Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Luisa Maia | -- | 4873 | 2023-12-27 15:28:25 | | | |
| 2 | Lindsay Dong | -6 word(s) | 4867 | 2023-12-28 02:34:14 | | | | |
| 3 | Lindsay Dong | Meta information modification | 4867 | 2023-12-29 09:40:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Maia, L.B.; Maiti, B.K.; Moura, I.; Moura, J.J.G. Role of Selenium in Redox Biology. Encyclopedia. Available online: https://encyclopedia.pub/entry/53184 (accessed on 07 February 2026).
Maia LB, Maiti BK, Moura I, Moura JJG. Role of Selenium in Redox Biology. Encyclopedia. Available at: https://encyclopedia.pub/entry/53184. Accessed February 07, 2026.
Maia, Luisa B., Biplab K. Maiti, Isabel Moura, José J. G. Moura. "Role of Selenium in Redox Biology" Encyclopedia, https://encyclopedia.pub/entry/53184 (accessed February 07, 2026).
Maia, L.B., Maiti, B.K., Moura, I., & Moura, J.J.G. (2023, December 27). Role of Selenium in Redox Biology. In Encyclopedia. https://encyclopedia.pub/entry/53184
Maia, Luisa B., et al. "Role of Selenium in Redox Biology." Encyclopedia. Web. 27 December, 2023.
Copy Citation
Living organisms use selenium mainly in the form of selenocysteine in the active site of oxidoreductases. Here, selenium’s unique chemistry is believed to modulate the reaction mechanism and enhance the catalytic efficiency of specific enzymes in ways not achievable with a sulfur-containing cysteine. However, despite the fact that selenium/sulfur have different physicochemical properties, several selenoproteins have fully functional cysteine-containing homologues and some organisms do not use selenocysteine at all.
selenium
biology
1. Introduction
Discovered in 1817, selenium was for long regarded as a toxic element [1][2][3] and only in the second half of the XX century was it demonstrated to be essential for all forms of life [4][5][6][7][8][9][10]. Living organisms have learned to harness the unique chemical features provided by selenium (over sulfur) and use this element mainly in the active site of oxidoreductases in the form of selenocysteine, an amino acid genetically encoded by a specific codon (UGA) that is considered the 21st amino acid.
Several selenocysteine-containing enzymes evolved to play essential roles in various biological processes. Still, some of those selenoproteins have fully functional cysteine-containing homologues and some organisms do not use selenocysteine at all. Hence, understanding the biological use of selenium is of considerable interest.
2. Selenium versus Sulfur
Selenium is a chemical element belonging to the chalcogens family of the Periodic Table (Group 16). It resembles the “lighter” sulfur in some chemical features and, in Biology, selenium can be found replacing sulfur in two amino acids: selenocysteine (Se-Cys) and selenomethionine (Se-Met). However, in spite of the similarities, many significant chemical differences exist between these two chalcogens [11][12][13][14][15]. As sulfur, selenium can display a wide range of oxidation states (from −2 to +6), but its preference for lower oxidation states and higher reactivity sets it apart from sulfur. Its reactions are often also faster than its sulfur counterparts because selenium is more polarizable (softer). Its larger spin—orbit coupling (compared to sulfur) probably facilitates spin-forbidden reactions, as the ones involved in the rapid oxidation of selenocysteine under air (compared to cysteine oxidation). Moreover, the selenocysteine selenol’s lower pKa value (5.2, compared to 8.3 of cysteine thiol) is expected to favor its deprotonation and nucleophilic character at physiological pH [16], while the weaker Se-H bond makes the selenocysteine less basic, compared to cysteine [17][18]. The biologically relevant redox chemistry is also significantly different in these two elements [19][20][21]. The selenocysteine one-electron oxidation-derived radical is more easily formed ((RSe•/RSeH) = 0.43 V versus (RS•/RSH) = 0.92 V [22]) and relatively more stable than the cisteine radical [22][23][24]. As a result, for example, while the cysteine radical can oxidize a tyrosine residue (to yield tyrosine radical), the selenocysteine radical can not [22]. Also noteworthy are the thiol/disulfide exchange reactions, where the selenocysteine reactions (Se-Cys/Cys-Se-Se-Cys) are faster than the cysteine ones [12][14][25][26].
3. Formate Dehydrogenase
FDH was one of the first enzymes demonstrated to contain selenium and a selenocysteine-specific codon (TGA) in its gene sequence (Clostridium thermoaceticum and E. coli enzymes) [27][28]. Those seminal works were essential to overcome the prevailing idea that selenium was (only) a toxic substance and lead to its recognition as an essential element (also for mammals and humans by contemporary works).
In spite of being one of the most widely distributed selenoproteins (probably due to its extensive lateral gene transfer, together with the corresponding selenocysteine synthesis and incorporation system) [29], FDH constitutes a key example where, as far as is presently known, selenium does not present any clear advantage over sulfur. Contrary to other selenoenzymes, living organisms hold both active selenocysteine- and cysteine-containing FDH homologues and, thus, the selenium role in FDH catalysis remains, so far, elusive.
3.1. The Current Picture
3.1.1. Enzymatic Machinery
FDHs catalyze the two-electron interconversion of formate and carbon dioxide (Equation (1)) in diverse metabolic pathways, operating in different subcellular locations, such as C1 metabolism, carbon dioxide fixation (carbon assimilation), and to derive energy (coupling formate oxidation to the reduction of different terminal electron acceptors) [30][31][32][33][34][35][36][37][38]. Since each pathway requires a specific “FDH enzymatic machinery” to accomplish the respective biological function, FDHs evolved as a highly heterogeneous group of enzymes, displaying diverse structural (subunits) organization and composition of redox-active centers [39][40][41][42][43][44][45][46][47][48].
