Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Armin Djamei | -- | 1765 | 2023-12-31 19:34:04 | | | |
| 2 | Catherine Yang | Meta information modification | 1765 | 2024-01-02 02:05:14 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Nagarajan, N.; Khan, M.; Djamei, A. Auxin Signaling by Smut Fungi during Plant Colonization. Encyclopedia. Available online: https://encyclopedia.pub/entry/53294 (accessed on 09 February 2026).
Nagarajan N, Khan M, Djamei A. Auxin Signaling by Smut Fungi during Plant Colonization. Encyclopedia. Available at: https://encyclopedia.pub/entry/53294. Accessed February 09, 2026.
Nagarajan, Nithya, Mamoona Khan, Armin Djamei. "Auxin Signaling by Smut Fungi during Plant Colonization" Encyclopedia, https://encyclopedia.pub/entry/53294 (accessed February 09, 2026).
Nagarajan, N., Khan, M., & Djamei, A. (2023, December 31). Auxin Signaling by Smut Fungi during Plant Colonization. In Encyclopedia. https://encyclopedia.pub/entry/53294
Nagarajan, Nithya, et al. "Auxin Signaling by Smut Fungi during Plant Colonization." Encyclopedia. Web. 31 December, 2023.
Copy Citation
A common feature of many plant-colonizing organisms is the exploitation of plant signaling and developmental pathways to successfully establish and proliferate in their hosts. Auxins are central plant growth hormones, and their signaling is heavily interlinked with plant development and immunity responses. Smuts, as one of the largest groups in basidiomycetes, are biotrophic specialists that successfully manipulate their host plants and cause fascinating phenotypes in so far largely enigmatic ways.
smut fungi
auxin signaling
biotrophic interactions
effectors
1. Auxin, the Master Regulator of Growth and Development in Plants
The word auxin originates from the Greek word “auxein”, which means “to grow”. Since its discovery in 1928 by Fritz Went [1], the roles of auxin have been implicated in almost all aspects of plant growth and development [2][3]. Auxin is involved in the control of processes such as cell division, elongation, differentiation, tropisms, flowering, apical dominance, lateral root formation, senescence, abscission, and responses to environmental stresses [4]. Indole-3-acetic acid (IAA) is the most prominent auxin and is widely investigated in plants, but other naturally occurring or synthetic molecules with auxin activity exist [5]. Auxin is synthesized primarily in the shoot meristem and young leaves from the amino acid tryptophan, but there are also tryptophan-independent pathways [6]. Once synthesized, auxin is transported in the plant through the phloem and a polar (directional) transport system [5]. Auxin concentrations in each tissue are tightly regulated by metabolism and polar transport [7], which, in turn, regulate auxin-mediated gene expression.
Auxin functions primarily by regulating gene expression through a pathway called canonical auxin signaling [7][8]. However, an increasing amount of data shows that auxin also functions through alternative mechanisms [9]. In this context, only a brief summary of canonical auxin signaling is presented, which has been highly conserved in plant evolution at least since the emergence of land plants [9]; noncanonical auxin signaling is reviewed elsewhere [3][9]. The core components of canonical auxin signaling include the auxin receptor of the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) family, the transcriptional repressors of the AUXIN/INDOLE-3-ACETIC ACID INDUCIBLE (Aux/IAA) family, and the transcription factors (TF) of the AUXIN RESPONSE FACTORS (ARF) family [2][7]. A TIR1/AFB F-box protein and an Aux/IAA transcriptional coregulator complex bind auxin, and this binding further promotes the interaction between TIR1/AFB and Aux/IAA, thus triggering ubiquitin-mediated degradation of Aux/IAA proteins via the proteasome (Figure 1). Aux/IAA proteins generally act as repressors to prevent auxin-responsive transcription by binding to ARFs [7][8]. The Aux/IAA proteins contain ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motifs through which they recruit TOPLESS (TPL)/TOPLESS-RELATED (TPR) corepressors that can themselves recruit histone deacetylases (HDACs) to target loci; thus, they are responsible for the repression [2][10]. The cocrystal structure between TPL and the Aux/IAA EAR domain demonstrated that Aux/IAAs bind through the EAR motif to the so-called CTLH region at the N-terminus of the TPL repressor [11]. The degradation of Aux/IAA proteins leads to the derepression of ARFs, and auxin-responsive genes are thus activated. Auxin response factors (ARFs) are transcriptional factors that are bound to the cis-regulatory regions of auxin-responsive genes. ARFs can act as transcriptional activators or repressors depending on their function [12]. Numerous auxin-responsive genes, comprising early auxin-responsive genes (like the SAUR, IAA, and GH3 genes) and late auxin-responsive genes (such as genes involved in cell wall remodeling and growth), are expressed as a consequence of ARF activation [13]. Among these genes, a distinct set of genes is regulated depending on the cell type, indicating responses to auxin that are space- and time-specific. The complexity of these interactions increases with the duration of exposure to auxin [2].
