Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Giuseppina Malcangi | -- | 3074 | 2024-01-04 13:43:23 | | | |
| 2 | Sirius Huang | Meta information modification | 3074 | 2024-01-05 03:08:26 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Bellocchio, L.; Dipalma, G.; Inchingolo, A.M.; Inchingolo, A.D.; Ferrante, L.; Del Vecchio, G.; Malcangi, G.; Palermo, A.; Qendro, A.; Inchingolo, F. COVID-19 on Oral Health. Encyclopedia. Available online: https://encyclopedia.pub/entry/53427 (accessed on 07 February 2026).
Bellocchio L, Dipalma G, Inchingolo AM, Inchingolo AD, Ferrante L, Del Vecchio G, et al. COVID-19 on Oral Health. Encyclopedia. Available at: https://encyclopedia.pub/entry/53427. Accessed February 07, 2026.
Bellocchio, Luigi, Gianna Dipalma, Angelo Michele Inchingolo, Alessio Danilo Inchingolo, Laura Ferrante, Gaetano Del Vecchio, Giuseppina Malcangi, Andrea Palermo, Andis Qendro, Francesco Inchingolo. "COVID-19 on Oral Health" Encyclopedia, https://encyclopedia.pub/entry/53427 (accessed February 07, 2026).
Bellocchio, L., Dipalma, G., Inchingolo, A.M., Inchingolo, A.D., Ferrante, L., Del Vecchio, G., Malcangi, G., Palermo, A., Qendro, A., & Inchingolo, F. (2024, January 04). COVID-19 on Oral Health. In Encyclopedia. https://encyclopedia.pub/entry/53427
Bellocchio, Luigi, et al. "COVID-19 on Oral Health." Encyclopedia. Web. 04 January, 2024.
Copy Citation
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and transmission are generally known to be produced by respiratory droplets and aerosols from the oral cavity (O.C.) of infected subjects, as stated by the World Health Organization. Saliva also retains the viral particles and aids in the spread of COVID-19.
COVID-19
SARS-CoV-2
saliva transmission
respiratory syndrome
infection
oral cavity (O.C.)
virus signs and symptoms
1. Introduction
In 2019, a new type of coronavirus, SARS-CoV-2 (Figure 1), the ethiopathological agent of COVID-19, was detected in Wuhan (China), and on 11 March 2020, it became a pandemic, according to the World Health Organization.
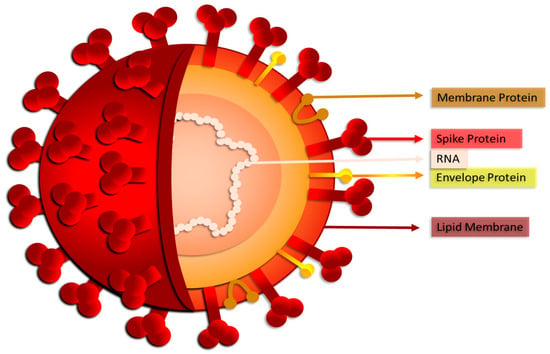
Figure 1. SARS-CoV-2: structure of the new coronavirus.
The manifestations of COVID-19 were mostly age-dependent, related to the clinical stage of the infection, and potentially more severe in cases already presenting co-morbidities. COVID-19 disease is characterized by runny nose and nasal congestion, anosmia, dysgeusia or hypogeusia, diarrhea, nausea/vomiting, respiratory distress, fatigue, ocular symptoms, diarrhea, vomiting, and abdominal pain. These systemic conditions were often accompanied by skin and mucosal lesions (Figure 2).
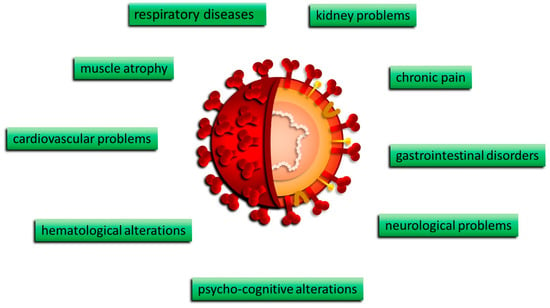
Figure 2. Clinical manifestations of COVID-19.
Several oral lesions were found in COVID-19 patients. Amongst them, the most commonly reported are: geographic tongue, herpes simplex, aphthous-like ulcers, candidiasis, hemorrhagic and necrotic ulcerations, erythematous surfaces, reddish macules, white hairy tongue, petechiae, and pustular enanthema [1][2]. In half of COVID-19 patients, viral infection was associated with taste loss, dry mouth, and mucosal lesions. Indeed, recent findings point out that the O.C. is not only the primary site of SARS-CoV-2 entrance and transmission but also a target for the disease’s clinical presentation [3][4]. The implication of oral tissue in COVID-19 pathogenesis is supported by growing evidence, which confirms the hypothesis of direct viral entry and replication of mucosal surfaces and the salivary glands [5][6][7].
SARS-CoV-2 penetrates cells in two different ways, such as endocytosis or host membrane-bound peptidases [8][9]. SARS-CoV-2 can start its viral envelope entrance by attaching its spike protein (S) to the metallopeptidase angiotensin-converting enzyme 2 (ACE2) that is present on the cellular membrane [10][11]. A protease derived from a host cell subsequently divides the spike into S1 and S2, respectively. S1 separates from the remainder related to the spike protein, and host cell-derived transmembrane serine protease 2 (TMPRSS2) further cleaves S2 (Figure 3) [12].
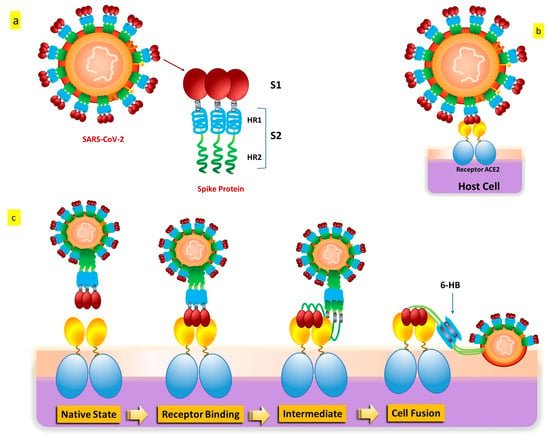
Figure 3. SARS-CoV-2 S protein: (a) The S protein’s schematic structure. (b) The S protein attaches itself to the ACE2 receptor. (c) The S protein-mediated binding and virus-cell fusion mechanisms.
This cleavage process results in exposure to the fusion peptide, which allows fusion of the membrane of the host cell as well as membrane and cell invasion [13]. Some in vitro evidence indicates that another entry factor that potentiates SARS-CoV-2’s infectivity [14] is represented by neuropilin-1 (NRP-1), a signaling protein highly present in the O.C. In addition to the well-known roles played by ACE2 and TMPRSS2, additional endosomal proteases (CTSB, CTSL, and BSG) and tissue-specific proteases (TMPRSS4 and TMPRSS11D) might facilitate the virus’s entrance into cells for intracellular reproduction [15][16]. Interaction of the cell membrane-based receptor ACE2 of the host cell with the spike protein of SARS-CoV-2 triggers the endosomal pathway internalization, which results in virus endocytosis [13][17]. After that, the endosome’s cathepsin L cleaves the spike protein into S1 and S2, allowing the viral capsid to merge with the membrane within the endosome [18][19][20]. The virus genome is therefore released because of endosomal processing, allowing it to begin replicating and producing new viral particles. Therefore, it appears that ACE2 binding and TMPRSS2 cleavage are the two primary crucial components in the SARS-CoV-2 infection process, even if several pathways and intracellular players are involved [21][22].
