Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Valya Vassileva | -- | 2518 | 2024-01-04 07:11:22 | | | |
| 2 | Lindsay Dong | Meta information modification | 2518 | 2024-01-05 06:31:08 | | | | |
| 3 | Lindsay Dong | -2 word(s) | 2516 | 2024-01-12 09:11:35 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Vassileva, V.; Georgieva, M.; Todorov, D.; Mishev, K. Nuclear Distribution C Proteins in Plants. Encyclopedia. Available online: https://encyclopedia.pub/entry/53405 (accessed on 07 February 2026).
Vassileva V, Georgieva M, Todorov D, Mishev K. Nuclear Distribution C Proteins in Plants. Encyclopedia. Available at: https://encyclopedia.pub/entry/53405. Accessed February 07, 2026.
Vassileva, Valya, Mariyana Georgieva, Dimitar Todorov, Kiril Mishev. "Nuclear Distribution C Proteins in Plants" Encyclopedia, https://encyclopedia.pub/entry/53405 (accessed February 07, 2026).
Vassileva, V., Georgieva, M., Todorov, D., & Mishev, K. (2024, January 04). Nuclear Distribution C Proteins in Plants. In Encyclopedia. https://encyclopedia.pub/entry/53405
Vassileva, Valya, et al. "Nuclear Distribution C Proteins in Plants." Encyclopedia. Web. 04 January, 2024.
Copy Citation
The family of Nuclear Distribution C (NudC) proteins plays a pivotal and evolutionarily conserved role in all eukaryotes. These proteins influence vital cellular processes like cell division, protein folding, nuclear migration and positioning, intracellular transport, and stress response.
Nuclear Distribution C (NudC)
plant
stress response
1. Nuclear Distribution Genes and Their Functional Significance across Eukaryotes
The nuclear distribution (Nud) genes encode evolutionary conserved proteins, which play an important role in various cellular processes. A notable example of Nud gene function, which inspired their name, is nuclear migration, a precisely regulated process that holds particular importance in highly elongated cells [1][2]. Nud genes were originally discovered in the filamentous fungus Aspergillus nidulans [3][4][5][6]. Xiang et al. [3] have identified four Nud genes, namely NudA, NudC, NudF, and NudG, encoding components of the cytoplasmic dynein–dynactin complex. NudC proteins are known for their compact structure and versatile functions, encompassing the facilitation of cell division, the regulation of gene expression, and contribution to various essential biological processes (Figure 1). Deletions in the NudC gene lead to a more severe phenotype compared to other nuclear distribution mutants, and can result in lethality [4]. NudC acts in concert with the molecular motor dynein and other Nud genes to regulate dynein-mediated processes, including vesicle transport in neurons [7][8], Golgi apparatus positioning, lysosomes and vesicles transport [9], kinetochore localization, spindle organization [10], phagosome movement [3], nuclear transport, and membrane organelle organization [7]. The NudA, NudI, and NudG genes encode the heavy, intermediate, and light chains of cytoplasmic dynein, respectively [5][11][12]. The NudF protein is required for nuclear migration through the fungal mycelium, and interacts with microtubule-related proteins, such as α-tubulin and dynein [13][14][15]. The NudF gene encodes a protein that exhibits a 42% sequence similarity with the human Lis1 gene, which is associated with Miller–Dieker syndrome. The hemizygous deletion or mutation of the Lis1 gene results in type I lissencephaly, a condition that obstructs proper neuronal migration during brain development and leads to a smooth brain surface and disorganized cortical layering [16][17].
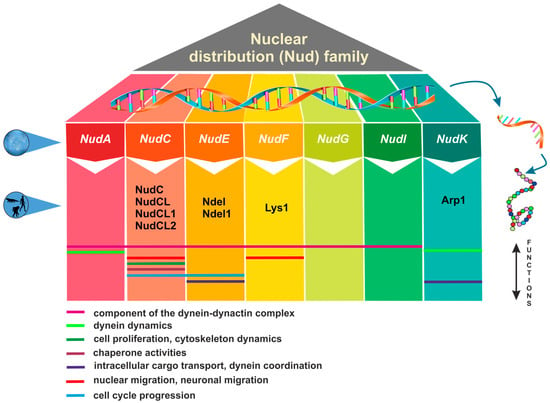
Figure 1. Overview of nuclear distribution (Nud) family members and their cellular functions.