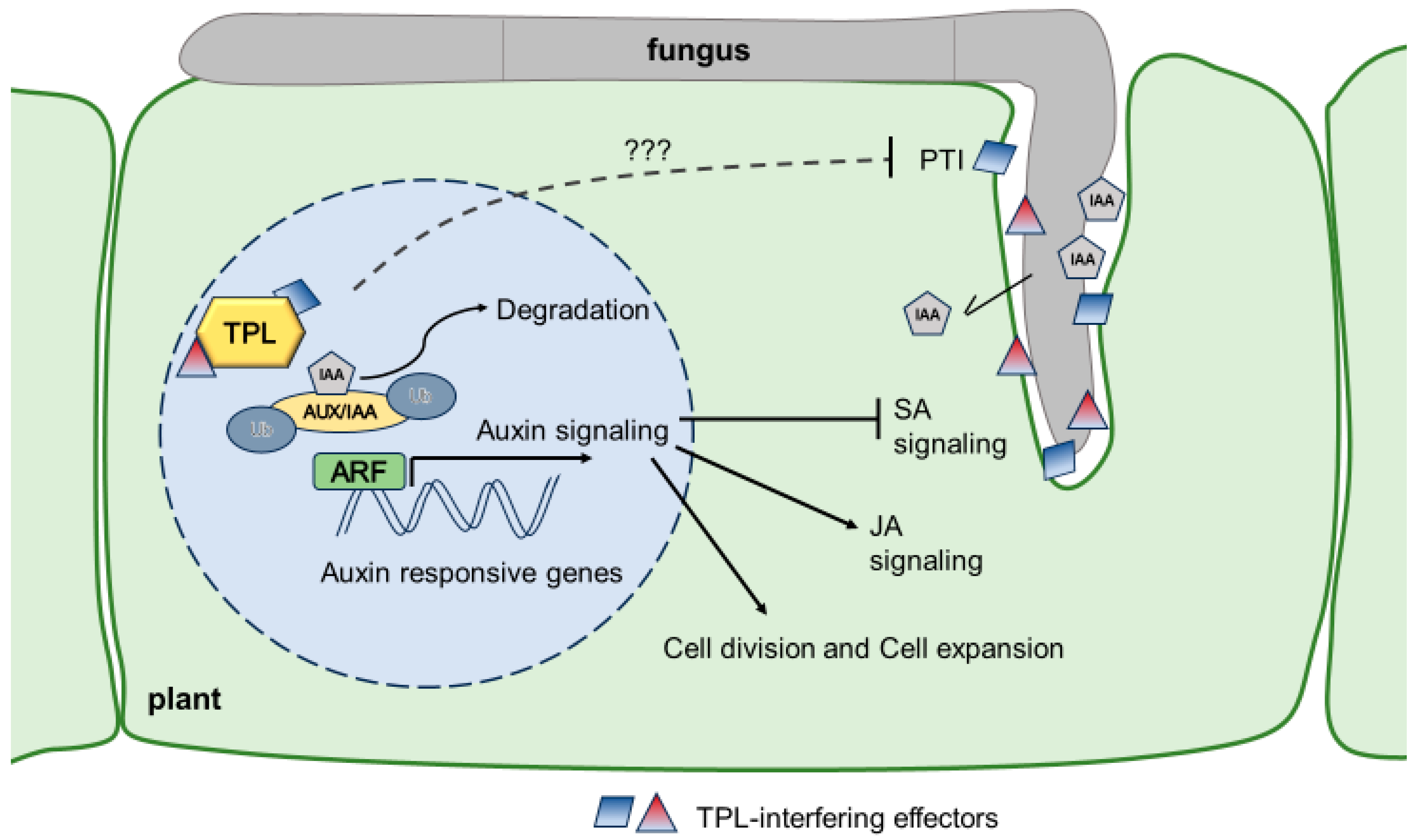
Figure 1. Model how U. maydis influences auxin signaling in maize. U. maydis produces IAA and releases it during the colonization of the host tissue. Additionally, it secretes at least ten translocated effectors (Tip1-8, Nkd1, Jsi1) which all interact with members of the TPL/TPR protein family and, among others, also lead to the derepression of auxin signaling and PTI suppression through unknown mechanisms (???) Figure 1 was created using Microsoft PowerPoint.
2. Auxin in Plant–Pathogen Interactions
Plant defense responses are tightly regulated to optimize the tradeoffs between growth and defense [14]. Crosstalk between phytohormones plays a significant role in this regulatory process. In addition to the growth–defense antagonism, plants have further evolved a signaling antagonism in their response to pathogens of different lifestyles. Generally speaking, plants depend on Jasmonate (JA)/ethylene-dependent defense responses when defending against necrotrophic invaders, whereas salicylic acid (SA)-dependent signaling is key in fighting off biotrophic or hemibiotrophic pathogens during the phase when they depend on living host cells during colonization. Due to their different roles against various lifestyle dependencies of invading pathogens, SA and JA/Ethylene act antagonistic to each other. Nevertheless, JA/ethylene and SA hormones can be considered as clearly defense-related phytohormones. Auxin and other growth hormones in plants are modulators of defense responses, and auxin has been reported to mainly act antagonistically to SA signaling and vice versa [15][16]. Therefore, it is not surprising that the vast majority of reports of auxin signaling modulation promoting plant susceptibility come from biotrophic and hemibiotrophic plant–pathogen interactions [17][18][19]. The interaction-promoting role of auxins in symbiotic interactions of plants with mycorrhizal fungi, nodulating bacteria, or plant-growth-promoting rhizobacteria might also partly rely on the defense-modulating effects of auxin [20]. The strategies that microbes have evolved to manipulate auxin signaling are fascinatingly versatile. Various plant-associated microbes produce and secrete auxins themselves [21], while others manipulate their distribution. Auxin is redistributed by PIN-FORMED, ABC, and AUX/LAX transporters within the plant body to shape morphogenesis. The relocalization of PIN transporters has been observed during cyst nematode infection, which likely causes an influx of auxins into the initial syncytial cell and is also involved in the expansion of the nematode-feeding structure [22]. The overexpression of the potyvirus suppressor of RNA silencing, HC-Pro, in the model plant