2. SARS-CoV-2 Transmission via the Saliva
The main paths for transmission for SARS-CoV-2 are Respiratory droplets, Flügge (Figure 4), which originate from the nose, O.C., and airways [23][24][25][26], making saliva the most significant droplet [27][28][29]. In the O.C., SARS-CoV-2 viral RNA titers range generally from 102 to 1010 copies/mL, reaching, in the first seven days, the highest concentrations of symptom appearance and declining with recovery over time [30][31].
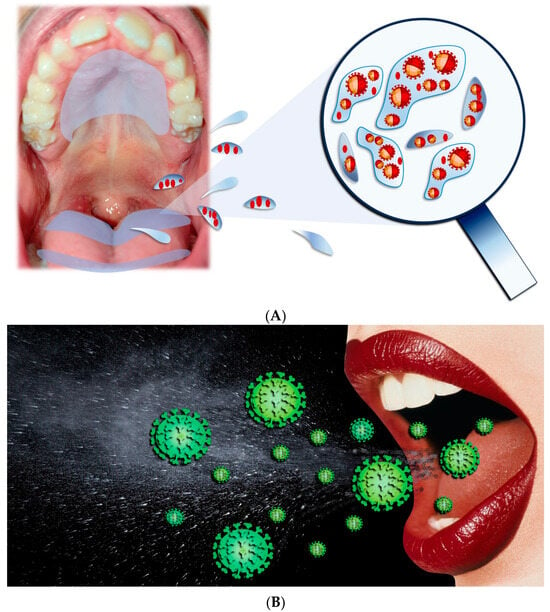
Figure 4. (A,B) Respiratory droplets originated from the O.C.
In saliva, fragments of oral mucosa, as well as salivary gland cells, are normally and mutually present. Indeed, in saliva samples obtained from light COVID-19 people, SARS-CoV-2 was reported to infect about 5–10% of salivary cells (pan-cytokeratin positive, pCK+) [27]. The observation that in lost cells in the salivary epithelium viral replication is happening is increasing the potential of saliva in the spread of infection and disease transmission. Also, suprabasal mucosal cells express all factors required for both SARS-CoV-2 entry and replication. These cells, which are normally shed as a potential protective phenomenon in oral tissue infection [32], are generated from the layers of tissue that are most terminally differentiated every 3 h and can represent a way SARS-CoV-2 is applicable to spread into saliva [8]. In vitro studies supported this hypothesis, showing that these cells were able to transmit a high viral load [27][33] from saliva to Vero cells in an ex vivo experimental setup. Moreover, a cell population normally present within the respiratory tract (i.e., pCK+ ciliated cells) was identified in the saliva while being positive for SARS-CoV-2. Thus SARS-CoV2 infection and propagation in the O.C. [34][35] with subsequent sustained COVID-19 in different body sites might be partly due to the high saliva viral load after the spreading of the cell population within the respiratory tract in this body fluid [34]. Furthermore, the lower respiratory tract being infected as the digestive apparatus might involve exfoliated epithelial cells present in saliva, containing active SARS-CoV-2 particles [36][37].
Refs. [27][38][39] a stable capacity for infection in fresh cell monolayers was also attributed to viral particles from culture supernatants with a cytopathic effect [40]. Altogether, these findings further strengthen the idea of saliva’s capacity to spread SARS-CoV-2, as pathogenic virus and diseased cells found in oral droplets that were ejected, including individuals without apparent symptoms or those in the early stages of the infection, represent the origin of airborne transmission [27][41][42]. SARS-CoV-2 can be detected in saliva for extended periods of time; for example, in asymptomatic subjects, weeks pass between the initial test and the negative saliva test result, a delay that is even longer in symptomatic COVID-19 patients [27][43]. The virus SARS-CoV-2 excludes the ability to be removed from the nasopharynx in saliva, as suggested by other observations, and this might indicate prolonged viral shedding from oral areas affected by SARS-CoV-2 [27][44][45].
Also, the periodontium has been cited as a possible site for SARS-CoV-2 replication and subsequent release. From this tissue, saliva and O.C. are highly accessible to the virus [46]. However, it can potentially disseminate to other distant organs by entering the local periodontal capillary network’s circulation. Thus, the mouth cavity is a key node not only as a potential external source of infection but also for the recurrence and development of systemic COVID-19 disease [47][48][49]. A recent postmortem investigation discovered the presence of SARS-CoV-2 RNA in the periodontal tissues of COVID-19-positive patients, perhaps indicating the virus’s existence within crevicular fluid several days after the beginning of the first symptoms [50]. Therefore, the periodontal pocket may serve as an advantageous reservoir for latent and active SARS-CoV-2 isoforms [5][47]. As a result, the mouth cavity is a crucial location and a possible pathway for the cellular and acellular particles of SARS-CoV-2 to become infected through saliva [2][27].
In addition to the reasons listed above that make saliva dangerous for the general public, other infectious material can expose dental practitioners during routine procedures in O.C. Components facilitating the entry and spread of SARS-CoV-2 were identified in dental pulp tissues, and lesions around the tooth root [27][51][52] suggest the possibility of the virus colonizing pulp tissues during pathological conditions such as caries or through a bloodstream-related infection in the pulp [53]. Dental and periodontal procedures may pose transmission risks of SARS-CoV-2 to dental professionals. Therefore, implementing specific preventive measures is crucial to mitigating the transmission of the disease during O.C.-related interventions [54][55]. We can therefore underline how much connection there is between SARS-CoV-2 infection and oral health. The O.C. stands as one of the initial points of entry for the virus; viral particles present in saliva can be widespread through direct contact or sharing contaminated objects. This has important implications for the propagation of the virus inside communities and highlights the importance of oral hygiene. People with pre-existing oral conditions, such as periodontitis or other gum disease, may be at potentially greater risk of becoming infected with COVID-19 or developing more severe forms of the disease. This may be attributable to local immune system impairment in inflamed areas of the mouth, making the body more susceptible to SARS-CoV-2 infection. The pandemic has led to changes in dental practices, including the adoption of more rigorous protocols for infection prevention. This included the use of protective equipment, changes in procedures to minimize the production of aerosols, and the implementation of social distancing measures in dental centers.
The connection between COVID-19 and oral health has been the subject of ongoing study to better understand the impact of the infection on the mouth and vice versa. Continuing to follow health guidelines, including good oral hygiene, is important not only for preventing COVID-19 infection but also for maintaining overall good oral and dental health [56][57].
3. SARS-CoV-2 and Oral Cavity: A New Entry Route in the Body
3.1. Salivary Glands
One major entry point for the infection of SARS-CoV-2 appears to be the epithelia of the glands that produce saliva. Here, virus entry factors are expressed at very high levels compared to other O.C. epithelial cells. The SARS-CoV-2 virus can infect the upper airways, including the mouth, in several ways:
-
The virus can enter the O.C. through the upper respiratory tract, such as the nose and throat, mainly via respiratory droplets released when an infected person coughs, sneezes, or talks. Viral particles can be inhaled or deposited on the surfaces of the mouth and nose.
-
Once in the O.C., the SARS-CoV-2 virus binds to ACE2 receptors present on host cells. This is the entry point of the virus into human cells.
-
The virus can penetrate the oral mucosa through adhesion and invasion of the epithelial cells present in this region. This process could be favored by lesions or microlesions in the mucosa, providing an entry route for the virus.