HCOO− ⇌ CO2 + 2e− + H+
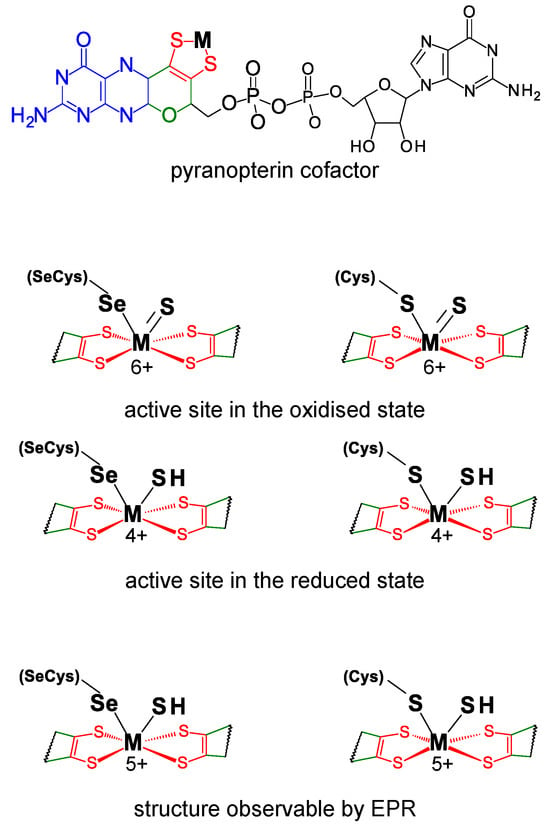
FDHs can be divided into two main classes. The metal-independent FDH class comprises enzymes, typically homodimers that have no metal ions or other redox-active centers, nor selenium [49][50][51][52][53][54]. These enzymes, found in bacteria, fungi, and plants, are NAD-dependent and belong to the D-specific dehydrogenases of the 2-oxyacids family. On the contrary, the metal-dependent FDH class, present only in prokaryotes, comprises enzymes that harbor different redox-active centers and display high structural diversity [41][42][43][45][46][48]. As the class name indicates, the active site of these enzymes holds one molybdenum or one tungsten ion in a very well conserved metal center (Figure 1). In its oxidized (6+) form, the metal (molybdenum or tungsten) is coordinated by the cis-dithiolene (–S–C=C–S–) group of two pyranopterin cofactor molecules, one terminal sulfido group (Mo6+/W6+=S), plus one selenium or one sulfur atom from a selenocysteine or cysteine residue (Mo6+/W6+-Se(Cys) or Mo6+/W6+-S(Cys)) (abbreviated as SeCys-Mo-FDH, SeCys-W-FDH, Cys-Mo-FDH, and Cys-W-FDH) [40][44][55][56]. Noteworthy, there is no apparent relation (as far as is presently known) between the metal (molybdenum or tungsten) and the presence of a selenocysteine or cysteine residue and catalytically efficient SeCys-Mo-FDH, SeCys-W-FDH, Cys-Mo-FDH, and Cys-W-FDH have been known for a long time.

Figure 1. Active site structure of metal-dependent FDHs and FMFDHs. Top: Structure of the pyranopterin cofactor. The pyranopterin cofactor molecule is formed by pyrano(green)–pterin(blue)–dithiolene(red)–methylphosphate(black) moieties; in all so far characterized enzymes, the cofactor is found esterified with a guanosine monophosphate (dark gray). The dithiolene (–S–C=C–S–) group forms a five-membered ene-1,2-dithiolene chelate ring with the molybdenum or tungsten ion, here indicated as M (from metal). Middle: Structure of the active site in the oxidized and reduced state. Bottom: Active site structure supported by EPR data. In middle and bottom structures, for simplicity, only the dithiolene moiety of the pyranopterin cofactor is represented.
Similar to FDHs, selenocysteine-containing and cysteine-containing N-formyl-methanofuran dehydrogenases (SeCys-FMFDH and Cys-FMFDH) exist and selenium’s role in FMFDH catalysis is unknown as well. FMFDHs are FDH-like enzymes that have two physically separated active sites: one catalyzes the reduction of carbon dioxide to formate, which is then intramolecularly transferred to the second active site, where it is condensed with methanofuran to form N-formyl-methanofuran [57][58][59][60].
3.1.2. Reaction Mechanism
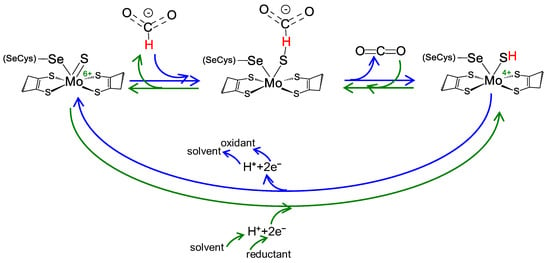
Regardless of the physiological function and structural complexity, the reaction mechanism of the interconversion of formate and carbon dioxide (Equation (1)) is believed to be similar in all these selenocysteine- and cysteine-containing FDH and FMFDH enzymes. As originally proposed by Niks et al. [61] for formate oxidation and shortly after also for carbon dioxide reduction by Maia et al. [62], it is currently well established that formate oxidation and carbon dioxide reduction proceed through hydride transfer, with the oxidized and reduced active site sulfido group, Mo/W6+=S and Mo/W4+-SH, acting as the direct hydride acceptor and donor, respectively (Figure 2) [63][64][65][66] (even though other atomic details of the reaction mechanism are not yet consensual; see, for example [67]). It is noteworthy that no direct role in the chemical transformations is presently ascribed to the selenocysteine or cysteine residue, in accordance with the existence of catalytically efficient SeCys enzymes and Cys enzymes (a similar situation occurs with molybdenum and tungsten).

Figure 2. Reversible FDH and FMFDH reaction mechanism, as proposed by Maia et al. [62]. Reaction mechanism proposed for formate oxidation (blue arrows) and carbon dioxide reduction (green arrows) for both metal-dependent FDHs and FMFDHs. For simplicity, the mechanism is represented for a molybdenum, selenocysteine-containing enzyme, but it should be similar for tungsten and cysteine-containing enzymes. See text for details.