Lis1 plays a role in mitosis both through direct mechanisms [18][19][20] as well as indirectly by impeding the movement of nuclei in radial glial progenitor cells towards the ventricular surface [21]. The NudE gene was subsequently identified as a multicopy suppressor of the NudF phenotype, demonstrating its capacity to rescue the aberrant characteristics associated with nudF mutations through the introduction of multiple NudE copies [22]. Furthermore, NudE possesses two homologs, Ndel and Ndel1, which play a role in recruiting dynein to mitotic kinetochores, facilitating the progression of mitosis [23][24]. Mutant mice lacking both Ndel and Ndel1 genes can exhibit varying neurological conditions, contingent upon the gene dosage. In particular, they may develop microcephaly due to mitotic abnormalities [25] or lissencephaly, which aligns with their role in the cytoplasmic dynein pathway [26]. Noteworthy, mutations in human NudE can lead to the co-occurrence of microcephaly and lissencephaly [27]. It has been shown that Lis1, in conjunction with NudE, orchestrates the assembly of an intricate molecular complex involving cytoplasmic dynein [28]. Within this assembly, Lis1, under the guidance of NudE, transforms dynein into a state of sustained force production, which proves to be a vital factor in facilitating the transport of heavy-load structures, including cell nuclei. Interestingly, NudE appears to be intricately connected within the same genetic pathway as NudF and NudA. The phenotypes observed in nudE homozygous mutants closely resemble those of nudF and nudA heterozygous mutants, suggesting that NudF, NudE, and NudA operate within the same functional pathways [29]. A crucial element in the complex network of Nud proteins is the actin-related protein Arp1, encoded by the NudK gene. Arp1 assumes a pivotal function in the dynactin complex [30] by activating dynein and consequently enhancing the intracellular cargo transport [31]. NudC also regulates the level of NudF protein in A. nidulans [6]. A nudC mutation leads to a significant decrease in NudF, primarily at the restrictive temperature. On the other hand, additional copies of the NudF gene can effectively complement the temperature-sensitive phenotype of nudC mutants [5][32][33].
2. Structural and Functional Features of NudC Proteins
NudC proteins display a remarkable degree of conservation across a broad spectrum of organisms, spanning from fungi and plants to animals [34] (Figure 2). The NudC homologous genes or proteins are found in a number of higher eukaryotes, including Caenorhabditis elegans [35], Drosophila melanogaster [36], newts [37], plants [38][39][40], and mammals [41]. The NudC family consists of four proteins: NudC [42][43][44], NudC-like (NudCL) also known as NudC domain-containing protein 3 (NUDCD3) [45][46], NudC-like 2 (NudCL2/NUDCD2) [47], and the more distant NudC domain-containing 1 (NudCD1) also known as CML66 (Chronic Myeloid Leukemia 66).
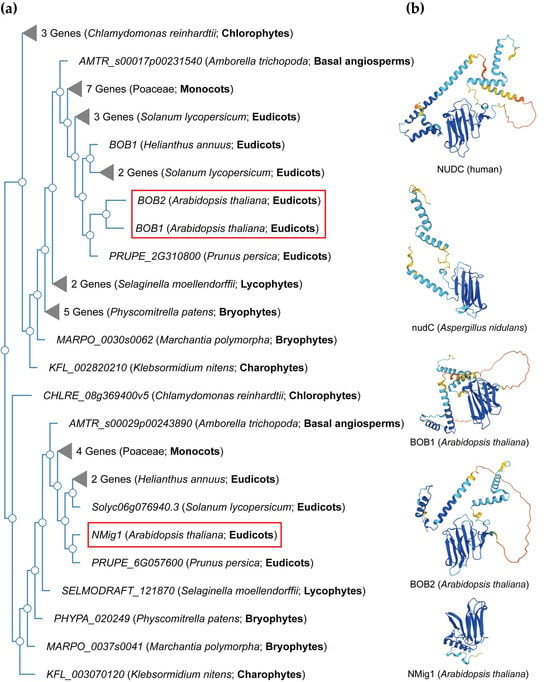
Figure 2. NudC protein conservation across eukaryotes. (a) Simplified phylogenetic tree showing the evolutionary relationships of plant and algal NudC proteins. The phylogenetic tree was generated using PhyloGenes (https://phylogenes.arabidopsis.org, accessed on 10 November 2023) with the following selected species (retrieved AtBOB1 gene homologs): Arabidopsis thaliana (3), Solanum lycopersicum (6), Prunus persica (2), Helianthus annuus (3), Hordeum vulgare (4), Oryza sativa (3), Zea mays (4), Amborella trichopoda (2), Selaginella moellendorffii (3), Physcomitrella patens (6), Marchantia polymorpha (2), Klebsormidium nitens (2), and Chlamydomonas reinhardtii (4). (b) Predicted protein structures of NudC family members in human, fungi, and Arabidopsis. The protein structures were generated and visualized with the help of the AlphaFold algorithm of EMBL-EBI (https://alphafold.ebi.ac.uk, accessed on 10 November 2023). Red rectangles outline the Arabidopsis NudC genes.
