Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Navdeep Kumar | -- | 1638 | 2024-01-05 06:01:14 | | | |
| 2 | Fanny Huang | Meta information modification | 1638 | 2024-01-09 08:59:30 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kumar, N.; Marée, R.; Geurts, P.; Muller, M. Imaging Techniques Used in Fish Bioimages. Encyclopedia. Available online: https://encyclopedia.pub/entry/53462 (accessed on 08 February 2026).
Kumar N, Marée R, Geurts P, Muller M. Imaging Techniques Used in Fish Bioimages. Encyclopedia. Available at: https://encyclopedia.pub/entry/53462. Accessed February 08, 2026.
Kumar, Navdeep, Raphaël Marée, Pierre Geurts, Marc Muller. "Imaging Techniques Used in Fish Bioimages" Encyclopedia, https://encyclopedia.pub/entry/53462 (accessed February 08, 2026).
Kumar, N., Marée, R., Geurts, P., & Muller, M. (2024, January 05). Imaging Techniques Used in Fish Bioimages. In Encyclopedia. https://encyclopedia.pub/entry/53462
Kumar, Navdeep, et al. "Imaging Techniques Used in Fish Bioimages." Encyclopedia. Web. 05 January, 2024.
Copy Citation
Detecting skeletal or bone-related deformities in model and aquaculture fish is vital for numerous biomedical studies. In biomedical research, model fish with bone-related disorders are potential indicators of various chemically induced toxins in their environment or poor dietary conditions. In aquaculture, skeletal deformities are affecting fish health, and economic losses are incurred by fish farmers.
fish
aquaculture
biomedical
image processing
1. Introduction
In the realm of biomedical research, model fish species like zebrafish (Danio rerio), and medaka (Oryzias latipes) are highly regarded as valuable vertebrate models. They are extensively used in a variety of biomedical applications, encompassing drug testing, morphometric screening, genome editing, toxicology assessments, and behavior analysis in vertebrates [1][2][3][4][5][6][7][8]. These model fish exhibit significant genetic and metabolic pathway similarities to both fish and mammals, sharing over 70% of their genes with humans [9][10][11][12]. Notably, zebrafish and medaka models are particularly advantageous due to their ease of maintenance and reproduction. Together with other technical advantages such as small size, low maintenance cost, high fecundity, and amenability to genetic engineering tools, the reason these fish are so popular among scientists is their suitability for in vivo imaging [13][14]. The embryonic and larval stages of these animals are translucent, allowing for the application of advanced imaging technologies to observe biological processes in a living animal. This property bears great potential for biomedical research when combined with the availability of transgenic and mutant lines that allow modeling human skeletal diseases and tracking specific organs and cell types with fluorescent markers [15]. Such characteristics not only offer an incredible tool for fundamental research, but also greatly benefit drug discovery. According to the Business Research Insights website, the global zebrafish model services market size was USD 434.4 million in the year 2022 and is projected to reach USD 618.23 million by the year 2031, with a compound annual growth rate (CAGR) of 14.4% during the forecast period.
Fish is recognized as a valuable source of high-quality protein and essential nutrients that are integral to a healthy human diet. Within the aquaculture industry, fish holds a primary position as the predominant source of cultivated seafood for human consumption. According to the European Commission’s Ocean and Fisheries website, marine and freshwater fish constitute approximately 49% of the total aquaculture production. Commonly consumed food fish species include gilthead seabream (Sparus aurata), meagre (Argyrosomus regius), and salmon (Salmo salar), which are saltwater species, while rainbow trout (Oncorhynchus mykiss) is a freshwater counterpart. In their natural habitats, such as the sea or rivers, healthy fish thrive without external interventions in terms of food and care. However, in fish farms, fish are reared within controlled or artificial environments, such as ponds, tanks, or cages, which necessitate external care and provisioning of food. Given the escalating global demand for aquaculture products, the industry faces significant pressure to enhance its supply. To meet this demand, fish farmers adopt intensive production practices, which can result in challenges like deteriorating water quality, higher fish density per unit of water volume, and limited food availability for the fish. These factors may contribute to stressed fish, the development of physical abnormalities, and susceptibility to serious diseases [16]. Fish with disease or deformities are rejected by the potential retailers or customers, thereby representing a significant economic loss to the fish farmers [17]. Major economic losses are directly due to the development of skeletal disorders altering the external shape of reared fish, i.e., opercular and vertebral column deformities [18]. Moreover, tedious technical effort and time are required to manually cull out the deformed fish from the productive cycle, which should be carried out as early as possible in order to not waste resources on growing suboptimal fish.