Briefly, formate oxidation (Figure 2, blue arrows) is initiated with the formate binding to the oxidized active site but not directly to the molybdenum/tungsten atom. Formate is suggested to bind in a binding pocket, where a conserved arginine residue “anchors” its oxygen atom(s) through hydrogen bond(s), and forces its Cα hydrogen to point towards the sulfido ligand (Mo6+/W6+=S). Subsequently, formate oxidation proceeds by a straightforward hydride transfer from formate to the sulfido group of the oxidized molybdenum/tungsten centre, leading to the formation of Mo/W4+-SH and CO2. The re-oxidation of Mo/W4+ to Mo/W6+ (via intramolecular electron transfer to the enzyme’s other redox center(s) and, eventually, to the physiological partner) and the release of carbon dioxide close the catalytic cycle. The now oxidized Mo/W6+ favors the sulfido group deprotonation (dictated by the ligand pKa [68][69][70]) and the initial oxidized metal centre, Mo/W6+=S, is regenerated. Under non-steady-state catalytic conditions (such as the ones created in EPR experiments described below), the molybdenum/tungsten one-electron oxidation should be favored (Mo/W4+→Mo/W5+), leading to the formation of the EPR detectable species.
The carbon dioxide reduction is suggested to follow the reverse reaction mechanism (Figure 2, green arrows) but starting with a reduced active site, holding a protonated sulfido group, Mo/W6+-SH (as is dictated by the ligands pKa [68][69][70]). Carbon dioxide is suggested to bind to the same binding pocket, where the arginine residue is key to anchor it in the correct position to orient its carbon atom towards the protonated sulfido. Afterwards, the reaction proceeds through straightforward hydride transfer from the protonated sulfido group. This yields a formate moiety and Mo/W6+=S. The subsequent re-reduction of Mo/W6+ to Mo/W4+ (via intramolecular electron transfer from the enzyme’s physiological partner, through its redox center(s)) and formate release closes the catalytic cycle. The now reduced Mo/W4+ favors the sulfido group protonation and the initial reduced molybdenum/tungsten center, Mo/W4+-SH, is regenerated.
3.2. How Was the Selenium Locus in Formate Dehydrogenases Established?
The presence and essentiality of selenium was demonstrated in pioneer works, mainly in the 1970s, following the incorporation in target enzymes of selenium-75 (present in the growth medium/feed). Actually, FDH was among the first enzymes shown to contain selenium [27][28].
The recognition of the presence of molybdenum or tungsten and selenium led to a series of spectroscopic studies that were decisive to the early characterization of the FDH active site. Electron paramagnetic resonance spectroscopy (EPR) was thoroughly explored (reviewed recently, for example, in [71][72][73]). In fact, the first evidence for the direct binding of selenium to a metal (molybdenum) active site center was obtained precisely with EPR [74][75]. The E. coli SeCys-Mo-FDH H was one of the first FDHs to be explored [75][76]. When reduced with formate, it gives rise to a nearly axial Mo5+ signal, with g1 = 2.094 and g2,3 = 2.001, 1.990, that displays coupling to one formate-derived solvent-exchangeable proton (A1,2,3(1H) = 7.5, 18.9, 20.9 MHz). When the EPR signal is generated from the 77Se-enriched enzyme, a very strong and anisotropic interaction is observed (A1,2,3(77Se) = 13.2, 75, 240 MHz [77], values that are almost five times higher than the ones observed in Mo-Se model compounds [77][78]). This strong interaction, observed simultaneously with the expected 95,97Mo hyperfine coupling, confirmed that the selenocysteine selenium atom is directly coordinated to the molybdenum (Figure 1) and suggested that the unpaired electron is delocalized 17–27% over the selenium (a finding in line with the expected covalency introduced by selenium in a Mo-Se bond) [77].
These original studies with E. coli FDH H were supported and consolidated with other selenium-containing FDHs, including Desulfovibrio desulfuricans [79], D. gigas [80][81], D. vulgaris Hildenborough [82][83][84][85], and Methylosinus trichosporium [86] FDHs. These enzymes display rhombic Mo5+/W5+ EPR signals with small anisotropy, a well-resolved hyperfine structure due to 95,97Mo/183W, and interaction with a solvent-exchangeable proton (for example: D. desulfuricans: g1,2,3 = 2.012, 1.996, 1.985, A1,2,3(solvent-exchangeable 1H) = 23.1, 29.9, 27.8 MHz [79]; D. vulgaris Hildenborough FDH 1: g1,2,3 = 1.995, 1.881, 1.852, A1,2,3(183W)= 225, 129, 134 MHz [82]; D. vulgaris Hildenborough FDH 2 (main component): g1,2,3 = 1.982, 1.876, 1.902, A1,2,3(183W)= 232, 119, 151 MHz [84]). The D. desulfuricans FDH displays also a hyperfine interaction with a second non-solvent-exchangeable proton (A1 = 35.1 MHz, A2,3 not detectable) that was assigned to the metal-bound selenocysteine Cβ hydrogen atoms [79]. Together, the EPR data suggest an FDH active site holding a stable selenocysteine–metal ligation. It also suggests that the active site holds a transient proton-accepting site (within the metal magnetic contact) that was assigned as the terminal sulfido group (please see Note above) [61][62].