NudC proteins typically exhibit a conserved domain architecture and contain specific motifs that are crucial for their functional roles in intracellular transport, nuclear positioning, cell cycle progression and stress responses (Figure 2b). They include a coiled-coil region at the N-terminus serving as the dimerization module, and also possess a central globular domain, a CS domain (a domain shared by CHORD-containing proteins and SGT1, PF04969) resembling p23 and other small heat shock proteins (sHSPs), and two conserved α-helices downstream [48][49][50]. This architecture is maintained across various species, albeit with some variations in the N-terminus. Compared to fungi, higher eukaryotes typically possess an extended N-terminal segment. In vertebrates, the most substantial amino acid conservation is observed in a short N-terminal segment and the C-terminal α-helices [50]. It is essential to note that NudC molecules operate as dimers, and their biological roles may depend on this dimeric state. Common structural features between NudC and NudCL imply a potential functional overlap, despite the relatively low amino acid sequence similarity. NudCL is unique to the animal kingdom, whereas NudCL2 is found across all eukaryotes, distinguishing itself by the absence of an N-terminal extension and featuring a distinct C-terminal fragment [50].
The architectural variations seen in NudC protein family across different species (Figure 2b) present an intriguing spectrum of structural diversity. This diversity highlights the remarkable adaptability of the NudC protein family to different biological contexts, providing compelling insights into the potential existence of variable functional roles played by the NudC protein family in various organisms.
The expression of NudC is markedly associated with the rate of proliferation in diverse cell types and tissues. The human NudC homolog displays robust expression in actively proliferating cells [51] with a central role in spindle formation during mitosis [12]. Depletion of NudC results in the presence of multiple spindles during metaphase and lagging chromosomes during anaphase [52].
o-chaperone activities [50][53]. The CS domain in the NudC family mirrors the molecular architecture of sHSPs, such as HSP20/α-crystallin and p23 proteins [49]. It is important to mention that HSP20 and p23 serve as central structures responsible for interacting with HSP90 and specific client proteins [49][53][54][55]. This implies that proteins containing these structures are instrumental in recruiting HSPs to multiprotein complexes [53].
3. NudC Proteins in Plants
3.1. Structural Features of Plant NudC Members
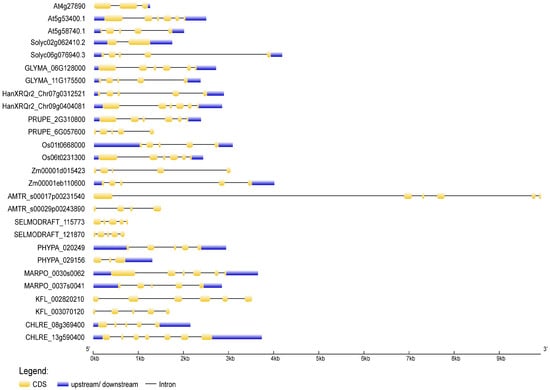
To deduce the evolutionary history of the NudC gene family, scholars traced the dynamics of intron–exon organization in selected NudC genes across a spectrum of plant species. The genomic and coding sequences (CDS) of NudC members were acquired from the Ensembl Plants (https://plants.ensembl.org, accessed on 10 November 2023) and the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov, accessed on 10 November 2023) databases. The analysis was conducted using the Gene Structure Display Server (GSDS) [56] available at http://gsds.gao-lab.org/, accessed on 10 November 2023. The positions of introns and exons in NudC genes were ascertained through the identification of gaps during the alignment of full-length cDNA transcripts with genomic sequences (Figure 3).

Figure 3. Exon–intron structures of selected plant and algal NudC genes. Untranslated regions are depicted by blue boxes, coding regions by yellow boxes, and introns by lines. The gene lengths can be estimated using the scale provided at the bottom. The exon–intron structures were generated and visualized using the Gene Structure Display Server (http://gsds.gao-lab.org, accessed on 11 November 2023).
