
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ivana Banic | -- | 2904 | 2024-01-08 10:00:10 | | | |
| 2 | Lindsay Dong | Meta information modification | 2904 | 2024-01-10 01:35:14 | | |
Video Upload Options
Allergic diseases are one of the most common chronic conditions and their prevalence is on the rise. Environmental exposure, primarily prenatal and early life influences, affect the risk for the development and specific phenotypes of allergic diseases via epigenetic mechanisms. Exposure to pollutants, microorganisms and parasites, tobacco smoke and certain aspects of diet are known to drive epigenetic changes that are essential for immune regulation (e.g., the shift toward T helper 2-Th2 cell polarization and decrease in regulatory T-cell (Treg) differentiation).
1. Introduction
2. Environmental Exposure and Epigenetic Regulation in Allergic Diseases
2.1. The Effect of Maternal and In Utero Exposure on Epigenetic Modifications
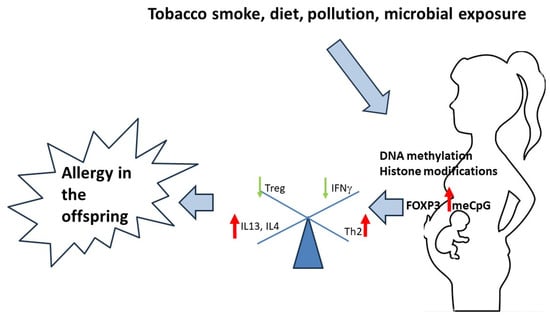
2.2. Exposure to Tobacco Smoke
2.3. Epigenetic Modifications and the Effects of Diet
2.4. Effects of Microbial Exposure to Epigenetic Regulation in Allergic Diseases
2.5. Exposure to Air Pollution and Epigenetic Effects in Allergic Diseases
2.6. Exposure to Persistent Organic Pollutants and Epigenetic Effects
2.7. Other Intrauterine Factors That Affect Epigenetic Modifications and Allergic Disease Risk
2.8. Early Life Events and Epigenetic Changes Affecting Allergic Diseases
2.9. Effects of Exposure to Heavy Metals on Epigenetic Changes in Allergy
2.10. The Changing Environment and Epigenetic Changes in Allergic Diseases
2.11. Expression of Small Non-Coding RNAs and Allergic Diseases
3. Epigenetic Changes and Treatment Outcomes in Allergic Diseases
4. Environmental Exposure, Epigenetic Changes and Protection from Allergy
Microbial exposure, both early in the postnatal development or in utero, seems to greatly affect the offspring’s susceptibility to allergic diseases. Other than microbial exposure, infections with other protozoa and parasites, such as helminths, seem to have an important role in immune modulation and development. This is reinforced by the fact that a great part of the immune responses involved in the defense from parasitic infections and allergic diseases overlap. Certain pattern recognition pattern members participate in the response to parasitic (e.g., helminth) infection and are responsible for driving T-cell polarization. Adult helminths secrete certain antigens that are known to activate a number of immune effector cells, including basophils, mast cells, eosinophils, Th2 cells and innate lymphocyte T cells 2 (ILC2), as well as induce the production of innate and adaptive cytokines. Additionally, lipids derived from helminths can stimulate the production of a number of inflammatory cytokines, such as TNFα, IL-6, IL-8, IL-10 and IL-12.
Other than differential DNA methylation, other epigenetic modifications have been associated with protection against the development of allergic diseases or the development of immune tolerance, such as decreased histone acetylation of Th2 genes in a murine food allergy study assessing acquired immune tolerance associated with raw milk consumption.
References
- Potaczek, D.P.; Harb, H.; Michel, S.; Alhamwe, B.A.; Renz, H.; Tost, J. Epigenetics and allergy: From basic mechanisms to clinical applications. Epigenomics 2017, 9, 539–571.
- Galli, S.J.; Tsai, M.; Piliponsky, A.M. The Development of Allergic Inflammation. Nature 2008, 454, 445–454.
- Shamji, M.H.; Valenta, R.; Jardetzky, T.; Verhasselt, V.; Durham, S.R.; Würtzen, P.A.; van Neerven, R.J. The role of allergen-specific IgE, IgG and IgA in allergic disease. Allergy 2021, 76, 3627–3641.