The SeCys-FDH active site was also explored by X-ray absorption spectroscopy (XAS) since early times [87]. XAS at the molybdenum and selenium K-edges of the most explored model FDH, E. coli SeCys-Mo-FDH H, revealed four Mo-S ligands at 2.35 Å, one (originally not assigned) Mo=S at 2.1 Å, and one Mo-Se ligand at 2.62 Å, in both oxidized and reduced enzyme [88]. In the D. desulfuricans SeCys-Mo-FDH, the molybdenum and selenium K-edges data also showed a hexa-coordinated active site, with one Mo-Se ligand at 2.57 Å in both oxidized and reduced enzyme [89]. It is noteworthy that the replacement of the E. coli SeCys-Mo-FDH H selenocysteine by a cysteine residue abolished the Mo-Se fingerprint and gave rise to a spectrum consistent with five Mo-S ligands and one Mo=O at 1.7 Å [88]. Comparatively, XAS studies of native Cys-FDHs (for example, oxidized Rhodobacter capsulatus Cys-Mo-FDH [90][91]) confirmed that the cysteine residue is bound to the metal, as expected. Hence, the XAS results are in excellent agreement with the EPR proposed FDH active site structure, Mo5+/W5+-Se(Cys)(-SH) (Figure 1).
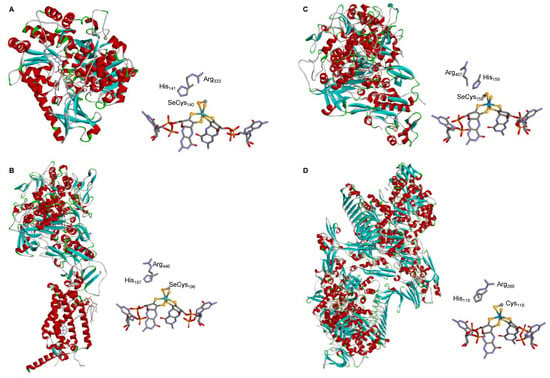
The crystallographic structure of different native SeCys- (and Cys-) FDHs entirely supports this active site structure. The first FDH 3D structure solved, in 1997, was the one of the model E. coli SeCys-Mo-FDH H [92] and this was the only one known for 5 years (2002), when the structure of two more enzymes were finally solved, the E. coli SeCys-Mo-FDH N [93] and Desulfovibrio gigas SeCys-W-FDH [94] (Figure 3). The first FMFDH structure (the Methanothermobacter wolfeii Cys-W-FMFDH) was revealed only 14 years after, in 2016 [59]. Presently, several structures are known [60][82][83][95][96][97][98][99] and the active site structure is firmly established to be the conserved Mo6+-Se(Cys)(=S), W6+-Se(Cys) (=S), Mo6+-S(Cys)(=S), or W6+-S(Cys)(=S) (Figure 1).

Figure 3. Three-dimensional structure view of some metal-dependent FDHs and FMFDHs and their active sites. (A) E. coli SeCys-Mo-FDH H [92]; (B) E. coli SeCys-Mo-FDH N [93]; (C) D. gigas SeCys-W-FDH [94]; (D) M. wolfeii Cys-W-FMFDH [59]. The structures shown are based on the PDB files 1FDO (A), 1KQF (B), 1H0H (C), and 5T5I (D) (α helices and β sheets are shown in red and cyan, respectively).
3.3. Why Do Some Formate Dehydrogenases Have a Selenocysteine and Not the Less “Expensive” Cysteine Residue?
Since its early identification as a selenium-containing enzyme, the role of selenium in FDH catalysis has intrigued the scientific community. A pioneer work in the late 1980s [100] with the model E. coli SeCys-Mo-FDH H showed that selenocysteine (SeCys140) replacement with a cysteine residue resulted in significant lower FDH activity, while replacement with a serine residue rendered the enzyme inactive. In a subsequent, more comprehensive work by the Stadtman group [101], it was clearly shown that selenocysteine replacement with a cysteine resulted in a marked decrease in FDH activity (kcat/Kmformate (SeCys-FDH) = 108 × 103 M−1s−1 to kcat/Kmformate (Cys-FDH) = 1 × 103 M−1s−1) and the Cys-FDH variant’s slower kinetics was suggested to be due to a lower rate of the hydrogen atom transfer step (deuterium (formate) isotope effect on kcat/Km). Simultaneously, the pH-dependent alkylation-induced inactivation of the native SeCys-FDH and variant Cys-FDH (reaction with iodoacetamide in the presence of formate) was shown to follow the trend of the expected pKa values of each amino acid (native SeCys-FDH was inactivated more than 80% at pH > 6 (pKa (SeCys) ≈ 5.2), while variant Cys-FDH was inactivated more than 80% only at pH > 7 (pKa (Cys) ≈ 8.2). Together, these results were taken to suggest that selenol (versus thiol) plays an essential role in catalysis.
As other variant enzymes are studied, it is becoming clear that it is not surprising that variants are less active than wild types. Most relevant to the present discussion was the recognition that several “wild-type variants” (native Cys-FDH) exist that are as catalytically efficient as the native SeCys-FDHs.
Selenocysteine incorporation is highly demanding (“expensive”) for the cell. It requires additional energy and dedicated machinery to uptake selenium and to synthesize and orchestrate different biomolecules that lead to the recognition of target UGA-codon by specific tRNA molecules (and not as the “opal” stop-codon), culminating in selenocysteine being incorporated in the target protein [102][103][104][105][106]. Therefore, it is generally accepted that the presence of selenocysteine should constitute an intrinsic advantage for the cell [14][107][108][109].
Regardless of the biological pressure behind the evolution of native SeCys-FDHs and native Cys-FDHs, it should be kept in mind that selenium is not a sulfur. Thus, it is reasonable that the presence of one or the other alters the reaction energy pattern, in spite of both enzyme types operating through the same general hydride transfer mechanism (same chemical transformations). Therefore, in order to be catalytically efficient, each enzyme type should have evolved a strategy to compensate for those Se/S physicochemical differences. Hence, more interesting and relevant than studying why some FDHs have selenium is to understand the strategies that allow both SeCys-FDH and Cys-FDH to be catalytically efficient.
4. Hydrogenases
Hases are crucial role as an alternative energy source as they have potential applications in green hydrogen production [110][111]. Hydrogenases are a heterogeneous group of enzymes that differ in size, subunit composition, metal content, and cellular location (periplasmic, cytoplasmic, and cytoplasmic membrane-bound) and catalyze the reversible two electron oxidation of hydrogen (Equation (2)).