In the Arabidopsis genome, three NudC genes have been identified. One is designated as BOBBER1 (BOB1), which has a paralog named BOBBER2 (BOB2) [38][39][57]. The third member is referred to as NUCLEAR MIGRATION 1 (NMig1), and displays a more distant relationship to the other two NudC members (Figure 2a).
In general, a considerable structural diversity was observed in the number, size, and distribution of exons and introns among the NudC members (Figure 3). The number of exons ranged from one to nine. In Z. mays, both investigated NudC genes exhibited a tendency for extended introns and shorter exons. The second intron of the BOB1-related A. trichopoda gene, which stood out as the longest, resulted in a genomic DNA sequence spanning 9918 kb. Conversely, in S. moellendorffii, A. thaliana, and the BOB1-related S. lycopersicum gene, the distribution leans towards more evenly sized introns and exons. Not all NudC genes comprised 5′ and 3′ untranslated regions (UTRs). The overall gene exon–intron organization indicated a lack of apparent correlation with phylogeny, suggesting that NudC genes may have undergone evolutionary changes such as gene duplication, exon shuffling, or other mechanisms influencing gene structure.
3.2. Developmental Role of Plant NudC Proteins
The A. thaliana gene BOB1 encodes a small 34.5 kDa protein featuring the NudC domain, which specifies a non-canonical sHSP with versatile functions, contributing to developmental processes and responses to elevated temperatures [38][39][57]. The other two NudC genes in the Arabidopsis genome, BOB2 and NMig1, have also been identified as CS domain-containing NudC family members.
The importance of the BOB1 gene becomes evident in its contribution to the precise partitioning and patterning of the apical domain in Arabidopsis embryos [38]. The loss of BOB1 results in embryo lethality, linked to developmental arrest at the globular stage without progressing to the transition stage. The bob1 null mutants exhibit an expansion of meristematic identity into the region that would typically give rise to cotyledons, leading to the failure of cotyledon formation. These developmental anomalies coincide with atypical gene expression patterns, particularly the overexpression of the meristem-specific SHOOTMERISTEMLESS (STM) gene in the upper half of the embryo.
These findings underscore the critical role of this NudC gene in orchestrating key aspects of embryonic development and its potential involvement in regulating auxin-related developmental pathways.
The importance of NudC genes extend beyond embryonic development to postembryonic phases. The hypomorphic bob1-3 allele introduces a broad spectrum of developmental abnormalities, including shorter roots, smaller serrated leaves, stunted branched inflorescences, and irregularities in inflorescence and floral meristem formation, leading to pin-formed meristems and abnormal floral organ numbers [39]. It is noteworthy to mention that many of these phenotypic anomalies closely correspond to those seen in mutants associated with impaired auxin signaling or transport.
In addition, the genetic interplay of BOB1 with AS1 and AS2 provides valuable insights into a developmental pathway that relies on the function of this NudC gene. AS1 and AS2 serve as transcriptional regulators, contributing to the establishment of meristem boundaries by suppressing KNOX gene expression and reinforcing adaxial polarity during leaf development [58][59][60]. An allelic variant of BOB1, known as eal-1 and sharing the same mutation as bob1-3, offers a unique perspective in this context. Remarkably, eal-1 represents the sole viable allele of BOB1 with well-documented phenotypes [39]. When combined with as1 and as2 mutations, eal-1 demonstrates leaf morphology characterized by abaxialized filamentous structures, along with an upregulation of KNOTTED-like homeobox (KNOX) and ETTIN (ETT)/AUXIN RESPONSE FACTOR3 (ARF3) genes. ETT plays a role in enhancing abaxial identity and is directly regulated by the AS1–AS2 complex [59]. The observed polarity abnormalities in eal-1; as2 plants are mitigated in an ett genetic background, suggesting that ETT acts downstream of BOB1, AS1, and AS2 [61].
3.3. Unveiling the Role of NudC Proteins in Plant Stress Response and Resilience
The structural similarities between the NudC domain and α-crystallin domain (ACD)/p23 proteins [49] suggest the potential for shared functions with ACD-containing sHSPs [62][63]. The ACD, initially identified in the chaperone of the vertebrate eye lens, spans approximately 80–100 amino acids and is primarily located in the C-terminal domain [64]. It encompasses two conserved regions that form a pleated β-sheet sandwich, separated by a variable-length hydrophilic domain [65][66]. These structural features closely resemble those observed in the human and mouse NudC homologs [50], suggesting the possibility of common functions with sHSPs. In plants, ACD proteins serve diverse roles, from responding to abiotic stresses and hormones to regulating transcription, virus movement, and DNA demethylation [67].