- Rothbauer, M.; Charwat, V.; Bachmann, B.; Sticker, D.; Novak, R.; Wanzenböck, H.; Mathies, R.A.; Ertl, P. Monitoring transient cell-to-cell interactions in a multi-layered and multi-functional allergy-on-a-chip system. Lab Chip 2019, 19, 1916–1921.
- Dierick, B.J.H.; van der Molen, T.; Blok, B.M.J.F.-D.; Muraro, A.; Postma, M.J.; Kocks, J.W.; van Boven, J.F. Burden and socioeconomics of asthma, allergic rhinitis, atopic dermatitis and food allergy. Expert Rev. Pharmacoeconomics Outcomes Res. 2020, 20, 437–453.
- Akdis, C.A.; Akdis, M. Mechanisms of allergen-specific immunotherapy. J. Allergy Clin. Immunol. 2011, 127, 18–27.
- Smith, D.H.; Malone, D.C.; Lawson, K.A.; Okamoto, L.J.; Battista, C.; Saunders, W.B. A National Estimate of the Economic Costs of Asthma. Am. J. Respir. Crit. Care Med. 1997, 156, 787–793.
- DeVries, A.; Wlasiuk, G.; Miller, S.J.; Bosco, A.; Stern, D.A.; Lohman, I.C.; Rothers, J.; Jones, A.C.; Nicodemus-Johnson, J.; Vasquez, M.M.; et al. Epigenome-wide analysis links SMAD3 methylation at birth to asthma in children of asthmatic mothers. J. Allergy Clin. Immunol. 2016, 140, 534–542.
- Hylkema, M.N.; Blacquiere, M.J. Intrauterine Effects of Maternal Smoking on Sensitization, Asthma, and Chronic Obstructive Pulmonary Disease. Proc. Am. Thorac. Soc. 2009, 6, 660–662.
- Noakes, P.S.; Holt, P.G.; Prescott, S.L. Maternal smoking in pregnancy alters neonatal cytokine responses. Allergy 2003, 58, 1053–1058.
- Dunstan, J.A.; Mori, T.A.; Barden, A.; Beilin, L.J.; Taylor, A.L.; Holt, P.G.; Prescott, S.L. Fish oil supplementation in pregnancy modifies neonatal allergen-specific immune responses and clinical outcomes in infants at high risk of atopy: A randomized, controlled trial. J. Allergy Clin. Immunol. 2003, 112, 1178–1184.
- Chatzi, L.; Torrent, M.; Romieu, I.; Garcia-Esteban, R.; Ferrer, C.; Vioque, J.; Kogevinas, M.; Sunyer, J. Mediterranean diet in pregnancy is protective for wheeze and atopy in childhood. Thorax 2008, 63, 507–513.
- Bobetsis, Y.A.; Barros, S.P.; Lin, D.M.; Weidman, J.R.; Dolinoy, D.C.; Jirtle, R.L.; Boggess, K.A.; Beck, J.D.; Offenbacher, S. Bacterial Infection Promotes DNA Hypermethylation. J. Dent. Res. 2007, 86, 169–174.
- Schaub, B.; Liu, J.; Höppler, S.; Schleich, I.; Huehn, J.; Olek, S.; Wieczorek, G.; Illi, S.; von Mutius, E. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J. Allergy Clin. Immunol. 2009, 123, 774–782.
- Liu, J.; Ballaney, M.; Al-Alem, U.; Quan, C.; Jin, X.; Perera, F.; Chen, L.-C.; Miller, R.L. Combined Inhaled Diesel Exhaust Particles and Allergen Exposure Alter Methylation of T Helper Genes and IgE Production In Vivo. Toxicol. Sci. 2008, 102, 76–81.
- Webster, R.B.; Rodriguez, Y.; Klimecki, W.T.; Vercelli, D. The Human IL-13 Locus in Neonatal CD4+ T Cells Is Refractory to the Acquisition of a Repressive Chromatin Architecture. J. Biol. Chem. 2007, 282, 700–709.
- Jones, B.; Chen, J. Inhibition of IFN-γ transcription by site-specific methylation during T helper cell development. EMBO J. 2006, 25, 2443–2452.