-
Once inside the oral mucosa, the virus can be transmitted to the salivary glands via the lymphatic or circulatory system. In saliva, the existence of SARS-CoV-2 has been observed, suggesting that the virus may be transported through the saliva itself or through the bloodstream.
ACE2 and TMPRSS2 immunoreactivity was found both in mucous and serous sacs within the labial gland, with stronger TMPRSS2 levels in the acini; moreover, ACE2 was detected in the striated ducts; however, TMPRSS2 staining seemed to show a negative result [58][59]. Thus, SARS-CoV-2 might primarily attach to the mucosa of O.C., the ductal opening of the salivary glands, and the small salivary glands scattered throughout the oral mucosa [58]. Zhu and colleagues analyzed the location of SARS-CoV-2 entry points, offering insights into the primary salivary glands of individuals with non-malignant conditions [60]. Specifically, within the submandibular and parotid glands, ACE2 and TMPRSS2 proteins were detected in the cytoplasm and cellular membrane of serous acinar cells, ductal epithelial cells, and mixed acini’s serous acinar cells within the sublingual glands [6][61]. Lower levels of ACE2 and TMPRSS2 were found in the submandibular, parotid, and sublingual glands, respectively, with Western blot analysis for protein quantification (particularly for TMPRSS2) [60][62].
Single-cell RNA sequencing revealed co-expression of ACE2 and protease TMPRSS2 (together with additional proteases CTSB and CTSL) in the salivary gland epithelial cells (parotid, labial minor, and submandibular), and co-in situ hybridization investigations confirmed these findings. It is interesting to note that the authors discovered distinct tissue-specific protease expression patterns, with TMPRSS2 abundant in the epithelia of salivary glands and TMPRSS11D rich in mucosal keratinocytes [63]. These differential patterns of protease expression might be indicative of tissue-specific infection routes [27][64][65]. On the other hand, the endosomal proteases CTSB and CTSL showed broader expression levels across epithelia [66][67]. Indeed, the entry factors were more highly expressed in the salivary glands (especially the minor ones) than in the O.C. mucosa; importantly, significant co-expression of the principal entry factors ACE2 and TMPRSS2 was found in acini and duct epithelial cells [58][68][69]. The levels of these factors in the minor salivary glands were comparable to those found in the respiratory tract and gastrointestinal tract [27][70][71]. In salivary gland homogenates, SARS-CoV-2 spike proteins were able to attach to human parotid, submandibular, and sublingual gland cells [60], confirming their infectious potential at this anatomical site [62][72][73]. Like the observations for the oral mucosa, Huang and colleagues reported SARS-CoV-2 replication in salivary glands. Indeed, using minor salivary glands from corpses and a person who was severely infected with COVID-19, the authors reported the presence of replicated viral particles in infected ducts and acini and a lower infection in parotid salivary glands [60][74]. Submandibular gland infection by SARS-CoV-2 was reported in two different studies [50][75][76]. In 60% of submandibular and parotid gland specimens, an electron microscopy study of postmortem biopsies of fatal COVID-19 patients revealed spherical 70–100 nm virus particles positive for SARS-CoV-2 RNA (consistent in size and shape with the Coronaviridae family) [50][77]. However, it is important to underline that scientific research in this field is continually developing, and there are still many questions without definitive answers. A complete understanding of the specific mechanisms by which the virus infects the O.C. and salivary glands requires further studies and insights.
It is always advisable to follow recommended health guidelines, such as hand hygiene, wearing masks, and social distancing, to reduce the risk of contracting or spreading the virus.
3.2. Tongue
Gustatory dysfunction affected around 40% of COVID-19 patients, and it mostly manifested two to fourteen days after exposure to SARS-CoV-2 [5][6]. This can be explained by the SARS-CoV-2 invasion of entry taste papillae cells, resulting in cellular harm and leading to the clinical symptom of dysgeusia [78]. In the mucous membrane of the tongue, immunohistochemistry studies showed ACE2 expression within the cell cytoplasm and across the cellular membrane within the non-keratinized area, alongside TMPRSS2 positioning specifically on the cellular membrane [79][80]. ACE2 was detected in minor quantities within the lamina propria of non-keratinized stratified squamous epithelia in mucosal structures. Conversely, TMPRSS2 expression was absent in the stratum basale or within the lamina propria [58][81][82]. Extended immunohistochemical examination revealed ACE2 presence within the nucleus and TMPRSS2 within the cytoplasm of taste cells in the papillae. These findings were additionally corroborated by RNA analysis conducted on human fungiform papillae taste cells [82]. Xu and colleagues further confirm ACE2 receptor presence in the tongue (especially abundant in epithelial cells) by using single-cell sequencing [83][84].
As already mentioned for the oral mucosa, the existence of SARS-CoV-2 entry components within the tongue might represent another infection gateway. Indeed, in SARS-CoV-2-infected subjects as well as autopsy patients, viral infection was found in the dorsal tongue [27][85][86]. Seventy-one percent of COVID-19 patients exhibited cytological smears from the tongue’s dorsum, with epithelial cells testing positive for the SARS-CoV-2 spike protein [87][88].
3.3. Oral Mucosa
Expression of TMPRSS and ACE2, together with other cellular factors for SARS-CoV-2 infection, has been identified in the tongue, oral mucosa, and salivary glands [81][89][90]. Immunohistochemistry studies showed ACE2 expression in the cytoplasm and on the cell membrane in the non-keratinized buccal mucosa, as well as TMPRSS2 localization on the cell membrane [91]. ACE2 was detected in both the cytoplasm and on the cell membrane, while TMPRSS2 was found solely on the cell membrane within the non-keratinized stratified squamous epithelia of the labial mucosa. Conversely, within the buccal mucosa, significant ACE2 levels were observed in the lamina propria, with no expression of TMPRSS2 noted in either the stratum basale or the lamina propria [58][92]. ACE2 was observed within the cytoplasm and on the cell membrane, while TMPRSS2 was specifically identified on the cell membrane. These were detected in the keratinized stratified squamous epithelia, primarily localized in the stratum granulosum and stratum spinosum, with no presence noted in the stratum basale [56]. Another study corroborated these findings, identifying immunoreactivity of both ACE2 and TMPRSS2 within the stratified squamous epithelium of the gingiva, specifically prevalent in the keratinized surface layer [85]. In the same sites, this study also showed the positioning of furin [82][93][94]. Furin is the second protease, in addition to TMPRSS2, used by SARS-CoV-2 to cleave the Spike protein that anchors to the cell membrane, without whose cleavage the cellular entry of the virus would not take place and therefore neither would replication and infection [95][96][97]. Another study by Okui and colleagues, utilizing gingival cells obtained from the gingival sulcus, demonstrated ACE2 immunoreactivity comparable to levels found in the tongue. Historically considered one of the primary entry routes for SARS-CoV-2, alongside the salivary glands, the gingiva showed consistent ACE2 expression in this study conducted by colleagues [36][98][99][100]. In the same study, low ACE2 expression levels were also found within the keratinized mucosa in the palate [100][101][102]. Through single-cell RNA sequencing analysis, SARS-CoV-2 entry factors were identified across various subtypes of oral epithelial cells. Specifically, mucosal keratinocytes were found to express ACE2, TMPRSS2, as well as the endosomal proteases CTSB and CTSL [21][27][103]. Xu and colleagues provided further single-cell sequencing data showing ACE2 receptor expression within the tissue of the inner cheek and the gums [83][104]. Interestingly, the co-expression levels of ACE2 and TMPRSS2 in the oral mucosa were found to be similar to those observed in nasal and intestinal epithelial cells [105][106][107], the best known sites of SARS-CoV-2 infection [108][109]. In healthy adult tissue samples of the inner cheek, both suprabasal and basal compartments (non-keratinized) displayed ACE2 and TMPRSS2 expression, identified through in situ hybridization. Similar findings were observed in the soft palate and palatine tissue [40][83][110].