To detect and classify the deformities in the reared or model fish, manual inspection or analysis is employed, which requires significant time and technical effort. Moreover, direct physical interaction with the fish can induce fear or stress that may reflect on its behavior. Due to abnormal behavior or stress, fish can not swim or take proper diet, which can lead to poor health of the fish [16]. To improve animal welfare both in aquaculture and biomedical research, scientists are looking for methods requiring minimal manual interaction with the animals, with more focus on their health and quality of life. Computer vision is one such area that is increasingly being adopted by fish farmers and biomedical researchers to monitor the health and/or behavioral changes of the animal/fish. It may be helpful in identifying the causes of fish stress or any health hazard with minimal interaction with the animal. Computer vision and image processing techniques can also be helpful to speed up other routine procedures such as animal feeding [19], animal sorting, and animal counting by automatizing these tasks. According to the website, https://this.fish/blog/ai-guide-tracking-ais-explosive-growth-in-aquaculture/ (accessed on 2 December 2023) “this.fish”, the top 10 artificial intelligence (AI) and software start-up companies for the aquaculture industry have raised USD 282 million in the past 5 years, illustrating the importance and prevalence of AI-based smart farming in aquaculture.
Nowadays, automatic or semi-automatic computer-vision-based image processing techniques are being used in aquaculture industries and biomedical research to speed up the detection and diagnosis of diseases in the fish under study. These computer-vision-based techniques employ artificial intelligence methods such as machine learning or deep learning, which not only speed up the diagnosis but are also helpful in improving the accuracy of the detection. Deep learning represents a cutting-edge AI approach that empowers computers to learn from data and perform tasks on par with human capabilities. It utilizes multi-layered neural networks to directly acquire task-specific features from the data and make informed decisions when confronted with unseen data after the learning process. This methodology avoids the use of manually engineered features (as used in older image processing approaches) and, instead, defines a learning process that autonomously extracts features from the data, reducing the need for human intervention. In scenarios involving image data, such as image analysis, specialized types of neural networks known as deep convolutional neural networks (CNNs) have been designed. Such architectures play a pivotal role in computer vision applications, including tasks like object classification, object recognition, segmentation, and object counting. For the analysis of biomedical images, deep-learning-based convolutional neural networks (CNNs) are widely employed and increasingly favored for their accuracy, which rivals human performance [20]. Although debatable, CNNs are considered an imitation of the working of neurons in the human brain for visual perception and understanding objects in the images. They use a set of consecutive convolutional blocks or layers in order to understand the useful patterns for recognizing the objects in the images. The fundamental component of a CNN is the convolutional layer, comprising a collection of filters (or kernels), whose parameters are trained during the learning process [21][22]. Convolutional layers excel in extracting features from images by addressing spatial redundancy through weight sharing, and the features become more distinct and informative while going deeper into the layers. The role of the activation layer is to fire or activate the particular neurons while processing the information, and spatial invariance is achieved using pooling layers. In the end, a condensed feature representation is generated, encapsulating the essential content of the image in fully connected layers [23]. A typical CNN architecture for object classification tasks in images is shown in Figure 1.