A. thaliana (L.) led to the downregulation of DNA methylation in the regulatory region of the YUCCA genes of the indole-3-pyruvic acid (IPyA) auxin biosynthesis pathway. As a consequence, changes in the methylation pattern lead to transcriptional upregulation and, therefore, increase auxin biosynthesis in planta [23]. Pseudomonas syringae (Van Hall) have been shown to secrete AvrRpt2 to interfere with auxin-signaling transcriptional repressor Aux/IAA proteins to increase auxin signaling for disease progression, as also shown for viral effectors [19][24][25][26]. The corepressor TOPLESS is manifold-manipulated by U. maydis, which will be described in more detail below. In contrast, the auxin response factor TF is targeted by various rice-infecting viruses. These viruses encode effectors, which block the dimerization of OSARF17 or, in other cases, direct DNA binding and thus render this specific auxin response factor inactive to block the transcription of antiviral gene responses under its control [27]. Similarly, several plant pathogenic bacteria encode IAA-Lys synthases. IAA-Lys is a less active form of IAA. The bacterial pathogen of oleander, Pseudomonas savastanoi pv. neri, produces IAA-LYS synthases and seems to control the free levels of IAA in infected tissue in this way [28]. There are not many reports in which auxin has been shown to play a general promoting role in plant immunity. In the fungal Rhizoctonia solani (J.G. Kühn)/rice interaction, external auxin treatment increased resistance [29]. Mutants of Arabidopsis thaliana (L.) affected in the auxin pathway are shown to be more susceptible than wild-type plants to the necrotrophic fungus Alternaria brassicicola (Schwein.). Upon infection, plants respond with the upregulation of auxin responses. The cotreatment of plants with both MeJA and IAA leads to the synergistic upregulation of typical JA marker genes [30]. This finding supports the hypothesis that the positive role of auxin signaling against necrotrophs might be through the enhancement of JA signaling.
3. Auxin Biosynthesis in Smut Fungi
Many plant pathogens possess the capacity to synthesize indole-3-acetic acid (IAA), the major form of auxin in plants. However, so far, the knowledge of auxin biosynthesis in smut fungi has been largely derived from the model smut fungus U. maydis. IAA is known to be synthesized through several pathways; the most common route is the conversion of L-tryptophan to IAA, but there are also tryptophan-independent pathways [31]. Although both pathways coexist in some organisms, most studies have focused on tryptophan-dependent pathways. At least six different tryptophan-dependent biosynthetic pathways have been described according to their key intermediates: indole-3- acetamide (IAM), indole-3-acetaldoxime (IAOx), indole-3-pyruvic acid (IPyA), indole-3-acetamide (IAM), tryptamine (TAM) and indole-3-acetonitrile (IAN), and Trp side chain oxidase (TSO) [32][33].