Thus, multiple regions of the O.C. are potential targets for SARS-CoV-2 infection, carrying the potential for viral transmission to both the respiratory and gastrointestinal tracts [100]. ACE2 and TMPRSS2 presence in the periodontal pocket and sulcular epithelia, coupled to the crevices, gingival sulcus, and periodontal pocket microenvironment, provide all conditions conducive to virus replication and sustainability [91][111].
Huang and colleagues clearly demonstrated the presence of SARS-CoV-2 colonization within the oral mucosa [27][112]. Spike protein was detected independently within shed epithelial cells and on their membrane surface. Infection and replication of SARS-CoV-2 were observed across all layers of the mucosa, with signs of infection also detected in mucosal scrapings [113].
3.4. Dental Pulp
SARS-CoV-2 entry components, namely ACE2 and TMPRSS2, exhibit substantial expression in both healthy and inflamed human dental pulp, as revealed by comprehensive transcriptomic analysis [114]. Accordingly, another study confirmed these findings by demonstrating RNA expression of ACE2, TMPRSS2, and NRP1 within healthy pulp tissues [52], indicating that SARS-CoV-2 infection can occur in pulp regardless of inflammatory status [115][116].
References
- Rusu, L.C.; Ardelean, L.C.; Tigmeanu, C.V.; Matichescu, A.; Sauciur, I.; Bratu, E.A. COVID-19 and Its Repercussions on Oral Health: A Review. Medicina 2021, 57, 1189. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8619825/ (accessed on 21 November 2023).
- Maggialetti, N.; Piemonte, S.; Sperti, E.; Inchingolo, F.; Greco, S.; Lucarelli, N.M.; De Chirico, P.; Lofino, S.; Coppola, F.; Catacchio, C.; et al. Iatrogenic Barotrauma in COVID-19-Positive Patients: Is It Related to the Pneumonia Severity? Prevalence and Trends of This Complication Over Time. Biomedicines 2022, 10, 2493.
- Villarroel-Dorrego, M.; Chacón, L.; Rosas, R.; Barrios, V.; Pernía, Y.; Vélez, H. Oral Findings in Patients With COVID-19. Actas Dermo-Sifiliográficas 2022, 113, T183–T186.
- Kusiak, A.; Cichońska, D.; Tubaja, M.; Skorek, A.; Jereczek-Fossa, B.A.; Corrao, G.; Marvaso, G.; Alterio, D. COVID-19 Manifestation in the Oral Cavity—A Narrative Literature Review. Acta Otorhinolaryngol. Ital. 2021, 41, 395–400.
- Brandini, D.A.; Takamiya, A.S.; Thakkar, P.; Schaller, S.; Rahat, R.; Naqvi, A.R. COVID-19 and Oral Diseases: Crosstalk, Synergy or Association? Rev. Med. Virol. 2021, 31, e2226.
- Tsuchiya, H. Characterization and Pathogenic Speculation of Xerostomia Associated with COVID-19: A Narrative Review. Dent. J. 2021, 9, 130.
- Dolci, C.; Cenzato, N.; Maspero, C.; Giannini, L.; Khijmatgar, S.; Dipalma, G.; Tartaglia, G.M.; Inchingolo, F. Skull Biomechanics and Simplified Cephalometric Lines for the Estimation of Muscular Lines of Action. J. Pers. Med. 2023, 13, 1569.
- Lee, W.S.; Yousefi, M.; Yan, B.; Yong, C.L.; Ooi, Y.S. Know Your Enemy and Know Yourself—The Case of SARS-CoV-2 Host Factors. Curr. Opin. Virol. 2021, 50, 159–170. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8379091/ (accessed on 21 November 2023).
- Barthe, M.; Hertereau, L.; Lamghari, N.; Osman-Ponchet, H.; Braud, V.M. Receptors and Cofactors That Contribute to SARS-CoV-2 Entry: Can Skin Be an Alternative Route of Entry? Int. J. Mol. Sci. 2023, 24, 6253. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10094153/ (accessed on 21 November 2023).
- Scarano, A.; Inchingolo, F.; Lorusso, F. Facial Skin Temperature and Discomfort When Wearing Protective Face Masks: Thermal Infrared Imaging Evaluation and Hands Moving the Mask. Int. J. Environ. Res. Public Health 2020, 17, 4624.
- Bohn, M.K.; Hall, A.; Sepiashvili, L.; Jung, B.; Steele, S.; Adeli, K. Pathophysiology of COVID-19: Mechanisms Underlying Disease Severity and Progression. Physiology 2020, 35, 288.
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149.
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. Available online: https://www.nature.com/articles/s41580-021-00418-x (accessed on 17 November 2023).
- Mayi, B.S.; Leibowitz, J.A.; Woods, A.T.; Ammon, K.A.; Liu, A.E.; Raja, A. The Role of Neuropilin-1 in COVID-19. PLoS Pathog. 2021, 17, e1009153. Available online: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1009153 (accessed on 8 November 2023).
- Singh, M.; Bansal, V.; Feschotte, C. A Single-Cell RNA Expression Map of Human Coronavirus Entry Factors. Cell Rep. 2020, 32, 108175.
- Balzanelli, M.G.; Distratis, P.; Lazzaro, R.; Dipalma, G.; Inchingolo, F.; Del Prete, R.; Hung Pham, V.; Aityan, S.K.; Nguyen, K.C.; Isacco Gargiulo, C. The Long COVID-19 Syndrome the Spike Protein and Stem Cells, the Underrated Role of Retrotransposons, a Working Hypothesis. Preprints 2023, 2023081130. Available online: https://www.preprints.org/manuscript/202308.1130/v1 (accessed on 14 November 2023).
- Demi, L.; Mento, F.; Di Sabatino, A.; Fiengo, A.; Sabatini, U.; Macioce, V.N.; Robol, M.; Tursi, F.; Sofia, C.; Di Cienzo, C.; et al. Lung Ultrasound in COVID-19 and Post-COVID-19 Patients, an Evidence-Based Approach. J. Ultrasound Med. 2022, 41, 2203–2215.
- Khan, U.; Afrakhteh, S.; Mento, F.; Fatima, N.; De Rosa, L.; Custode, L.L.; Azam, Z.; Torri, E.; Soldati, G.; Tursi, F.; et al. Benchmark Methodological Approach for the Application of Artificial Intelligence to Lung Ultrasound Data from COVID-19 Patients: From Frame to Prognostic-Level. Ultrasonics 2023, 132, 106994.
- Ghosh, S.; Dhobley, A.; Avula, K.K.; Joseph, S.; Gavali, N.; Sinha, S. Role of Saliva as a Non-Invasive Diagnostic Method for Detection of COVID-19. Cureus 2022, 14, e27471. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9421123/ (accessed on 26 November 2023).
- Daniell, H.; Nair, S.K.; Esmaeili, N.; Wakade, G.; Shahid, N.; Ganesan, P.K.; Islam, M.R.; Shepley-McTaggart, A.; Feng, S.; Gary, E.N.; et al. Debulking SARS-CoV-2 in Saliva Using Angiotensin Converting Enzyme 2 in Chewing Gum to Decrease Oral Virus Transmission and Infection. Mol. Ther. 2022, 30, 1966.
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 Entry Factors Are Highly Expressed in Nasal Epithelial Cells Together with Innate Immune Genes. Nat. Med. 2020, 26, 681–687.