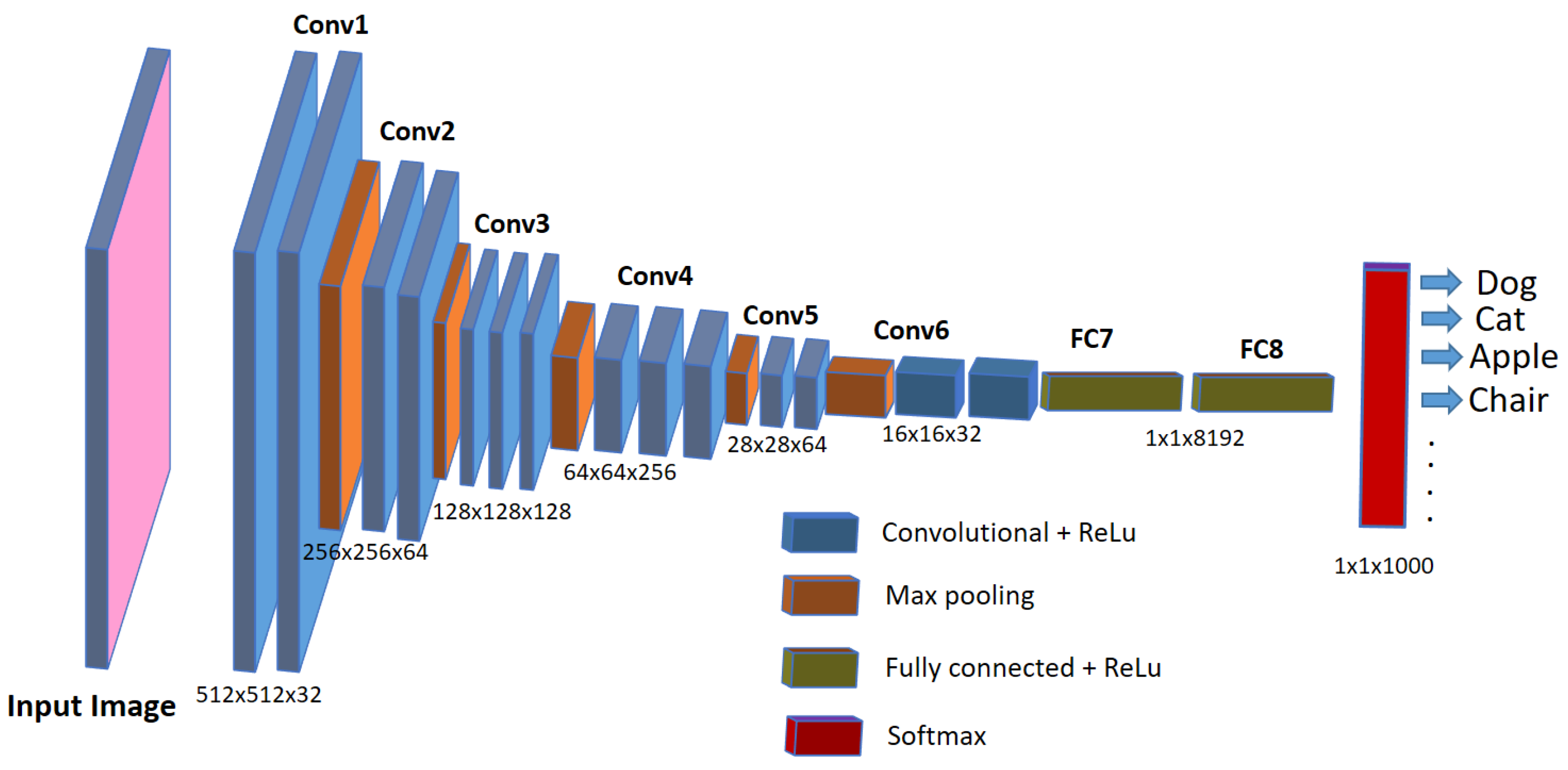
Figure 1. A typical CNN architecture for image classification tasks.
