–613.
- Shen, Y.; Kim, J.I.; Song, P.S. Ndpk2 as a signal transducer in the phytochrome-mediated light signaling. J. Biol. Chem. 2005, 280, 5740–5749.
- Shen, Y.; Kim, J.I.; Song, P.S. Autophosphorylation of Arabidopsis nucleoside diphosphate kinase 2 occurs only on its active histidine residue. Biochemistry 2006, 45, 1946–1949.
- Hetmann, A.; Kowalczyk, S. Nucleoside diphosphate kinase isoforms regulated by phytochrome a isolated from oat coleoptiles. Acta Biochim. Pol. 2009, 56, 143–154.
- Han, X.Z.; Tohge, T.; Lalor, P.; Dockery, P.; Devaney, N.; Esteves-Ferreira, A.A.; Fernie, A.R.; Sulpice, R. Phytochrome a and b regulate primary metabolism in Arabidopsis leaves in response to light. Front. Plant Sci. 2017, 8, 1394.
- Fankhauser, C.; Yeh, K.C.; Lagarias, J.C.; Zhang, H.; Elich, T.D.; Chory, J. Pks1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 1999, 284, 1539–1541.
- de Carbonnel, M.; Davis, P.; Roelfsema, M.R.G.; Inoue, S.; Schepens, I.; Lariguet, P.; Geisler, M.; Shimazaki, K.; Hangarter, R.; Fankhauser, C. The Arabidopsis phytochrome kinase substrate2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol. 2010, 152, 1391–1405.
- Sheerin, D.J.; Menon, C.; Oven-Krockhaus, S.Z.; Enderle, B.; Zhu, L.; Johnen, P.; Schleifenbaum, F.; Stierhof, Y.D.; Huq, E.; Hiltbrunner, A. Light-activated phytochrome a and b interact with members of the spa family to promote photomorphogenesis in Arabidopsis by reorganizing the cop1/spa complex. Plant Cell 2015, 27, 189–201.
More
Information
Subjects:
Biochemistry & Molecular Biology; Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
879
Revisions:
2 times
(View History)
Update Date:
10 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