H2 ⇌ 2H+ + 2e−
4.1. Enzymatic Machineries
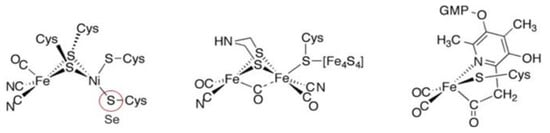
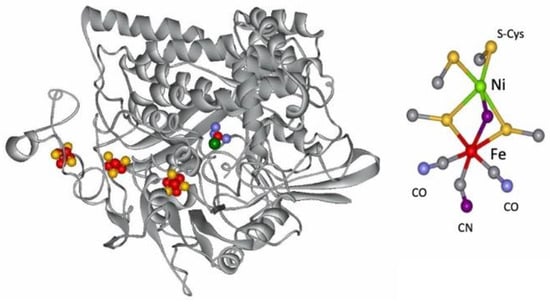
The metal-containing hydrogenases are subdivided into three classes: [Fe]-, [FeFe]-, and [NiFe]-hydrogenases (Figure 4) [110][111][112][113][114][115]. [Fe]-hydrogenases only contain one Fe ion in their active site and are designated as “Fe-only” hydrogenases. [FeFe]-hydrogenases contain an unusual iron-sulfur cluster termed the H-cluster that consists of an [Fe4S4] subcluster bridged via a cysteine (Cys) thiolate to the binuclear iron subcluster, also coordinated by inorganic ligands: two S atoms and one CO or CN ligand. [NiFe]-hydrogenases are heterodimeric proteins constituted by a small and a large subunit (Figure 5). The small subunit accommodates three iron-sulfur clusters (two [4Fe-4S] clusters and one [3Fe-4S] cluster) involved in the electron transport to/from the active site ([NiFe] cluster); the large subunit contains the catalytic site: the nickel-iron center. In some [NiFe]-hydrogenases, one of the Ni-bound cysteines is replaced by a selenocysteine, and [NiFe]- and [NiFeSe]-hydrogenases represent a single superfamily, and the Ni-Fe core contains unusual ligands: carbon monoxide (CO) and cyanide (CN−).

Figure 4. Active site structure of [NiFe]-, [FeFe]-, and [Fe]-hydrogenases [110].

Figure 5. Structure of the D. gigas hydrogenase enzyme and of its active site.
The [NiFe-Se] hydrogenases are found in some species of Desulfovibrio sp. The genes encoding the large and small subunits of the periplasmic hydrogenase from Desulfovibrio (D.) baculatus (DSM 1743) exhibit homology (40%) to the [NiFe] hydrogenases. The gene for the large subunit contains a codon (TGA) for selenocysteine in a position homologous to a codon (TGC) for cysteine in the [NiFe] hydrogenase. Spectroscopic studies support that selenium is a ligand to the nickel site (see below) [116][117][118][119][120][121].
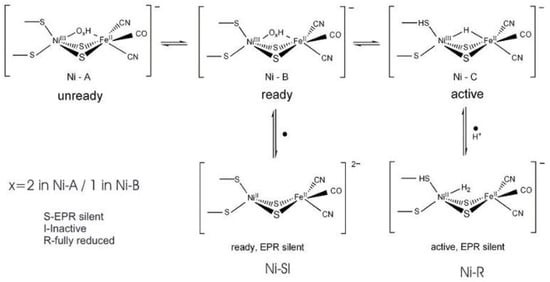
As isolated, the active [NiFe] cluster contains a Ni(III) and a low-spin Fe(II) (diamagnetic) that remain unchanged during the enzyme mechanism. Different oxidized inactive states are attained by the enzyme. In general, the isolated states are mixtures of “unready” Ni-A and “ready” Ni-B states (Figure 6). These states show delocalized electron density between nickel and iron, attributed to a third bridging oxygenated ligand. Both oxidized states are paramagnetic and characterized by different EPR g-values. The bridging ligand in the Ni-B state has been assigned to an OH− ligand and a water molecule is probably present in the Ni-A state [122][123][124][125]. After the reaction with the substrate (hydrogen), (Ni-C) develops with a bridging hydride (H−) ligand.

Figure 6. Redox and catalytic intermediates in [NiFe] hydrogenases. Adapted from [126].
4.2. Selenium and the Hydrogenase Reaction Mechanism
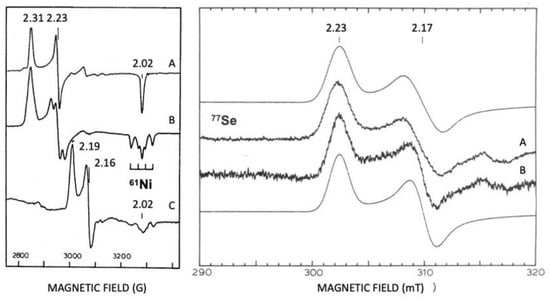
Isotopic substitutions are crucial for the identification of Ni and Se in hydrogenases. A 61Ni isotope was used for assigning EPR signals to Ni (Figure 7) [127]. Selenium contains six isotopes, and five of them are stable (atomic numbers 74, 76, 77, 78, and 80). The sixth isotope, with an atom abundance of 8.73%, is selenium-82 (82Se), a beta emitter which is weakly radioactive. The 77Se isotope (7.5%) is a useful EPR marker, with an I = 1/2. 33Se and 77Se are useful markers for spectroscopic studies (EXAFS and EPR) [127][128][129][130].

Figure 7. Revealing EPR, Ni, and Se at the active site of hydrogenases. Isotopic substitutions with 61Ni and 77Se. Left panel D. gigas [Ni-Fe] Hase. (A) Ni-A 61Ni un-enriched; (B) Ni-A 61Ni enriched; (C) Ni-C 61Ni enriched. Right panel D. baculatus [Ni-Fe-Se] Hase. (A) Ni-C 77Se enriched and (B) Ni-C 77Se un-enriched; mooth lines are simulations of spectra A and B. Adapted from refs [127][129].