2. Imaging Techniques Used in Fish Bioimages
One of the main advantages of using zebrafish as a model animal over other animals is its transparent body during early, external development life stages, especially from 0 to 10 days post-fertilization (dpf). The transparent body of the larva makes it easy for the biologists to see through its developing organs and bones during in vivo studies and also helps to produce bioimage datasets using various image acquisition equipment [24][25]. Given that image acquisition precedes image analysis, it is crucial to employ suitable imaging methods and protocols to ensure effective and accurate image analysis, particularly when conducting AI-based image analysis. Due to the small size of the zebrafish and medaka embryos and larvae, advanced optical microscopy imaging methods are employed to capture maximum information at the microscopic level [26]. Microscopic imaging methods necessitate a meticulous pipeline to be adhered to, ensuring the prevention of unwarranted variations in acquisition adjustments and parameters that might introduce artifacts capable of influencing the outcomes of image analysis algorithms [27]. Beyond fundamental considerations like luminosity and focus control, special attention to the fish’s positioning and the characteristics of the glass plates is also needed to mitigate potential issues related to light refraction. This precautionary approach aims to prevent problems like shadowed areas in the images that could disrupt the subsequent analysis [28]. Since most phenotype and morphometric studies in biomedical research require capturing the fine-grained information at the sub-cellular level, microscopy methods such as bright-field or fluorescence microscopy are prevalent compared to other imaging approaches [29][30]. More recently, confocal and light-sheet microscopy deliver three-dimensional images [31], while Raman spectroscopy, Fourier-transform infrared spectroscopy, or mass spectrometry imaging are able to reveal the spatial distribution of individual (bio)molecules or classes of molecules [32][33][34][35], resulting in ever more high-content and demanding analysis requirements.
Apart from microscopy methods, X-ray radiography techniques are also popular in biomedical and aquaculture research for analyzing the skeletal structures of the juvenile and adult fish, including microCT imaging [36][37][38]. While microscopy imaging methods are employed in the early life stages (embryonic and larval) of the model fish due to its body’s optical clarity and small size, radiography methods are employed in the later life stages to visualize hard tissues. The adult model fish serves as a distinct and valuable resource for studying pathogenic and therapeutic aspects of adult human bone diseases. This is attributed to the fact that certain functions such as bone turnover, repair, degeneration, and metabolic responses are not fully mature in embryos [39]. Similarly, in aquaculture research, radiography imaging methods are utilized for juvenile and adult fish for several types of phenotype and morphometric studies [40][41].
References
- Selderslaghs, I.W.; Hooyberghs, J.; Blust, R.; Witters, H.E. Assessment of the developmental neurotoxicity of compounds by measuring locomotor activity in zebrafish embryos and larvae. Neurotoxicology Teratol. 2013, 37, 44–56.
- Rihel, J.; Prober, D.A.; Arvanites, A.; Lam, K.; Zimmerman, S.; Jang, S.; Haggarty, S.J.; Kokel, D.; Rubin, L.L.; Peterson, R.T.; et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 2010, 327, 348–351.
- Bugel, S.M.; Tanguay, R.L. Multidimensional chemobehavior analysis of flavonoids and neuroactive compounds in zebrafish. Toxicol. Appl. Pharmacol. 2018, 344, 23–34.
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721–731.
- Love, D.R.; Pichler, F.B.; Dodd, A.; Copp, B.R.; Greenwood, D.R. Technology for high-throughput screens: The present and future using zebrafish. Curr. Opin. Biotechnol. 2004, 15, 564–571.
- Scholz, S.; Fischer, S.; Gündel, U.; Küster, E.; Luckenbach, T.; Voelker, D. The zebrafish embryo model in environmental risk assessment—Applications beyond acute toxicity testing. Environ. Sci. Pollut. Res. 2008, 15, 394–404.
- Peterson, R.T.; MacRae, C.A. Systematic approaches to toxicology in the zebrafish. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 433–453.
- Braunbeck, T.; Kais, B.; Lammer, E.; Otte, J.; Schneider, K.; Stengel, D.; Strecker, R. The fish embryo test (FET): Origin, applications, and future. Environ. Sci. Pollut. Res. 2015, 22, 16247–16261.
- Hill, A.J.; Teraoka, H.; Heideman, W.; Peterson, R.E. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol. Sci. 2005, 86, 6–19.
- Ali, S.; Champagne, D.L.; Spaink, H.P.; Richardson, M.K. Zebrafish embryos and larvae: A new generation of disease models and drug screens. Birth Defects Res. Part Embryo Today Rev. 2011, 93, 115–133.