- Schoenborn, J.R.; Dorschner, M.O.; Sekimata, M.; Santer, D.M.; Shnyreva, M.; Fitzpatrick, D.R.; A Stamatoyannopoulos, J.; Wilson, C.B. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-γ. Nat. Immunol. 2007, 8, 732–742.
- Martino, D.J.; Prescott, S.L. Silent mysteries: Epigenetic paradigms could hold the key to conquering the epidemic of allergy and immune disease. Allergy 2010, 65, 7–15.
- Han, P.; Gu, J.-Q.; Li, L.-S.; Wang, X.-Y.; Wang, H.-T.; Wang, Y.; Chang, C.; Sun, J.-L. The Association Between Intestinal Bacteria and Allergic Diseases—Cause or Consequence? Front. Cell. Infect. Microbiol. 2021, 11, 650893.
- Breton, C.V.; Byun, H.-M.; Wenten, M.; Pan, F.; Yang, A.; Gilliland, F.D. Prenatal Tobacco Smoke Exposure Affects Global and Gene-specific DNA Methylation. Am. J. Respir. Crit. Care Med. 2009, 180, 462–467.
- Zakarya, R.; Adcock, I.; Oliver, B.G. Epigenetic impacts of maternal tobacco and e-vapour exposure on the offspring lung. Clin. Epigenetics 2019, 11, 32.
- Neophytou, A.M.; Oh, S.S.; Hu, D.; Huntsman, S.; Eng, C.; Rodríguez-Santana, J.R.; Kumar, R.; Balmes, J.R.; Eisen, E.A.; Burchard, E.G. In utero tobacco smoke exposure, DNA methylation, and asthma in Latino children. Environ. Epidemiology 2019, 3, e048.
- Kabesch, M.; Tost, J. Recent findings in the genetics and epigenetics of asthma and allergy. Semin. Immunopathol. 2020, 42, 43–60.
- Ulrey, C.L.; Liu, L.; Andrews, L.G.; Tollefsbol, T.O. The impact of metabolism on DNA methylation. Hum. Mol. Genet. 2005, 14, R139–R147.
- Prescott, S.; Saffery, R. The role of epigenetic dysregulation in the epidemic of allergic disease. Clin. Epigenetics 2011, 2, 223–232.
- Håberg, S.E.; London, S.J.; Stigum, H.; Nafstad, P.; Nystad, W. Folic acid supplements in pregnancy and early childhood respiratory health. Arch. Dis. Child. 2009, 94, 180–184.
- Whitrow, M.J.; Moore, V.M.; Rumbold, A.R.; Davies, M.J. Effect of Supplemental Folic Acid in Pregnancy on Childhood Asthma: A Prospective Birth Cohort Study. Am. J. Epidemiology 2009, 170, 1486–1493.
- Idris, F.S.; Shaikh, H.A.; Vahora, I.; Moparthi, K.P.; Al Rushaidi, M.T.; Muddam, M.; Obajeun, O.A.; Abaza, A.; Jaramillo, A.P.; Hamid, P. Maternal Diet and Infant Risk of Eczema and Food Allergy: A Systematic Review. Cureus 2023, 15, e45114.
- Wesemann, D.R.; Nagler, C.R. The Microbiome, Timing, and Barrier Function in the Context of Allergic Disease. Immunity 2016, 44, 728–738.
- Savage, J.H.; Lee-Sarwar, K.A.; Sordillo, J.; Bunyavanich, S.; Zhou, Y.; O’Connor, G.; Sandel, M.; Bacharier, L.B.; Zeiger, R.; Sodergren, E.; et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018, 73, 145–152.
- Huang, Y.J.; Marsland, B.J.; Bunyavanich, S.; O’Mahony, L.; Leung, D.Y.M.; Muraro, A.; Fleisher, T.A. The microbiome in allergic disease: Current understanding and future opportunities—2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J. Allergy Clin. Immunol. 2017, 139, 1099–1110.
- Woo, V.; Alenghat, T. Host–microbiota interactions: Epigenomic regulation. Curr. Opin. Immunol. 2017, 44, 52–60.
- Dai, Z.; Ramesh, V.; Locasale, J.W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 2020, 21, 737–753.