The Arabidopsis protein BOB1 demonstrates characteristics that align with typical sHSPs. It is induced under heat stress, possesses a NudC domain that shares structural homology with ACD-containing sHSPs, and exhibits in vitro chaperone activity, effectively preventing the aggregation of model protein substrates [39][65][68]. Under normal conditions, BOB1 is primarily localized in the plant cell cytoplasm, but during heat stress, it translocates to heat shock granules in association with HSP17.6 [39][69][70].
BOB1, with its sHSP-like characteristics and crucial role in heat stress response, is not only involved in thermotolerance but also contributes to proteostasis. In conjunction with the 26S proteasome (26SP), it forms part of a genetic network linking proteostasis to the AS1-AS2 developmental pathway [57]. The interactions within this network are believed to rely on BOB1 chaperone activities. Importantly, this network plays a vital role in repressing KNOX gene expression, which is critical for normal plant development.
The second NudC gene in Arabidopsis, NMig1, also shares structural homology with ACD-containing sHSPs, and exhibits significant upregulation in response to various abiotic stressors, including heat shock [40]. Constitutive overexpression of NMig1 results in enhanced root growth and lateral root development, even under adverse abiotic stress conditions. NMig1-overexpressing plants display reduced susceptibility to the inhibitory effects of abiotic stress on root morphology.
The involvement of tomato NudC homologs in the context of tomato immune responses is notably intriguing, given the broader context of the role of NudC genes in enhancing plant stress resilience as discussed earlier. In a study conducted by Liu et al. [71], a compelling connection between tomato orthologues of NudC domain proteins and SlSAP3, a member of the stress-associated protein family, came to light. SlSAP3 serves as a positive regulator of tomato immunity against Pseudomonas syringae pv. tomato (Pst) DC3000. The three identified tomato SlBOB proteins, namely SlBOB1, SlBOB2, and SlBOB3, share a common NudC domain at their C termini, with variations in regions outside the NudC domain [71]. Silencing SlBOB1 or the simultaneous silencing of all three SlBOB genes, which act as negative regulators of immunity, leads to enhanced resistance against Pst DC3000. Hence, it appears that plant BOB proteins play diverse roles in responding to biotic and abiotic stress.
These findings further emphasize the vital role played by NudC genes in modulating plant stress resilience, and provide valuable insights into their potential as targets for the development of stress-tolerant crops.
References
- Morris, N.R. Nuclear migration. From fungi to the mammalian brain. J. Cell Biol. 2000, 148, 1097–1101.
- Zhang, J.; Li, S.; Fischer, R.; Xiang, X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol. Biol. Cell 2003, 14, 1479–1488.
- Xiang, X.; Beckwith, S.M.; Morris, N.R. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 1994, 91, 2100–2104.
- Chiu, Y.H.; Xiang, X.; Dawe, A.L.; Morris, N.R. Deletion of nudC, a nuclear migration gene of Aspergillus nidulans, causes morphological and cell wall abnormalities and is lethal. Mol. Biol. Cell 1997, 8, 1735–1749.
- Xiang, X.; Roghi, C.; Morris, N.R. Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 1995, 92, 9890–9894.
- Osmani, A.H.; Osmani, S.A.; Morris, N.R. The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J. Cell Biol. 1990, 111, 543–551.
- Fath, K.R.; Trimbur, G.M.; Burgess, D.R. Molecular motors and a spectrin matrix associate with Golgi membranes in vitro. J. Cell Biol. 1997, 139, 1169–1181.
- Kamal, A.; Goldstein, L.S. Principles of cargo attachment to cytoplasmic motor proteins. Curr. Opin. Cell Biol. 2002, 14, 63–68.
- Aniento, F.; Emans, N.; Griffiths, G.; Gruenberg, J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J. Cell Biol. 1993, 123, 1373–1387.
- Pfarr, C.M.; Coue, M.; Grissom, P.M.; Hays, T.S.; Porter, M.E.; McIntosh, J.R. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature 1990, 345, 263–265.
- Beckwith, S.M.; Roghi, C.H.; Liu, B.; Morris, N.R. The “8-kD” cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J. Cell Biol. 1998, 143, 1239–1247.