The gall-forming corn smut fungus U. maydis has been reported to produce indole acetic acid, which is also associated with elevated auxin levels during its biotrophy in maize [34][35]. Several genes involved in the IAA biosynthesis pathway from U. maydis have been identified and characterized [36][37], including two NAD-dependent IAAld dehydrogenases (iad1 and iad2) and two predicted aromatic amino acid aminotransferase genes (tam1 and tam2). Aminotransferases Tam1 and Tam2 participate in the conversion of tryptophan to IPyA. Downstream of this, the dehydrogenases Iad1 and Iad2 are shown to be involved in the formation of IAA from indole-3-acetamide (IAAld). The increase in host IAA levels upon U. maydis infection was significantly reduced in tissue infected with quadruple Δiad1Δiad2Δtam1Δtam2 U. maydis mutants, suggesting a critical contribution of fungal IAA production to elevated IAA levels in infected tissue. However, a reduction in fungal auxin production did not have any effect on gall formation and the development of host plant disease symptoms, suggesting alternative strategies for this phenomenon [37]. In the genome of the head smut fungus Sporisorium scitamineum (Syd.), a gene coding for the tryptophan aminotransferase SsAro8 has been identified. The tryptophan aminotransferase SsAro8 has been shown to catalyze the first step of tryptophan-dependent IAA production, but also for the Ehrlich pathway for tryptophol. It was found to be essential for mating/filamentation and proper biofilm formation in S. scitamineum [38]. As SsAro8 is at the beginning of several pathways with different intermediates and products, it is not clear whether any of the fungal phenotypes identified are related to altered IAA production or due to the lack of other Aro8-dependent metabolites. Furthermore, in U. scitaminea and U. esculenta, the ability to synthesize IAA from tryptophan was found through the indole-pyruvate (IPA) pathway. U. esculenta apparently could convert IAAld and indole-lactic acid (ILA) into IAA in addition to tryptophan [39]. The finding that auxin biosynthesis genes are expressed in a carbon-source-dependent manner in axenic culture in U. maydis [37], but also findings in other fungi, like the ascomycete M. oryzae (T.T. Hebert), show that IAA acts as a quorum-based modulator of virulence [40]. These findings raise questions about whether, beyond its obvious impact as a phytohormone, fungal auxin possibly also plays a role as an intraspecific signaling molecule of IAA-producing smuts.