- Strähle, U.; Scholz, S.; Geisler, R.; Greiner, P.; Hollert, H.; Rastegar, S.; Schumacher, A.; Selderslaghs, I.; Weiss, C.; Witters, H.; et al. Zebrafish embryos as an alternative to animal experiments—A commentary on the definition of the onset of protected life stages in animal welfare regulations. Reprod. Toxicol. 2012, 33, 128–132.
- Pruvot, B.; Curé, Y.; Djiotsa, J.; Voncken, A.; Muller, M. Developmental defects in zebrafish for classification of EGF pathway inhibitors. Toxicol. Appl. Pharmacol. 2014, 274, 339–349.
- Spoorendonk, K.M.; Hammond, C.L.; Huitema, L.F.; Vanoevelen, J.; Schulte-Merker, S. Zebrafish as a unique model system in bone research: The power of genetics and in vivo imaging. J. Appl. Ichthyol. 2010, 26, 219–224.
- Zon, L.I.; Peterson, R.T. In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 2005, 4, 35–44.
- Driever, W.; Solnica-Krezel, L.; Schier, A.; Neuhauss, S.; Malicki, J.; Stemple, D.; Stainier, D.; Zwartkruis, F.; Abdelilah, S.; Rangini, Z.; et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 1996, 123, 37–46.
- Martos-Sitcha, J.A.; Mancera, J.M.; Prunet, P.; Magnoni, L.J. Welfare and stressors in fish: Challenges facing aquaculture. Front. Physiol. 2020, 11, 162.
- Llorente, I.; Fernández-Polanco, J.; Baraibar-Diez, E.; Odriozola, M.D.; Bjørndal, T.; Asche, F.; Guillen, J.; Avdelas, L.; Nielsen, R.; Cozzolino, M.; et al. Assessment of the economic performance of the seabream and seabass aquaculture industry in the European Union. Mar. Policy 2020, 117, 103876.
- Verhaegen, Y.; Adriaens, D.; De Wolf, T.; Dhert, P.; Sorgeloos, P. Deformities in larval gilthead sea bream (Sparus aurata): A qualitative and quantitative analysis using geometric morphometrics. Aquaculture 2007, 268, 156–168.
- Hu, W.C.; Chen, L.B.; Huang, B.K.; Lin, H.M. A computer vision-based intelligent fish feeding system using deep learning techniques for aquaculture. IEEE Sens. J. 2022, 22, 7185–7194.
- Santosh, K.; Das, N.; Ghosh, S. Deep Learning Models for Medical Imaging; Academic Press: Cambridge, MA, USA, 2021.
- Mostafa, S.; Wu, F.X. Diagnosis of autism spectrum disorder with convolutional autoencoder and structural MRI images. In Neural Engineering Techniques for Autism Spectrum Disorder; Elsevier: Amsterdam, The Netherlands, 2021; pp. 23–38.
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248.
- Guo, Y.; Liu, Y.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep learning for visual understanding: A review. Neurocomputing 2016, 187, 27–48.
- Hussain, S.; Aponte-Rivera, R.; Barghout, R.M.; Trapani, J.G.; Kindt, K.S. In Vivo Analysis of Hair Cell Sensory Organs in Zebrafish: From Morphology to Function. In Developmental, Physiological, and Functional Neurobiology of the Inner Ear; Humana: New York, NY, USA, 2022; pp. 175–220.
- Bauer, B.; Mally, A.; Liedtke, D. Zebrafish embryos and larvae as alternative animal models for toxicity testing. Int. J. Mol. Sci. 2021, 22, 13417.
- Mikut, R.; Dickmeis, T.; Driever, W.; Geurts, P.; Hamprecht, F.A.; Kausler, B.X.; Ledesma-Carbayo, M.J.; Marée, R.; Mikula, K.; Pantazis, P.; et al. Automated processing of zebrafish imaging data: A survey. Zebrafish 2013, 10, 401–421.
- Shamir, L.; Delaney, J.D.; Orlov, N.; Eckley, D.M.; Goldberg, I.G. Pattern recognition software and techniques for biological image analysis. PLoS Comput. Biol. 2010, 6, e1000974.