- Zhang, M.Y.; Huang, N.N.; Clawson, G.A.; Osmani, S.A.; Pan, W.; Xin, P.; Razzaque, M.S.; Miller, B.A. Involvement of the fungal nuclear migration gene nudC human homolog in cell proliferation and mitotic spindle formation. Exp. Cell Res. 2002, 273, 73–84.
- Willins, D.A.; Xiang, X.; Morris, N.R. An alpha tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans. Genetics 1995, 141, 1287–1298.
- Willins, D.A.; Liu, B.; Xiang, X.; Morris, N.R. Mutations in the heavy chain of cytoplasmic dynein suppress the nudF nuclear migration mutation of Aspergillus nidulans. Mol. Gen. Genet. 1997, 255, 194–200.
- Morris, N.R.; Efimov, V.P.; Xiang, X. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 1998, 8, 467–470.
- Dobyns, W.B.; Reiner, O.; Carrozzo, R.; Ledbetter, D.H. Lissencephaly. A human brain malformation associated with deletion of the LIS1 gene located at chromosome 17p13. JAMA 1993, 270, 2838–2842.
- Hirotsune, S.; Fleck, M.W.; Gambello, M.J.; Bix, G.J.; Chen, A.; Clark, G.D.; Ledbetter, D.H.; McBain, C.J.; Wynshaw-Boris, A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 1998, 19, 333–339.
- Faulkner, N.E.; Dujardin, D.L.; Tai, C.Y.; Vaughan, K.T.; O’Connell, C.B.; Wang, Y.L.; Vallee, R.B. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000, 2, 784–791.
- Siller, K.H.; Serr, M.; Steward, R.; Hays, T.S.; Doe, C.Q. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell 2005, 16, 5127–5140.
- Yingling, J.; Youn, Y.H.; Darling, D.; Toyo-Oka, K.; Pramparo, T.; Hirotsune, S.; Wynshaw-Boris, A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell 2008, 132, 474–486.
- Tsai, J.W.; Lian, W.N.; Kemal, S.; Kriegstein, A.R.; Vallee, R.B. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 2010, 13, 1463–1471.
- Efimov, V.P.; Morris, N.R. The Lis1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J. Cell Biol. 2000, 150, 681–688.
- Ma, L.; Tsai, M.Y.; Wang, S.; Lu, B.; Chen, R.; Iii, J.R.; Zhu, X.; Zheng, Y. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat. Cell Biol. 2009, 11, 247–256.
- Stehman, S.A.; Chen, Y.; McKenney, R.J.; Vallee, R.B. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J. Cell Biol. 2007, 178, 583–594.
- Feng, Y.; Walsh, C.A. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 2004, 44, 279–293.
- Sasaki, S.; Mori, D.; Toyo-Oka, K.; Chen, A.; Garrett-Beal, L.; Muramatsu, M.; Miyagawa, S.; Hiraiwa, N.; Yoshiki, A.; Wynshaw-Boris, A.; et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol. Cell. Biol. 2005, 25, 7812–7827.
- Alkuraya, F.S.; Cai, X.; Emery, C.; Mochida, G.H.; Al-Dosari, M.S.; Felie, J.M.; Hill, R.S.; Barry, B.J.; Partlow, J.N.; Gascon, G.G.; et al. Human mutations in NDE1 cause extreme microcephaly with lissencephaly. Am. J. Med. Genet. 2011, 88, 536–547.
- McKenney, R.J.; Vershinin, M.; Kunwar, A.; Vallee, R.B.; Gross, S.P. LIS1 and NudE induce a persistent dynein force-producing state. Cell 2010, 141, 304–314.
- Feng, Y.; Olson, E.C.; Stukenberg, P.T.; Flanagan, L.A.; Kirschner, M.W.; Walsh, C.A. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 2000, 28, 665–679.
- Xiang, X.; Zuo, W.; Efimov, V.P.; Morris, N.R. Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr. Genet. 1999, 35, 626–630.
- Xiang, X.; Han, G.; Winkelmann, D.A.; Zuo, W.; Morris, N.R. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr. Biol. 2000, 10, 603–606.
- Chiu, Y.H.; Morris, N.R. Extragenic suppressors of nudC3, a mutation that blocks nuclear migration in Aspergillus nidulans. Genetics 1995, 141, 453–464.
- Chiu, Y.H.; Morris, N.R. Genetic and molecular analysis of a tRNALeu missense suppressor of nudC3, a mutation that blocks nuclear migration in Aspergillus nidulans. Genetics 1997, 145, 707–714.