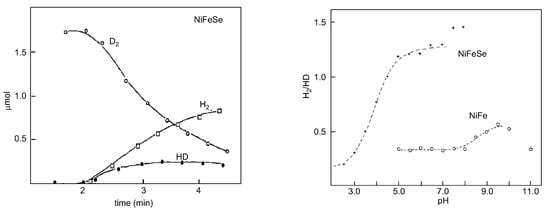
Proton–deuterium exchange measurements are quite appropriate to probe the influence of the Se–cysteine ligand in the mechanism of hydrogen handling. An important clue was the observation that the H2/HD ratios were higher for [NiFeSe] hydrogenases than those observed for the [NiFe] ones, which is related to the activation of the hydrogen molecule (Figure 8).

Figure 8. D2/HD exchange activity of D. salexigens [NiFeSe] hydrogenase (left panel) and variation in the experimental ratios H2/HD as a function of pH (right panel) of D. baculatus (cytoplasmic) [NiFeSe] hydrogenase and D. gigas [NiFe] (periplasmic). Left panel adapted from [131]; right panel adapted from [132].
4.3. Overview
Hydrogenases are a clear case study of the influence of Selenium (as a Se-Cys) on the modulating or fine-tuning of enzyme catalytic properties through an acid-base equilibrium at the proton acceptor site or at the hydride site and should be explored for protein design and molecular modelling [115].
The [NiFeSe] hydrogenases clearly emerge as a subgroup of [NiFe] and there is a structural homology between [NiFe] and [NiFeSe]. However, [NiFeSe] is distinct in terms of its catalytic and active-site composition. Electrochemical studies help to reveal the interplay between the catalytic intermediates [133]. These enzymes display very interesting catalytic properties for biological hydrogen production and bio-electrochemical applications: high H2 production activity, low H2 inhibition, and O2 tolerance [134].
The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis has also been discussed. An expression system for the production of recombinant [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough and study of a selenocysteine–to-cysteine variant (Sec489Cys) in which, for the first time, a [NiFeSe] hydrogenase was converted to a [NiFe] type, reveal the direct involvement of this residue in the maturation process. It was proposed that selenium plays a crucial role in protecting against oxidative damage and the high catalytic activities of [NiFeSe] hydrogenases [135].
5. Glutathione Peroxidases
GPx is a multiple-isozyme family which protects the cellular organism from oxidative stress by the reductive transformation of hydroperoxide (H2O2) or organic hydroperoxide substrates (ROOH) to the product of H2O or alcohol, respectively, using cellular glutathione (GSH) as an electron source [136][137]. In 1952, Mills and Co-workers first noticed that GPX protected hemoglobin from oxidative degradation [138]. After that, in the 1960s, GPX activity was also observed in the lungs and kidneys [139]. In the 1970s, GPx was characterized and discovered selenocysteine amino acid, which played a vital role in enzymatic activity [140][141][142]. In the GPx family, only one GPx1 member was known until the 1980s. Then, this family grew to eight members [143].
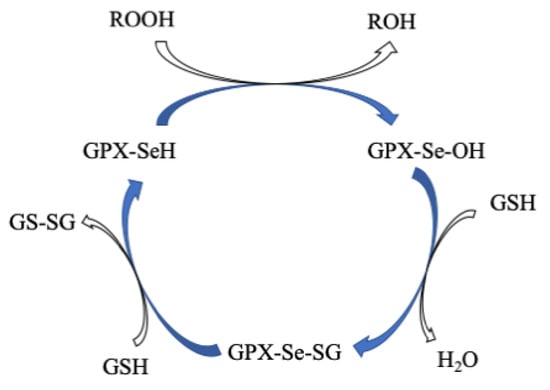
All GPxs display two steps of redox reactions in their catalytic cycle (Figure 9) [144][145]. In the first step, the selenocysteine (Sec-SeH) is oxidized to selenic acid (Sec-SeOH), which is a key intermediate product in the catalytic cycle. Simultaneously, the toxic hydroperoxide is reduced to the corresponding alcohol. In the second step, the reduction of oxidized Sec-SeOH proceeds into two subsequent 1 e− reduction steps. The Sec-SeOH is converted into GPx-SeGS by interacting with one equivalent reduced GSH, followed by the reduction of GPx-SeGS into GPx-Se by a second equivalent GSH for the next catalytic cycle [138][140][146][147][148]. The intermediate Sec-SeOH is stabilized by Gln and Trp, which are in the catalytic tetrad site [149], and additional Asn in tetrad contributes to the catalytic reaction [150].

Figure 9. Proposed catalytic cycle of GPxs. Modified from [138].
6. Thioredoxin Reductases
TrxR belongs to the pyridine nucleotide–disulfide oxidoreductases family, of which some members are glutathione reductase, mercuric ion reductase, and lipoamide dehydrogenase [151]. A homodimeric flavoenzyme, it contains one redox-active dithiol/disulfide motif, FAD prosthetic group, and an NADPH binding site in each monomeric subunit [152]. TrxRs are distributed in all living systems and are generally classified into two major classes: (a) low molecular weight (LMW~35 kDa) TrxRs that are present in both lower eukaryotes and prokaryotes, and (b) high molecular weight (HMW~55 kDa) TrxRs that are present in higher eukaryotes [151][153]. Both classes of TrxR utilize NADPH as an electron source to reduce the oxidized state of TrxR that plays a vital role in cell proliferation. Due to large differences in structures, both classes of TrxRs have different catalytic paths to execute the same biochemical reaction. The LMW TrxRs have two redox centers such as an N-terminal dithiol/disulfide pair and an FAD prosthetic group [154][155], whereas HMW TrxRs contain three redox centers such as an N-terminal dithiol/disulfide pair and an FAD prosthetic group and sixteen additional amino acid residues with penultimate selenocysteine (Sec) in the catalytic site (-Cys-Secys-Gly sequence) at the end of the C-terminal [156][157][158][159].