- Fiuza, B.S.D.; Fonseca, H.F.; Meirelles, P.M.; Marques, C.R.; da Silva, T.M.; Figueiredo, C.A. Understanding Asthma and Allergies by the Lens of Biodiversity and Epigenetic Changes. Front. Immunol. 2021, 12, 623737.
- Baskara-Yhuellou, I.; Tost, J. The Impact of MicroRNAs on Alterations of Gene Regulatory Networks in Allergic Diseases. Adv. Protein Chem. Struct. Biol. 2020, 120, 237–312.
- Martino, D.; Prescott, S. Epigenetics and Prenatal Influences on Asthma and Allergic Airways Disease. Chest 2011, 139, 640–647.
- Rodrigues, M.d.B.; de Carvalho, D.S.; Chong-Silva, D.C.; Urrutia-Pereira, M.; de Albuquerque, G.S.C.; Cieslak, F.; Chong-Neto, H.J. Association between exposure to pesticides and allergic diseases in children and adolescents: A systematic review with meta-analysis. J. Pediatr. 2022, 98, 551–564.
- Daniel, V.; Huber, W.; Bauer, K.; Suesal, C.; Conradt, C.; Opelz, G. Associations of blood levels of PCB, HCHS, and HCB with numbers of lymphocyte subpopulations, in vitro lymphocyte response, plasma cytokine levels, and immunoglobulin autoantibodies. Environ. Health Perspect. 2001, 109, 173–178.
- Baccarelli, A.; Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009, 21, 243–251.
- Hoang, T.T.; Qi, C.; Paul, K.C.; Lee, M.; White, J.D.; Richards, M.; Auerbach, S.S.; Long, S.; Shrestha, S.; Wang, T.; et al. Epigenome-Wide DNA Methylation and Pesticide Use in the Agricultural Lung Health Study. Environ. Health Perspect. 2021, 129, 97008.
- Kim, K.-Y.; Kim, D.-S.; Lee, S.-K.; Lee, I.-K.; Kang, J.-H.; Chang, Y.-S.; Jacobs, D.R.; Steffes, M.; Lee, D.-H. Association of Low-Dose Exposure to Persistent Organic Pollutants with Global DNA Hypomethylation in Healthy Koreans. Environ. Health Perspect. 2010, 118, 370–374.
- Prescott, S.L.; Holt, P.G.; Jenmalm, M.; Björkstén, B. Effects of maternal allergen-specific IgG in cord blood on early postnatal development of allergen-specific T-cell immunity. Allergy 2000, 55, 469–474.
- Prescott, S.L.; Breckler, L.A.; Witt, C.S.; Smith, L.; Dunstan, J.A.; Christiansen, F.T. Allergic women show reduced T helper type 1 alloresponses to fetal human leucocyte antigen mismatch during pregnancy. Clin. Exp. Immunol. 2010, 159, 65–72.
- Gheorghe, C.P.; Goyal, R.; Mittal, A.; Longo, L.D. Gene expression in the placenta: Maternal stress and epigenetic responses. Int. J. Dev. Biol. 2010, 54, 507–523.
- Prescott, S.L.; Clifton, V. Asthma and pregnancy: Emerging evidence of epigenetic interactions in utero. Curr. Opin. Allergy Clin. Immunol. 2009, 9, 417–426.
- Scott, N.M.; Hodyl, N.A.; Murphy, V.E.; Osei-Kumah, A.; Wyper, H.; Hodgson, D.M.; Smith, R.; Clifton, V.L. Placental Cytokine Expression Covaries with Maternal Asthma Severity and Fetal Sex. J. Immunol. 2009, 182, 1411–1420.
- Tulic, M.K.; Hodder, M.; Forsberg, A.; McCarthy, S.; Richman, T.; D’Vaz, N.; Van Den Biggelaar, A.H.; Thornton, C.A.; Prescott, S.L. Differences in innate immune function between allergic and nonallergic children: New insights into immune ontogeny. J. Allergy Clin. Immunol. 2011, 127, 470–478.e1.
- Kumar, R.K.; Hitchins, M.P.; Foster, P.S. Epigenetic changes in childhood asthma. Dis. Model. Mech. 2009, 2, 549–553.