- Fu, Q.; Wang, W.; Zhou, T.; Yang, Y. Emerging roles of NudC family: From molecular regulation to clinical implications. Sci. China Life Sci. 2016, 59, 455–462.
- Dawe, A.L.; Caldwell, K.A.; Harris, P.M.; Morris, N.R.; Caldwell, G.A. Evolutionarily conserved nuclear migration genes required for early embryonic development in Caenorhabditis elegans. Dev. Genes Evol. 2001, 211, 434–441.
- Cunniff, J.; Chiu, Y.H.; Morris, N.R.; Warrior, R. Characterization of DnudC, the Drosophila homolog of an Aspergillus gene that functions in nuclear motility. Mech Dev. 1997, 66, 55–68.
- Moreau, N.; Aumais, J.P.; Prudhomme, C.; Morris, S.M.; Yu-Lee, L.Y. NUDC expression during amphibian development. Int. J. Dev. Biol. 2001, 45, 839–843.
- Jurkuta, R.J.; Kaplinsky, N.J.; Spindel, J.E.; Barton, M.K. Partitioning the apical domain of the Arabidopsis embryo requires the BOBBER1 NudC domain protein. Plant Cell 2009, 21, 1957–1971.
- Perez, D.E.; Hoyer, J.S.; Johnson, A.I.; Moody, Z.R.; Lopez, J.; Kaplinsky, N.J. BOBBER1 is a noncanonical Arabidopsis small heat shock protein required for both development and thermotolerance. Plant Physiol. 2009, 151, 241–252.
- Velinov, V.; Vaseva, I.; Zehirov, G.; Zhiponova, M.; Georgieva, M.; Vangheluwe, N.; Beeckman, T.; Vassileva, V. Overexpression of the NMig1 gene encoding a NudC domain protein enhances root growth and abiotic stress tolerance in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 815.
- Riera, J.; Lazo, P.S. The mammalian NudC-like genes: A family with functions other than regulating nuclear distribution. Cell. Mol. Life Sci. 2009, 66, 2383–2390.
- Axtell, S.M.; Truong, T.M.; O’Neal, K.D.; Yu-Lee, L.Y. Characterization of a prolactin-inducible gene, clone 15, in T cells. Mol. Endocrinol. 1995, 9, 312–318.
- Miller, B.A.; Zhang, M.Y.; Gocke, C.D.; De Souza, C.; Osmani, A.H.; Lynch, C.; Davies, J.; Bell, L.; Osmani, S.A. A homolog of the fungal nuclear migration gene nudC is involved in normal and malignant human hematopoiesis. Exp. Hematol. 1999, 27, 742–750.
- Matsumoto, N.; Ledbetter, D.H. Molecular cloning and characterization of the human NUDC gene. Hum Genet. 1999, 104, 498–504.
- Shu, T.; Tseng, H.C.; Sapir, T.; Stern, P.; Zhou, Y.; Sanada, K.; Fischer, A.; Coquelle, F.M.; Reiner, O.; Tsai, L.H. Doublecortin-like kinase controls neurogenesis by regulating mitotic spindles and M phase progression. Neuron 2006, 49, 25–39.
- Zhou, T.; Zimmerman, W.; Liu, X.; Erikson, R.L. A mammalian NudC-like protein essential for dynein stability and cell viability. Proc. Natl. Acad. Sci. USA 2006, 103, 9039–9044.
- Yang, Y.; Yan, X.; Cai, Y.; Lu, Y.; Si, J.; Zhou, T. NudC-like protein 2 regulates the LIS1/dynein pathway by stabilizing LIS1 with Hsp90. Proc. Natl. Acad. Sci. USA 2010, 107, 3499–3504.
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164.
- Garcia-Ranea, J.A.; Mirey, G.; Camonis, J.; Valencia, A. p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 2002, 529, 162–167.
- Zheng, M.; Cierpicki, T.; Burdette, A.J.; Utepbergenov, D.; Janczyk, P.; Derewenda, U.; Stukenberg, P.T.; Caldwell, K.A.; Derewenda, Z.S. Structural features and chaperone activity of the NudC protein family. J. Mol. Biol. 2011, 409, 722–741.
- Gocke, C.D.; Osmani, S.A.; Miller, B.A. The human homologue of the Aspergillus nuclear migration gene nudC is preferentially expressed in dividing cells and ciliated epithelia. Histochem. Cell Biol. 2000, 114, 293–301.
- Zhou, T.; Aumais, J.P.; Liu, X.; Yu-Lee, L.Y.; Erikson, R.L. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev. Cell 2003, 5, 127–138.