There are three types of Mammals’ TrxRs: (a) the cytosolic form, TrxR1 [160], (b) the mitochondrial form, TrxR2 [161][162], and (c) the testis-specific thioredoxin glutathione reductase (TGR) [163]. The overall protein fold of TrxR1 [164] is similar to other TrxR2 [165] and TGR [166]. Among them, TrxR1 is well-characterized. In 2001, the first three-dimensional (3D) structure of rat TrxR1 (Sec to Cys mutant) [164], followed by a large number of 3D structures (Sec-substituted mutants) of human TrxR1 [167] and mouse TrxR2 were published [165][168].
TrxR is an important biological redox mediator for the two-electron reduction of substrates. The catalytic cycle of mammalian TrxR involves three redox centers: N-terminal dithiol (Cys59–Cys64), adjacent FAD/NADH, and C-terminal selenolthiol pair (Cys498–Sec497) in the other subunit), which relay e− from N-terminal dithiol to the substrate, thioredoxin via FAD/NADH. The human TrxR1 substrate–thioredoxin (Trx) complex is identified and the 3D structure of that complex reveals that the C-terminal arm binds with the substrate Trx through the disulphide bond (TrxR-Cys-S-S-Cys-Trx) [169].
7. Iodothyronine Deiodinases
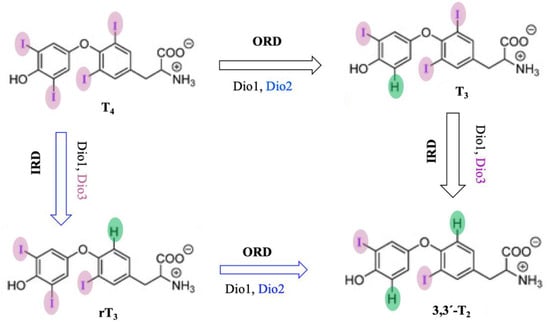
Dios are selenocysteine-dependent mammalian deiodinase enzymes that regulate thyroid hormones by deiodination of iodothyronine [170][171][172][173]. Dios have been classified into three isoforms, Dio1, Dio2, and Dio3, based on their sequence of amino acids and specificity of substrates. Dio1 enzymes non-selectively catalyze both inner- (phenolic group) and outer-ring (tyrosine group) deiodination of thyroid hormones, but Dio2 and Dio3 both selectively catalyze outer-ring and inner-ring deiodination (ORD and IRD) of thyroid hormones, respectively (Figure 10) [174][175][176][177][178][179][180].

Figure 10. Probable mechanism of deiodination by deiodinase with thyroid hormone substrates.
8. Selenoproteins and Human Health
8.1. Cancer
To date, several studies have attempted to analyze the role of GPxs, as well as changes in GPxs levels, in different types of tumors [181][182], but it remains controversial [183]. Indeed, GPx1 inhibits the oxidation of DNA mutations and, therefore, it may inhibit tumorigenesis [184], and overexpressed GPx1 reduces tumor growth, suggesting its protective effect in tumorigenesis [185]. However, reduced expression of GPx1 is detected in thyroid cancer [186], gastric cancer [187], and colorectal cancer [188], whereas GPx1 is highly expressed in kidney cancer [189] and pancreatic cancer [190]. Similar to GPx1, unusual expression of GPx2 is also observed in different tumors; for example, GPx2 is overexpressed in colorectal cancer [191], whereas a lower expression of GPx2 is detected in prostate intraepithelial neoplasia [192][193].
Polymorphism of human GPxs gene is a common phenomenon and it is associated with various diseases, especially tumors [194]. The GPx1 gene has various genetic polymorphisms and its most common polymorphism is the substitution of cytosine (C) to thymine (T) in DNA, resulting in the alteration of amino acid from proline (Pro) to leucine (Leu); thereby, the activity of GPx1 reduces by 5% [195]. Pro198Leu GPx1 polymorphism is associated with various types of cancer, mainly breast [196], prostate [197], lung [198], bladder [199], leukemia [200], and colon cancers [201]. However, the connection between GPx1 polymorphism and cancer vulnerability is controversial and inconclusive.
8.2. Diabetes
Diabetes mellitus (DM), a common human health problem around the globe, is a metabolic disorder and it is characterized by high levels of blood sugar (hyperglycemia), causing dysfunction in insulin secretion and/or sensitivity [202][203][204][205][206]. Insulin is a hormone synthesized in the β-cell of the pancreas and its action is also regulated by the pancreatic β-cell [206]. The most common type diabetes is Type 2 diabetes mellitus (T2DM) which is characterized by insulin resistance, caused by impairment of the pancreatic β-cell [206]. However, oxidative stress is believed to be the main cause of the onset and development of T2DM [207][208]. So, generation of ROS is a crucial factor in β-cell function [207].
8.3. Viral Infections
Viral infections occur when the human body is invaded by viruses, such as human immunodeficiency virus (HIV) and severe acute respiratory syndrome—coronavirus 2 (SARSCoV2), that lead to many diseases. Viral infection often alters the intracellular redox homeostasis in the host cell by increasing ROS production, which enhances the viral replication [209][210][211][212]. Several selenoproteins, like glutathione peroxidases (GPxs) and thioredoxin reductase (TrxR), are important host antioxidants that may play an important role against viral infections by consuming ROS.