References
- Went, F. Wuchsstoff und Wachstum. Recl. Trav. Bot. Néerl. 1928, 25, 1–16.
- Caumon, H.; Vernoux, T. A matter of time: Auxin signaling dynamics and the regulation of auxin responses during plant development. J. Exp. Bot. 2023, 74, 3887–3902.
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr Plant Biol. 2022, 64, 371–392.
- Enders, T.A.; Strader, L.C. Auxin activity: Past, present, and future. Am. J. Bot. 2015, 102, 180–196.
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904.
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343.
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479.
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574.
- Ang, A.C.H.; Ostergaard, L. Save your TIRs—More to auxin than meets the eye. New Phytol. 2023, 238, 971–976.
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386.
- Ke, J.; Ma, H.; Gu, X.; Thelen, A.; Brunzelle, J.S.; Li, J.; Xu, H.E.; Melcher, K. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 2015, 1, e1500107.
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971.
- Eckardt, N.A. Auxin and the power of the proteasome in plants. Plant Cell 2001, 13, 2161–2163.
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287.
- Iglesias, M.J.; Terrile, M.C.; Casalongué, C.A. Auxin and salicylic acid signalings counteract the regulation of adaptive responses to stress. Plant Signal. Behav. 2011, 6, 452–454.
- Singh, D.; Dhiman, V.K.; Pandey, H.; Dhiman, V.K.; Pandey, D. Crosstalk Between Salicylic Acid and Auxins, Cytokinins and Gibberellins Under Biotic Stress. In Auxins, Cytokinins and Gibberellins Signaling in Plants; Aftab, T., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 249–262.
- Kunkel, B.N.; Johnson, J.M.B. Auxin Plays Multiple Roles during Plant-Pathogen Interactions. Cold Spring Harb. Perspect. Biol. 2021, 13, a040022.
- Oosterbeek, M.; Lozano-Torres, J.L.; Bakker, J.; Goverse, A. Sedentary Plant-Parasitic Nematodes Alter Auxin Homeostasis via Multiple Strategies. Front. Plant Sci. 2021, 12, 668548.
- Chen, Z.; Agnew, J.L.; Cohen, J.D.; He, P.; Shan, L.; Sheen, J.; Kunkel, B.N. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA 2007, 104, 20131–20136.
- Hilbert, M.; Nostadt, R.; Zuccaro, A. Exogenous auxin affects the oxidative burst in barley roots colonized by Piriformospora indica. Plant Signal. Behav. 2013, 8, e23572.
- Tang, J.; Li, Y.; Zhang, L.; Mu, J.; Jiang, Y.; Fu, H.; Zhang, Y.; Cui, H.; Yu, X.; Ye, Z. Biosynthetic Pathways and Functions of Indole-3-Acetic Acid in Microorganisms. Microorganisms 2023, 11, 2077.
- Grunewald, W.; Cannoot, B.; Friml, J.; Gheysen, G. Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog. 2009, 5, e1000266.
- Yang, L.; Meng, D.; Wang, Y.; Wu, Y.; Lang, C.; Jin, T.; Zhou, X. The viral suppressor HCPro decreases DNA methylation and activates auxin biosynthesis genes. Virology 2020, 546, 133–140.
- Padmanabhan, M.S.; Goregaoker, S.P.; Golem, S.; Shiferaw, H.; Culver, J.N. Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J. Virol. 2005, 79, 2549–2558.