- Le, T.H.; Balzanelli, M.G.; Le, T.V.; le Quoc, T.; Thuy, D.D.; Distratis, P.; Lazzaro, R.; Prete, R.D.; Triggiano, F.; Gargiulo, C.I.; et al. A Different Perspective on SARS-CoV-2 Pandemic: Data, Outcomes and Demographic Analysis of a Study Conducted at General Hospital Ninh Thuan Province in Vietnam in 2022. Preprint 2023.
- Balzanelli, M.G.; Distratis, P.; Lazzaro, R.; D’Ettorre, E.; Nico, A.; Inchingolo, F.; Dipalma, G.; Tomassone, D.; Serlenga, E.M.; Dalagni, G.; et al. New Translational Trends in Personalized Medicine: Autologous Peripheral Blood Stem Cells and Plasma for COVID-19 Patient. J. Pers. Med. 2022, 12, 85.
- Vaz, S.N.; de Santana, D.S.; Netto, E.M.; Pedroso, C.; Wang, W.-K.; Santos, F.D.A.; Brites, C. Saliva Is a Reliable, Non-Invasive Specimen for SARS-CoV-2 Detection. Braz. J. Infect. Dis. 2020, 24, 422–427.
- Scarano, A.; Inchingolo, F.; Lorusso, F. Environmental Disinfection of a Dental Clinic during the COVID-19 Pandemic: A Narrative Insight. BioMed Res. Int. 2020, 2020, 8896812.
- Lo Giudice, R. The Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) in Dentistry. Management of Biological Risk in Dental Practice. Int. J. Environ. Res. Public Health 2020, 17, 3067.
- Huang, N.; Pérez, P.; Kato, T.; Mikami, Y.; Okuda, K.; Gilmore, R.C.; Conde, C.D.; Gasmi, B.; Stein, S.; Beach, M.; et al. SARS-CoV-2 Infection of the Oral Cavity and Saliva. Nat. Med. 2021, 27, 892–903.
- Demi, L.; Wolfram, F.; Klersy, C.; De Silvestri, A.; Ferretti, V.V.; Muller, M.; Miller, D.; Feletti, F.; Wełnicki, M.; Buda, N.; et al. New International Guidelines and Consensus on the Use of Lung Ultrasound. J. Ultrasound Med. 2023, 42, 309–344.
- Cazzolla, A.P.; Lovero, R.; Spirito, F.; Di Cosola, M.; Santacroce, L.; Muzio, E.L.; Ciavarella, D.; Dioguardi, M.; Crincoli, V.; Pepe, M.; et al. Evaluation of Qualitative and Quantitative Taste Alterations in COVID-19. Biomol. Biomed. 2023, 23, 344. Available online: https://pubmed.ncbi.nlm.nih.gov/35801415/ (accessed on 26 November 2023).
- Dioguardi, M.; Cazzolla, A.P.; Arena, C.; Sovereto, D.; Caloro, G.A.; Dioguardi, A.; Crincoli, V.; Laino, L.; Troiano, G.; Lo Muzio, L. Innate Immunity in Children and the Role of ACE2 Expression in SARS-CoV-2 Infection. Pediatr. Rep. 2021, 13, 363–382.
- Cazzolla, A.P.; Lovero, R.; Lo Muzio, L.; Testa, N.F.; Schirinzi, A.; Palmieri, G.; Pozzessere, P.; Procacci, V.; Di Comite, M.; Ciavarella, D.; et al. Taste and Smell Disorders in COVID-19 Patients: Role of Interleukin-6. ACS Chem. Neurosci. 2020, 11, 2774–2781.
- Dawes, C. Estimates, from Salivary Analyses, of the Turnover Time of the Oral Mucosal Epithelium in Humans and the Number of Bacteria in an Edentulous Mouth. Arch. Oral Biol. 2003, 48, 329–336.
- Pham, V.H.; Pham, H.T.; Balzanelli, M.G.; Distratis, P.; Lazzaro, R.; Nguyen, Q.V.; Tran, V.Q.; Tran, D.K.; Phan, L.D.; Pham, S.M.; et al. Multiplex RT Real-Time PCR Based on Target Failure to Detect and Identify Different Variants of SARS-CoV-2: A Feasible Method That Can Be Applied in Clinical Laboratories. Diagnostics 2023, 13, 1364.
- Silva, J.; Lucas, C.; Sundaram, M.; Israelow, B.; Wong, P.; Klein, J.; Tokuyama, M.; Lu, P.; Venkataraman, A.; Liu, F.; et al. Saliva Viral Load Is a Dynamic Unifying Correlate of COVID-19 Severity and Mortality. MedRxiv Prepr. Serv. Health Sci. 2021.
- Bustamante-Marin, X.M.; Ostrowski, L.E. Cilia and Mucociliary Clearance. Cold Spring Harb. Perspect. Biol. 2017, 9, a028241. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5378048/ (accessed on 20 December 2023).
- Xiang, Z.; Koo, H.; Chen, Q.; Zhou, X.; Liu, Y.; Simon-Soro, A. Potential Implications of SARS-CoV-2 Oral Infection in the Host Microbiota. J. Oral Microbiol. 2021, 13, 1853451.
- Mohapatra, R.K.; Dhama, K.; Mishra, S.; Sarangi, A.K.; Kandi, V.; Tiwari, R.; Pintilie, L. The Microbiota-Related Coinfections in COVID-19 Patients: A Real Challenge. Beni-Suef Univ. J. Basic Appl. Sci. 2021, 10, 47.
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg. Infect. Dis. 2020, 26, 1266–1273.
- Marchesan, J.T.; Warner, B.M.; Byrd, K.M. The “Oral” History of COVID-19: Primary Infection, Salivary Transmission, and Post-acute Implications. J. Periodontol. 2021, 92, 1357–1367.
- Drozdzik, A.; Drozdzik, M. Oral Pathology in COVID-19 and SARS-CoV-2 Infection-Molecular Aspects. Int. J. Mol. Sci. 2022, 23, 1431.
- Tang, S.; Mao, Y.; Jones, R.M.; Tan, Q.; Ji, J.S.; Li, N.; Shen, J.; Lv, Y.; Pan, L.; Ding, P.; et al. Aerosol Transmission of SARS-CoV-2? Evidence, Prevention and Control. Environ. Int. 2020, 144, 106039.
- Balzanelli, M.G.; Distratis, P.; Lazzaro, R.; Pham, V.H.; Tran, T.C.; Dipalma, G.; Bianco, A.; Serlenga, E.M.; Aityan, S.K.; Pierangeli, V.; et al. Analysis of Gene Single Nucleotide Polymorphisms in COVID-19 Disease Highlighting the Susceptibility and the Severity towards the Infection. Diagn. Basel 2022, 12, 2824.
- De Maria, L.; Sponselli, S.; Caputi, A.; Stefanizzi, P.; Pipoli, A.; Giannelli, G.; Delvecchio, G.; Tafuri, S.; Inchingolo, F.; Migliore, G.; et al. SARS-CoV-2 Breakthrough Infections in Health Care Workers: An Italian Retrospective Cohort Study on Characteristics, Clinical Course and Outcomes. J. Clin. Med. 2023, 12, 628.
- Zhu, J.; Guo, J.; Xu, Y.; Chen, X. Viral Dynamics of SARS-CoV-2 in Saliva from Infected Patients. J. Infect. 2020, 81, e48–e50.
- Puhach, O.; Meyer, B.; Eckerle, I. SARS-CoV-2 Viral Load and Shedding Kinetics. Nat. Rev. Microbiol. 2023, 21, 147–161.