- Lee, Y.T.; Jacob, J.; Michowski, W.; Nowotny, M.; Kuznicki, J.; Chazin, W.J. Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J. Biol. Chem. 2004, 279, 16511–16517.
- Botër, M.; Amigues, B.; Peart, J.; Breuer, C.; Kadota, Y.; Casais, C.; Moore, G.; Kleanthous, C.; Ochsenbein, F.; Shirasu, K.; et al. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 2007, 19, 3791–3804.
- Echtenkamp, F.J.; Zelin, E.; Oxelmark, E.; Woo, J.I.; Andrews, B.J.; Garabedian, M.; Freeman, B.C. Global functional map of the p23 molecular chaperone reveals an extensive cellular network. Mol. Cell 2011, 43, 229–241.
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297.
- Silverblatt-Buser, E.W.; Frick, M.A.; Rabeler, C.; Kaplinsky, N.J. Genetic interactions between BOB1 and multiple 26S proteasome subunits suggest a role for proteostasis in regulating Arabidopsis development. G3-Genes Genom. Genet. 2018, 8, 1379–1390.
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. 2002. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478.
- Iwasaki, M.; Takahashi, H.; Iwakawa, H.; Nakagawa, A.; Ishikawa, T.; Tanaka, H.; Matsumura, Y.; Pekker, I.; Eshed, Y.; Vial-Pradel, S.; et al. Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 2013, 140, 1958–1969.
- Machida, C.; Nakagawa, A.; Kojima, S.; Takahashi, H.; Machida, Y. The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 655–671.
- Ishibashi, N.; Kanamaru, K.; Ueno, Y.; Kojima, S.; Kobayashi, T.; Machida, C.; Machida, Y. ASYMMETRIC-LEAVES2 and an ortholog of eukaryotic NudC domain proteins repress expression of AUXIN RESPONSE-FACTOR and class 1 KNOX homeobox genes for development of flat symmetric leaves in Arabidopsis. Biol. Open 2012, 1, 197–207.
- Studer, S.; Narberhaus, F. Chaperone activity and homo-and hetero-oligomer formation of bacterial small heat shock proteins. J. Biol. Chem. 2000, 275, 37212–37218.
- Eisenhardt, B.D. Small heat shock proteins: Recent developments. BioMol. Concepts 2013, 4, 583–595.
- Boelens, W.C.; Croes, Y.; de Ruwe, M.; de Reu, L.; de Jong, W.W. Negative charges in the C-terminal domain stabilize the αB-crystallin complex. J. Biol. Chem. 1998, 273, 28085–28090.
- Haslbeck, M.; Franzmann, T.; Weinfurtner, D.; Buchner, J. Some like it hot: The structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 2005, 12, 842–846.
- Bondino, G.H.; Valle, M.E.; Ten Have, A. Evolution and functional diversification of the small heat shock protein/α-crystallin family in higher plants. Planta 2012, 235, 1299–1313.
- Paul, A.; Rao, S.; Mathur, S. The α-crystallin domain containing genes: Identification, phylogeny and expression profiling in abiotic stress, phytohormone response and development in tomato (Solanum lycopersicum). Front. Plant Sci. 2016, 7, 426.
- Wallace, E.W.J.; Kear-Scott, J.L.; Pilipenko, E.V.; Schwartz, M.H.; Laskowski, P.R.; Rojek, A.E.; Katanski, C.D.; Riback, J.A.; Dion, M.F.; Franks, A.M.; et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 2015, 162, 1286–1298.
- Kirschner, M.; Winkelhaus, S.; Thierfelder, J.M.; Nover, L. Transient expression and heat-stress-induced co-aggregation of endogenous and heterologous small heat-stress proteins in tobacco protoplasts. Plant J. 2000, 24, 397–412.
- Miroshnichenko, S.; Tripp, J.; Nieden, U.Z.; Neumann, D.; Conrad, U.; Manteuffel, R. Immunomodulation of function of small heat shock proteins prevents their assembly into heat stress granules and results in cell death at sublethal temperatures. Plant J. 2005, 41, 269–281.
- Liu, S.; Wang, J.; Jiang, S.; Wang, H.; Gao, Y.; Zhang, H.; Li, D.; Song, F. Tomato SlSAP3, a member of the stress-associated protein family, is a positive regulator of immunity against Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 2019, 20, 815–830.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
628
Revisions:
3 times
(View History)
Update Date:
12 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