- Jin, L.; Qin, Q.; Wang, Y.; Pu, Y.; Liu, L.; Wen, X.; Ji, S.; Wu, J.; Wei, C.; Ding, B.; et al. Rice Dwarf Virus P2 Protein Hijacks Auxin Signaling by Directly Targeting the Rice OsIAA10 Protein, Enhancing Viral Infection and Disease Development. PLoS Pathog. 2016, 12, e1005847.
- Cui, F.; Wu, S.; Sun, W.; Coaker, G.; Kunkel, B.; He, P.; Shan, L. The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 2013, 162, 1018–1029.
- Zhang, H.; Li, L.; He, Y.; Qin, Q.; Chen, C.; Wei, Z.; Tan, X.; Xie, K.; Zhang, R.; Hong, G.; et al. Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc. Natl. Acad. Sci. USA 2020, 117, 9112–9121.
- Cerboneschi, M.; Decorosi, F.; Biancalani, C.; Ortenzi, M.V.; Macconi, S.; Giovannetti, L.; Viti, C.; Campanella, B.; Onor, M.; Bramanti, E.; et al. Indole-3-acetic acid in plant-pathogen interactions: A key molecule for in planta bacterial virulence and fitness. Res. Microbiol. 2016, 167, 774–787.
- Qiao, L.; Zheng, L.; Sheng, C.; Zhao, H.; Jin, H.; Niu, D. Rice siR109944 suppresses plant immunity to sheath blight and impacts multiple agronomic traits by affecting auxin homeostasis. Plant J. 2020, 102, 948–964.
- Qi, L.; Yan, J.; Li, Y.; Jiang, H.; Sun, J.; Chen, Q.; Li, H.; Chu, J.; Yan, C.; Sun, X.; et al. Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 2012, 195, 872–882.
- Wang, B.; Chu, J.; Yu, T.; Xu, Q.; Sun, X.; Yuan, J.; Xiong, G.; Wang, G.; Wang, Y.; Li, J. Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 4821–4826.
- Fu, J.; Wang, S. Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2011, 2, 74.
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735.
- Turian, G.; Hamilton, R.H. Chemical detection of 3-indolylacetic acid in Ustilago zeae tumors. Biochim. Biophys. Acta 1960, 41, 148–150.
- Wolf, F.T. The Production of Indole Acetic Acid by Ustilago Zeae, and Its Possible Significance in Tumor Formation. Proc. Natl. Acad. Sci. USA 1952, 38, 106–111.
- Basse, C.W.; Lottspeich, F.; Steglich, W.; Kahmann, R. Two potential indole-3-acetaldehyde dehydrogenases in the phytopathogenic fungus Ustilago maydis. Eur. J. Biochem. 1996, 242, 648–656.
- Reineke, G.; Heinze, B.; Schirawski, J.; Buettner, H.; Kahmann, R.; Basse, C.W. Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol. Plant Pathol. 2008, 9, 339–355.
- Cui, G.; Huang, C.; Bi, X.; Wang, Y.; Yin, K.; Zhu, L.; Jiang, Z.; Chen, B.; Deng, Y.Z. Aminotransferase SsAro8 Regulates Tryptophan Metabolism Essential for Filamentous Growth of Sugarcane Smut Fungus Sporisorium scitamineum. Microbiol. Spectr. 2022, 10, e0057022.
- Chung, K.R.; Tzeng, D. Biosynthesis of Indole-3-Acetic Acid by the Gall-inducing Fungus Ustilago esculenta. J. Biol. Sci. 2004, 4, 744–750.
- Dong, L.; Shen, Q.; Chen, C.-Y.; Shen, L.; Yang, F.; Naqvi, N.I.; Deng, Y.Z. Fungal auxin is a quorum-based modulator of blast disease severity. bioRxiv 2021.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
483
Revisions:
2 times
(View History)
Update Date:
02 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