- Drozdzik, A. COVID-19 and SARS-CoV-2 Infection in Periodontology: A Narrative Review. J. Periodontal Res. 2022, 57, 933–941.
- Badran, Z.; Gaudin, A.; Struillou, X.; Amador, G.; Soueidan, A. Periodontal Pockets: A Potential Reservoir for SARS-CoV-2? Med. Hypotheses 2020, 143, 109907.
- Botros, N.; Iyer, P.; Ojcius, D.M. Is There an Association between Oral Health and Severity of COVID-19 Complications? Biomed. J. 2020, 43, 325–327.
- Inchingolo, A.D.; Gargiulo, C.I.; Malcangi, G.; Ciocia, A.M.; Patano, A.; Azzollini, D.; Piras, F.; Barile, G.; Settanni, V.; Mancini, A.; et al. Diagnosis of SARS-CoV-2 during the Pandemic by Multiplex RT-rPCR hCoV Test: Future Perspectives. Pathogens 2022, 11, 1378.
- Fernandes Matuck, B.; Dolhnikoff, M.; Maia, G.V.A.; Isaac Sendyk, D.; Zarpellon, A.; Costa Gomes, S.; Duarte-Neto, A.N.; Rebello Pinho, J.R.; Gomes-Gouvêa, M.S.; Sousa, S.C.O.M.; et al. Periodontal Tissues Are Targets for SARS-CoV-2: A Post-Mortem Study. J. Oral Microbiol. 2020, 13, 1848135.
- Inchingolo, A.D.; Malcangi, G.; Ceci, S.; Patano, A.; Corriero, A.; Azzollini, D.; Marinelli, G.; Coloccia, G.; Piras, F.; Barile, G.; et al. Antispike Immunoglobulin-G (IgG) Titer Response of SARS-CoV-2 mRNA-Vaccine (BNT162b2): A Monitoring Study on Healthcare Workers. Biomedicines 2022, 10, 2402.
- Altaie, A.M.; Hamdy, R.; Venkatachalam, T.; Hamoudi, R.; Soliman, S.S.M. Estimating the Viral Loads of SARS-CoV-2 in the Oral Cavity When Complicated with Periapical Lesions. BMC Oral Health 2021, 21, 567.
- Amante, L.F.L.S.; Afonso, J.T.M.; Skrupskelyte, G. Dentistry and the COVID-19 Outbreak. Int. Dent. J. 2021, 71, 358–368.
- Izzetti, R.; Nisi, M.; Gabriele, M.; Graziani, F. COVID-19 Transmission in Dental Practice: Brief Review of Preventive Measures in Italy. J. Dent. Res. 2020, 99, 1030–1038. Available online: https://journals.sagepub.com/doi/full/10.1177/0022034520920580 (accessed on 15 November 2023).
- Atukorallaya, D.S.; Ratnayake, R.K. Oral Mucosa, Saliva, and COVID-19 Infection in Oral Health Care. Front. Med. 2021, 8, 656926.
- Di Spirito, F.; Amato, A.; Di Palo, M.P.; Contaldo, M.; D’Ambrosio, F.; Lo Giudice, R.; Amato, M. Oral Lesions Following Anti-SARS-CoV-2 Vaccination: A Systematic Review. Int. J. Environ. Res. Public. Health 2022, 19, 10228.
- Coke, C.J.; Davison, B.; Fields, N.; Fletcher, J.; Rollings, J.; Roberson, L.; Challagundla, K.B.; Sampath, C.; Cade, J.; Farmer-Dixon, C.; et al. SARS-CoV-2 Infection and Oral Health: Therapeutic Opportunities and Challenges. J. Clin. Med. 2021, 10, 156.
- Sawa, Y.; Ibaragi, S.; Okui, T.; Yamashita, J.; Ikebe, T.; Harada, H. Expression of SARS-CoV-2 Entry Factors in Human Oral Tissue. J. Anat. 2021, 238, 1341–1354.
- Inchingolo, A.D.; Malcangi, G.; Ceci, S.; Patano, A.; Corriero, A.; Vimercati, L.; Azzollini, D.; Marinelli, G.; Coloccia, G.; Piras, F.; et al. Effectiveness of SARS-CoV-2 Vaccines for Short- and Long-Term Immunity: A General Overview for the Pandemic Contrast. Int. J. Mol. Sci. 2022, 23, 8485.
- Zhu, F.; Zhong, Y.; Ji, H.; Ge, R.; Guo, L.; Song, H.; Wu, H.; Jiao, P.; Li, S.; Wang, C.; et al. ACE2 and TMPRSS2 in Human Saliva Can Adsorb to the Oral Mucosal Epithelium. J. Anat. 2022, 240, 398–409.
- Inchingolo, A.D.; Ceci, S.; Limongelli, L.; Corriero, A.; Curatoli, L.; Azzollini, D.; Mezzapesa, P.P.; Marinelli, G.; Malcangi, G.; Coloccia, G.; et al. Cavernous Sinus Involvement and Near Miss Mediastinitis Following Mandibular Tooth Infection Treated during the COVID-19 Pandemic: Clinical Diagnosis and Treatment. Case Rep. Dent. 2022, 2022, 8650099.
- Matuck, B.F.; Dolhnikoff, M.; Duarte-Neto, A.N.; Maia, G.; Gomes, S.C.; Sendyk, D.I.; Zarpellon, A.; De Andrade, N.P.; Monteiro, R.A.; Pinho, J.R.R.; et al. Salivary Glands Are a Target for SARS-CoV-2: A Source for Saliva Contamination. J. Pathol. 2021, 254, 239–243.
- Tanaka, J.; Senpuku, H.; Ogawa, M.; Yasuhara, R.; Ohnuma, S.; Takamatsu, K.; Watanabe, T.; Mabuchi, Y.; Nakamura, S.; Ishida, S.; et al. Human Induced Pluripotent Stem Cell-Derived Salivary Gland Organoids Model SARS-CoV-2 Infection and Replication. Nat. Cell Biol. 2022, 24, 1595–1605.
- Anahtar, M.; Chan, L.W.; Ko, H.; Rao, A.; Soleimany, A.P.; Khatri, P.; Bhatia, S.N. Host Protease Activity Classifies Pneumonia Etiology. Proc. Natl. Acad. Sci. USA 2022, 119, e2121778119.
- Maggialetti, N.; Villanova, I.; Castrì, A.; Greco, C.N.; Inchingolo, F.; Virgilio, D.; Moschetta, M.; Sardaro, A.; Stabile Ianora, A.A.; Scardapane, A. COVID-19 in Italy: Comparison of CT Findings from Time Zero to the Delta Variant. Microorganisms 2022, 10, 796.
- Parigiani, M.A.; Ketscher, A.; Timme, S.; Bronsert, P.; Schlimpert, M.; Kammerer, B.; Jacquel, A.; Chaintreuil, P.; Reinheckel, T. Conditional Gene Targeting Reveals Cell Type-Specific Roles of the Lysosomal Protease Cathepsin L in Mammary Tumor Progression. Cancers 2020, 12, 2004.
- Sudhan, D.R.; Siemann, D.W. Cathepsin L Targeting in Cancer Treatment. Pharmacol. Ther. 2015, 155, 105–116.
- Pascolo, L.; Zupin, L.; Melato, M.; Tricarico, P.; Crovella, S. TMPRSS2 and ACE2 Coexpression in SARS-CoV-2 Salivary Glands Infection. J. Dent. Res. 2020, 99, 002203452093358.