- Jeanray, N.; Marée, R.; Pruvot, B.; Stern, O.; Geurts, P.; Wehenkel, L.; Muller, M. Phenotype classification of zebrafish embryos by supervised learning. PLoS ONE 2015, 10, e0116989.
- Kuchmiy, A.; Efimov, G.; Nedospasov, S. Methods for in vivo molecular imaging. Biochemistry 2012, 77, 1339–1353.
- Abu-Siniyeh, A.; Al-Zyoud, W. Highlights on selected microscopy techniques to study zebrafish developmental biology. Lab. Anim. Res. 2020, 36, 12.
- Bruneel, B.; Witten, P.E. Power and challenges of using zebrafish as a model for skeletal tissue imaging. Connect. Tissue Res. 2015, 56, 161–173.
- Høgset, H.; Horgan, C.C.; Armstrong, J.P.; Bergholt, M.S.; Torraca, V.; Chen, Q.; Keane, T.J.; Bugeon, L.; Dallman, M.J.; Mostowy, S.; et al. In vivo biomolecular imaging of zebrafish embryos using confocal Raman spectroscopy. Nat. Commun. 2020, 11, 6172.
- Bennet, M.; Akiva, A.; Faivre, D.; Malkinson, G.; Yaniv, K.; Abdelilah-Seyfried, S.; Fratzl, P.; Masic, A. Simultaneous Raman microspectroscopy and fluorescence imaging of bone mineralization in living zebrafish larvae. Biophys. J. 2014, 106, L17–L19.
- Fiedler, I.A.; Schmidt, F.N.; Wölfel, E.M.; Plumeyer, C.; Milovanovic, P.; Gioia, R.; Tonelli, F.; Bale, H.A.; Jähn, K.; Besio, R.; et al. Severely impaired bone material quality in chihuahua zebrafish resembles classical dominant human osteogenesis imperfecta. J. Bone Miner. Res. 2018, 33, 1489–1499.
- da Silva, K.M.; Iturrospe, E.; Bars, C.; Knapen, D.; Van Cruchten, S.; Covaci, A.; van Nuijs, A.L. Mass spectrometry-based zebrafish toxicometabolomics: A review of analytical and data quality challenges. Metabolites 2021, 11, 635.
- Ding, Y.; Vanselow, D.J.; Yakovlev, M.A.; Katz, S.R.; Lin, A.Y.; Clark, D.P.; Vargas, P.; Xin, X.; Copper, J.E.; Canfield, V.A.; et al. Computational 3D histological phenotyping of whole zebrafish by X-ray histotomography. Elife 2019, 8, e44898.
- Merrifield, G.D.; Mullin, J.; Gallagher, L.; Tucker, C.; Jansen, M.A.; Denvir, M.; Holmes, W.M. Rapid and recoverable in vivo magnetic resonance imaging of the adult zebrafish at 7T. Magn. Reson. Imaging 2017, 37, 9–15.
- Babaei, F.; Hong, T.L.C.; Yeung, K.; Cheng, S.H.; Lam, Y.W. Contrast-enhanced X-ray micro-computed tomography as a versatile method for anatomical studies of adult zebrafish. Zebrafish 2016, 13, 310–316.
- Carnovali, M.; Banfi, G.; Mariotti, M. Zebrafish models of human skeletal disorders: Embryo and adult swimming together. BioMed Res. Int. 2019, 2019, 1253710.
- Dellacqua, Z.; Di Biagio, C.; Costa, C.; Pousão-Ferreira, P.; Ribeiro, L.; Barata, M.; Gavaia, P.J.; Mattei, F.; Fabris, A.; Izquierdo, M.; et al. Distinguishing the Effects of Water Volumes versus Stocking Densities on the Skeletal Quality during the Pre-Ongrowing Phase of Gilthead Seabream (Sparus aurata). Animals 2023, 13, 557.
- Beckmann, M.C.; Gilliam, J.F.; Langerhans, R.B. X-ray imaging as a time-saving, non-invasive technique for diet analysis. Fish. Res. 2015, 161, 1–7.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
548
Revisions:
2 times
(View History)
Update Date:
09 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