- Ho, S.-M.; Johnson, A.; Tarapore, P.; Janakiram, V.; Zhang, X.; Leung, Y.-K. Environmental Epigenetics and Its Implication on Disease Risk and Health Outcomes. ILAR J. 2012, 53, 289–305.
- Ijomone, O.M.; Ijomone, O.K.; Iroegbu, J.D.; Ifenatuoha, C.W.; Olung, N.F.; Aschner, M. Epigenetic influence of environmentally neurotoxic metals. NeuroToxicology 2020, 81, 51–65.
- Stepanyan, A.; Petrackova, A.; Hakobyan, S.; Savara, J.; Davitavyan, S.; Kriegova, E.; Arakelyan, A. Long-term environmental metal exposure is associated with hypomethylation of CpG sites in NFKB1 and other genes related to oncogenesis. Clin. Epigenetics 2023, 15, 126.
- Cardenas, A.; Fadadu, R.; Bunyavanich, S. Climate change and epigenetic biomarkers in allergic and airway diseases. J. Allergy Clin. Immunol. 2023, 152, 1060–1072.
- Barnes, P.J. Targeting the Epigenome in the Treatment of Asthma and Chronic Obstructive Pulmonary Disease. Proc. Am. Thorac. Soc. 2009, 6, 693–696.
- Heffler, E.; Allegra, A.; Pioggia, G.; Picardi, G.; Musolino, C.; Gangemi, S. MicroRNA Profiling in Asthma: Potential Biomarkers and Therapeutic Targets. Am. J. Respir. Cell Mol. Biol. 2017, 57, 642–650.
- Pinkerton, M.; Chinchilli, V.; Banta, E.; Craig, T.; August, A.; Bascom, R.; Cantorna, M.; Harvill, E.; Ishmael, F.T. Differential expression of microRNAs in exhaled breath condensates of patients with asthma, patients with chronic obstructive pulmonary disease, and healthy adults. J. Allergy Clin. Immunol. 2013, 132, 217–219.e2.
- Panganiban, R.P.L.; Pinkerton, M.H.; Maru, S.Y.; Jefferson, S.J.; Roff, A.N.; Ishmael, F.T. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am. J. Clin. Exp. Immunol. 2012, 1, 154–165.
- Dong, X.; Xu, M.; Ren, Z.; Gu, J.; Lu, M.; Lu, Q.; Zhong, N. Regulation of CBL and ESR1 expression by microRNA-22-3p, 513a-5p and 625-5p may impact the pathogenesis of dust mite-induced pediatric asthma. Int. J. Mol. Med. 2016, 38, 446–456.
- Jardim, M.J.; Dailey, L.; Silbajoris, R.; Diaz-Sanchez, D. Distinct MicroRNA Expression in Human Airway Cells of Asthmatic Donors Identifies a Novel Asthma-Associated Gene. Am. J. Respir. Cell Mol. Biol. 2012, 47, 536–542.
- Yang, I.V.; Pedersen, B.S.; Liu, A.H.; O’Connor, G.T.; Pillai, D.; Kattan, M.; Misiak, R.T.; Gruchalla, R.; Szefler, S.J.; Hershey, G.K.K.; et al. The nasal methylome and childhood atopic asthma. J. Allergy Clin. Immunol. 2016, 139, 1478–1488.
- Nicodemus-Johnson, J.; Myers, R.A.; Sakabe, N.J.; Sobreira, D.R.; Hogarth, D.K.; Naureckas, E.T.; Sperling, A.I.; Solway, J.; White, S.R.; Nobrega, M.A.; et al. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight 2016, 1, e90151.
- Tost, J. A translational perspective on epigenetics in allergic diseases. J. Allergy Clin. Immunol. 2018, 142, 715–726.
- Lovinsky-Desir, S.; Miller, R.L. Epigenetics, Asthma, and Allergic Diseases: A Review of the Latest Advancements. Curr. Allergy Asthma Rep. 2012, 12, 211–220.
- Li, J.; Panganiban, R.; Kho, A.T.; McGeachie, M.J.; Farnam, L.; Chase, R.P.; Weiss, S.T.; Lu, Q.; Tantisira, K.G. Circulating MicroRNAs and Treatment Response in Childhood Asthma. Am. J. Respir. Crit. Care Med. 2020, 202, 65–72.