- Inchingolo, A.D.; Malcangi, G.; Inchingolo, A.M.; Piras, F.; Settanni, V.; Garofoli, G.; Palmieri, G.; Ceci, S.; Patano, A.; De Leonardis, N.; et al. Benefits and Implications of Resveratrol Supplementation on Microbiota Modulations: A Systematic Review of the Literature. Int. J. Mol. Sci. 2022, 23, 4027.
- GTEx Consortium. The GTEx Consortium Atlas of Genetic Regulatory Effects across Human Tissues. Science 2020, 369, 1318–1330.
- Li, N.; Ye, Y.; Wu, Y.; Li, L.; Hu, J.; Luo, D.; Li, Y.; Yang, J.; Gao, Y.; Hai, W.; et al. Alterations in Histology of the Aging Salivary Gland and Correlation with the Glandular Inflammatory Microenvironment. iScience 2023, 26, 106571.
- Shang, Y.F.; Shen, Y.Y.; Zhang, M.C.; Lv, M.C.; Wang, T.Y.; Chen, X.Q.; Lin, J. Progress in Salivary Glands: Endocrine Glands with Immune Functions. Front. Endocrinol. 2023, 14, 88.
- Pedrosa, M.d.S.; Sipert, C.R.; Nogueira, F.N. Salivary Glands, Saliva and Oral Findings in COVID-19 Infection. Pesqui. Bras. Odontopediatria Clínica Integr. 2020, 20, e0104.
- Santacroce, L.; Charitos, I.A.; Ballini, A.; Inchingolo, F.; Luperto, P.; De Nitto, E.; Topi, S. The Human Respiratory System and Its Microbiome at a Glimpse. Biology 2020, 9, 318.
- Schurink, B.; Roos, E.; Radonic, T.; Barbe, E.; Bouman, C.S.C.; de Boer, H.H.; de Bree, G.J.; Bulle, E.B.; Aronica, E.M.; Florquin, S.; et al. Viral Presence and Immunopathology in Patients with Lethal COVID-19: A Prospective Autopsy Cohort Study. Lancet Microbe 2020, 1, e290–e299.
- Malcangi, G.; Inchingolo, A.D.; Inchingolo, A.M.; Piras, F.; Settanni, V.; Garofoli, G.; Palmieri, G.; Ceci, S.; Patano, A.; Mancini, A.; et al. COVID-19 Infection in Children and Infants: Current Status on Therapies and Vaccines. Child. Basel Switz. 2022, 9, 249.
- Balzanelli, M.G.; Distratis, P.; Dipalma, G.; Vimercati, L.; Catucci, O.; Amatulli, F.; Cefalo, A.; Lazzaro, R.; Palazzo, D.; Aityan, S.K.; et al. Immunity Profiling of COVID-19 Infection, Dynamic Variations of Lymphocyte Subsets, a Comparative Analysis on Four Different Groups. Microorganisms 2021, 9, 2036.
- Harikrishnan, P. Gustatory Dysfunction as an Early Symptom in COVID-19 Screening. J. Craniofac. Surg. 2020.
- Gupta, A.; Bhanushali, S.; Sanap, A.; Shekatkar, M.; Kharat, A.; Raut, C.; Bhonde, R.; Shouche, Y.; Kheur, S.; Sharma, A. Oral Dysbiosis and Its Linkage with SARS-CoV-2 Infection. Microbiol. Res. 2022, 261, 127055.
- Lin, W.; Gao, F.; Wang, X.; Qin, N.; Chen, X.; Tam, K.Y.; Zhang, C.; Zhang, M.; Sha, O. The Oral Manifestations and Related Mechanisms of COVID-19 Caused by SARS-CoV-2 Infection. Front. Cell. Neurosci. 2023, 16, 1006977.
- Tamiya, J.; Sakaguchi, W.; Nakagawa, K.; Yamamoto, T.; Saruta, J.; Kubota, N.; Kawata, A.; Hasegawa, I.; Hamada, N.; Tsukinoki, K. Detection of SARS-CoV-2 and Its Related Factors on the Mucosal Epithelium of the Tongue. Acta Histochem. Cytochem. 2023, 56, 29–37. Available online: https://www.jstage.jst.go.jp/article/ahc/56/2/56_22-00089/_html/-char/ja (accessed on 20 November 2023).
- Sakaguchi, W.; Kubota, N.; Shimizu, T.; Saruta, J.; Fuchida, S.; Kawata, A.; Yamamoto, Y.; Sugimoto, M.; Yakeishi, M.; Tsukinoki, K. Existence of SARS-CoV-2 Entry Molecules in the Oral Cavity. Int. J. Mol. Sci. 2020, 21, 6000.
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High Expression of ACE2 Receptor of 2019-nCoV on the Epithelial Cells of Oral Mucosa. Int. J. Oral Sci. 2020, 12, 8.
- Marouf, N.; Cai, W.; Said, K.N.; Daas, H.; Diab, H.; Chinta, V.R.; Hssain, A.A.; Nicolau, B.; Sanz, M.; Tamimi, F. Association between Periodontitis and Severity of COVID-19 Infection: A Case-Control Study. J. Clin. Periodontol. 2021, 48, 483–491.
- Orozco, M.F.S.; Niño-Martínez, N.; Martínez-Castañón, G.-A.; Marín, N.P.; Valencia, C.S.; Velázquez, F.A.D.; Munguía, P.d.C.S.; Santana, M.A.C. Presence of SARS-CoV-2 and Its Entry Factors in Oral Tissues and Cells: A Systematic Review. Medicina 2021, 57, 523.
- Balzanelli, M.G.; Distratis, P.; Lazzaro, R.; Cefalo, A.; Catucci, O.; Aityan, S.K.; Dipalma, G.; Vimercati, L.; Inchingolo, A.D.; Maggiore, M.E.; et al. The Vitamin D, IL-6 and the eGFR Markers a Possible Way to Elucidate the Lung–Heart–Kidney Cross-Talk in COVID-19 Disease: A Foregone Conclusion. Microorganisms 2021, 9, 1903.
- Marques, B.B.F.; Guimarães, T.C.; Fischer, R.G.; Tinoco, J.M.M.; Pires, F.R.; Lima Junior, J.D.C.; Stevens, R.H.; Tinoco, E.M.B. Morphological Alterations in Tongue Epithelial Cells Infected by SARS-CoV-2: A Case–Control Study. Oral Dis. 2022, 28, 2417–2422. Available online: https://onlinelibrary.wiley.com/doi/10.1111/odi.13988 (accessed on 20 November 2023).
- Cui, J.-Y.; Ting, L.; Cao, Y.-X.; Sun, D.-X.; Bing, L.; Wu, X.-P. Morphology Changes of Maxillary Molar Distalization by Clear Aligner Therapy. Int. J. Morphol. 2022, 40, 920–926.
- ACE2 and Spike Protein Cell Lines for SARS-CoV-2 Investigation|GenScript. Available online: https://www.genscript.com/ace2-stable-cell-lines.html?src=google&utm_source=google&utm_medium=cpc&utm_campaign=PRD-SC.-ACE2_Stable_Cell_Line-TCPA&gad_source=1&gclid=EAIaIQobChMItoyc3ZPSggMV0JKDBx0mpAUaEAAYASAAEgKlv_D_BwE (accessed on 20 November 2023).
- Zhong, M.; Lin, B.; Pathak, J.L.; Gao, H.; Young, A.J.; Wang, X.; Liu, C.; Wu, K.; Liu, M.; Chen, J.-M.; et al. ACE2 and Furin Expressions in Oral Epithelial Cells Possibly Facilitate COVID-19 Infection via Respiratory and Fecal-Oral Routes. Front. Med. 2020, 7, 580796.
- Tsuchiya, H. The Oral Cavity Potentially Serving as a Reservoir for SARS-CoV-2 but Not Necessarily Facilitating the Spread of COVID-19 in Dental Practice. Eur. J. Dent. 2023, 17, 310–318.
- Zipeto, D.; Palmeira, J.D.F.; Argañaraz, G.A.; Argañaraz, E.R. ACE2/ADAM17/TMPRSS2 Interplay May Be the Main Risk Factor for COVID-19. Front. Immunol. 2020, 11, 576745.
- Bosshart, H.; Humphrey, J.; Deignan, E.; Davidson, J.; Drazba, J.; Yuan, L.; Oorschot, V.; Peters, P.J.; Bonifacino, J.S. The Cytoplasmic Domain Mediates Localization of Furin to the Trans- Golgi Network En Route to the Endosomal/Lysosomal System. J. Cell Biol. 1994, 126, 1157–1172.
- Braun, E.; Sauter, D. Furin-mediated Protein Processing in Infectious Diseases and Cancer. Clin. Transl. Immunol. 2019, 8, e1073.
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 2020, 581, 221–224.
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5.
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734.
- De Oliveira, G.L.V.; Oliveira, C.N.S.; Pinzan, C.F.; de Salis, L.V.V.; Cardoso, C.R.d.B. Microbiota Modulation of the Gut-Lung Axis in COVID-19. Front. Immunol. 2021, 12, 214.
- Ballini, A.; Signorini, L.; Inchingolo, A.M.; Saini, R.; Gnoni, A.; Scacco, S.; Cantore, S.; Dipalma, G.; Inchingolo, F.; Santacroce, L. Probiotics May Improve Serum Folate Availability in Pregnant Women: A Pilot Study. Open Access Maced. J. Med. Sci. 2020, 8, 1124–1130.
- Okui, T.; Matsuda, Y.; Karino, M.; Hideshima, K.; Kanno, T. Oral Mucosa Could Be an Infectious Target of SARS-CoV-2. Healthc. Basel Switz. 2021, 9, 1068.
- Niknam, Z.; Jafari, A.; Golchin, A.; Danesh Pouya, F.; Nemati, M.; Rezaei-Tavirani, M.; Rasmi, Y. Potential Therapeutic Options for COVID-19: An Update on Current Evidence. Eur. J. Med. Res. 2022, 27, 6.
- Bellocchio, L.; Bordea, I.R.; Ballini, A.; Lorusso, F.; Hazballa, D.; Isacco, C.G.; Malcangi, G.; Inchingolo, A.D.; Dipalma, G.; Inchingolo, F.; et al. Environmental Issues and Neurological Manifestations Associated with COVID-19 Pandemic: New Aspects of the Disease? Int. J. Environ. Res. Public Health 2020, 17, 8049. Available online: https://pubmed.ncbi.nlm.nih.gov/33139595/ (accessed on 15 November 2023).
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8.
- Balzanelli, M.G.; Distratis, P.; Dipalma, G.; Vimercati, L.; Inchingolo, A.D.; Lazzaro, R.; Aityan, S.K.; Maggiore, M.E.; Mancini, A.; Laforgia, R.; et al. SARS-CoV-2 Virus Infection May Interfere CD34+ Hematopoietic Stem Cells and Megakaryocyte-Erythroid Progenitors Differentiation Contributing to Platelet Defection towards Insurgence of Thrombocytopenia and Thrombophilia. Microorganisms 2021, 9, 1632.
- Patano, A.; Cirulli, N.; Beretta, M.; Plantamura, P.; Inchingolo, A.D.; Inchingolo, A.M.; Bordea, I.R.; Malcangi, G.; Marinelli, G.; Scarano, A.; et al. Education Technology in Orthodontics and Paediatric Dentistry during the COVID-19 Pandemic: A Systematic Review. Int. J. Environ. Res. Public Health 2021, 18, 6056.
- Vieira Braga, F.A.; Kar, G.; Berg, M.; Carpaij, O.A.; Polanski, K.; Simon, L.M.; Brouwer, S.; Gomes, T.; Hesse, L.; Jiang, J.; et al. A Cellular Census of Human Lungs Identifies Novel Cell States in Health and in Asthma. Nat. Med. 2019, 25, 1153–1163.
- Smillie, C.S.; Biton, M.; Ordovas-Montanes, J.; Sullivan, K.M.; Burgin, G.; Graham, D.B.; Herbst, R.H.; Rogel, N.; Slyper, M.; Waldman, J.; et al. Intra- and Inter-Cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019, 178, 714–730.e22.
- Mather, M.W.; Jardine, L.; Talks, B.; Gardner, L.; Haniffa, M. Complexity of Immune Responses in COVID-19. Semin. Immunol. 2021, 55, 101545.
- Sarode, S.C.; Sarode, G.S. Saliva Sample and SARS-CoV-2 Detection: More Complexities than Clarity. J. Oral Biol. Craniofacial Res. 2021, 11, 500–501.
- Inchingolo, A.D.; Dipalma, G.; Inchingolo, A.M.; Malcangi, G.; Santacroce, L.; D’Oria, M.T.; Isacco, C.G.; Bordea, I.R.; Candrea, S.; Scarano, A.; et al. The 15-Months Clinical Experience of SARS-CoV-2: A Literature Review of Therapies and Adjuvants. Antioxidants 2021, 10, 881.
- Inchingolo, A.D.; Inchingolo, A.M.; Bordea, I.R.; Malcangi, G.; Xhajanka, E.; Scarano, A.; Lorusso, F.; Farronato, M.; Tartaglia, G.M.; Isacco, C.G.; et al. SARS-CoV-2 Disease through Viral Genomic and Receptor Implications: An Overview of Diagnostic and Immunology Breakthroughs. Microorganisms 2021, 9, 793.
- Alsulami, M.; Kattan, W.; Alsamadani, L.; Alahmari, G.; Al Juhani, W.; Almabadi, M. An Outlook on Dental Practices to Avoid the Oral Transmission of COVID-19. Microorganisms 2023, 11, 146.
- Knyazev, E.; Nersisyan, S.; Tonevitsky, A. Endocytosis and Transcytosis of SARS-CoV-2 Across the Intestinal Epithelium and Other Tissue Barriers. Front. Immunol. 2021, 12, 636966.
- Galicia, J.C.; Guzzi, P.H.; Giorgi, F.M.; Khan, A.A. Predicting the Response of the Dental Pulp to SARS-CoV2 Infection: A Transcriptome-Wide Effect Cross-Analysis. Genes Immun. 2020, 21, 360–363.
- Karnik, M.; Beeraka, N.M.; Uthaiah, C.A.; Nataraj, S.M.; Bettadapura, A.D.S.; Aliev, G.; Madhunapantula, S.V. A Review on SARS-CoV-2-Induced Neuroinflammation, Neurodevelopmental Complications, and Recent Updates on the Vaccine Development. Mol. Neurobiol. 2021, 58, 4535–4563.
- Scarano, A.; Inchingolo, F.; Rapone, B.; Festa, F.; Tari, S.R.; Lorusso, F. Protective Face Masks: Effect on the Oxygenation and Heart Rate Status of Oral Surgeons during Surgery. Int. J. Environ. Res. Public Health 2021, 18, 2363.
More
Information
Subjects:
Dentistry, Oral Surgery & Medicine
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
594
Revisions:
2 times
(View History)
Update Date:
05 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