8.4. Gestational Disorders
Selenium also plays an essential role in gestational health or during pregnancy, being one of the most important phases of a woman’s life and human reproduction [213]. It is reported that during pregnancy, the mother and fetus both demand more oxygen, resulting in the formation of more ROS, which is associated with miscarriage, premature rupture of membranes, preeclampsia, and intrauterine growth restriction [214][215]. Se performs its antioxidant activity by including Se as selenocysteine in the active sites of selenoproteins such as GPx and TrxR. Many studies have reported that Se deficiency enables poor levels of GPx and TrxR expression, leading to gestational disorders [213][216][217]. Therefore, supplementation of Se during pregnancy can reduce oxidative stress, resulting in a decrease in pregnancy complications [218][219].
References
- Painter, E.P. The Chemistry and Toxicity of Selenium Compounds, with Special Reference to the Selenium Problem. Chem. Rev. 1941, 28, 179–213.
- Hadrup, N.; Ravn-Haren, G. Acute human toxicity and mortality after selenium ingestion: A review. J. Trace Elem. Med. Biol. 2020, 58, 126435.
- Weekley, C.M.; Harris, H.H. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem. Soc. Rev. 2013, 42, 8870–8894.
- Schwarz, K.; Foltz, C.M. Selenium as an integral part of factor 3 against dietary necrotic liver degeneration. J. Am. Chem. Soc. 1957, 79, 3292–3293.
- Fairweather-Tait, S.J.; Bao, Y.; Broadley, M.R.; Collings, R.; Ford, D.; Hesketh, J.E.; Hurst, R. Selenium in human health and disease. Antioxid. Redox Signal. 2011, 14, 1337–1383.
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777.
- Kieliszek, M. Selenium–Fascinating Microelement, Properties and Sources in Food. Molecules 2019, 24, 1298.
- Roman, M.; Jitaru, p.; Barbante, C. Selenium biochemistry and its role for human health. Metallomics 2014, 6, 25–54.
- Driscoll, D.M.; Copeland, P.R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003, 23, 17–40.
- Hatfield, D.L.; Gladyshev, V.N. How selenium has altered our understanding of the genetic code. Mol. Cell Biol. 2002, 22, 3565–3576.
- Jacob, C.; Giles, G.I.; Giles, N.M.; Helmut Sies, H. Sulfur and Selenium: The Role of Oxidation State in Protein Structure and Function. Angew. Chem. Int. Ed. 2003, 42, 4742–4758.
- Wessjohann, L.A.; Schneider, A.; Abbas, M.; Brandt, W. Selenium in chemistry and biochemistry in comparison to sulfur. Biol. Chem. 2007, 388, 997–1006.
- Boyd, R. Selenium stories. Nat. Chem. 2011, 3, 570.
- Reich, H.J.; Hondal, R.J. Why Nature Chose Selenium. ACS Chem. Biol. 2016, 11, 821–841.
- Maiti, B.K. Cross-talk Between (Hydrogen)Sulfite and Metalloproteins: Impact on Human Health. Chem.–A Eur. J. 2022, 28, e202104342.
- Poole, L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015, 80, 148–157.
- Yukio Sugiura, Y.; Hojo, Y.; Tamai, Y.; Hisashi Tanaka, H. Selenium protection against mercury toxicity. Binding of methylmercury by the selenohydryl-containing ligand. J. Am. Chem. Soc. 1976, 98, 2339–2341.
- Huber, R.E.; Criddle, R.S. Comparison of the chemical properties of selenocysteine and selenocystine with their sulfur analogs. Arch. Biochem. Biophys. 1967, 122, 164–173.
- Bell, I.M.; Fisher, M.L.; Wu, Z.P.; Hilvert, D. Kinetic studies on the peroxidase activity of selenosubtilisin. Biochemistry 1993, 32, 3754–3762.
- Ruggles, E.L.; Snider, G.W.; Hondal, R.J. Chemical basis for the use of selenocysteine. In Selenium: Its Molecular Biology and Role in Human Health, 3rd ed.; Hatfield, D.L., Berry, M.J., Gladyshev, V.N., Eds.; Springer: New York, NY, USA, 2012; pp. 73–83.
- Abdo, M.; Knapp, S. Biomimetic Seleninates and Selenonates. J. Am. Chem. Soc. 2008, 130, 9234–9235.
- Nauser, T.; Steinmann, D.; Grassi, G.; Koppenol, W.H. Why selenocysteine replaces cysteine in thioredoxin reductase: A radical hypothesis. Biochemistry 2014, 53, 5017–5022.
- Nauser, T.; Dockheer, S.; Kissner, R.; Koppenol, W.H. Catalysis of Electron Transfer by Selenocysteine. Biochemistry 2006, 45, 6038–6043.
- Nauser, T.; Steinmann, D.; Koppenol, W.H. Why do proteins use selenocysteine instead of cysteine? Amino Acids 2012, 42, 39–44.
- Joan, C.; Pleasants, J.C.; Guo, W.; Rabenstein, D.L. A comparative study of the kinetics of selenol/diselenide and thiol/disulfide exchange reactions. J. Am. Chem. Soc. 1989, 111, 6553–6558.
- Hondal, R.J.; Marino, S.M.; Gladyshev, V.N. Selenocysteine in thiol/disulfide-like exchange reactions. Antioxid Redox Signal. 2013, 18, 1675–1689.
- Andreesen, J.R.; Ljungdahl, L.G. Formate dehydrogenase of Clostridium thermoaceticum: Incorporation of selenium-75, and the effects of selenite, molybdate, and tungstate on the enzyme. J. Bacteriol. 1973, 116, 867–873.
- Zinoni, F.; Birkmann, A.; Stadtman, T.C.; Böck, A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc. Natl. Acad. Sci. USA 1986, 83, 4650–4654.
- Zhang, Y.; Romero, H.; Salinas, G.; Gladyshev, V.N. Dynamic evolution of selenocysteine utilization in bacteria: A balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006, 7, R94.
- Thauer, R.K.; Fuchs, G.; Jungermann, K. Role of iron-sulfur proteins in formate metabolism. In Iron–Sulfur Proteins; Lovenber, W., Ed.; Academic: New York, NY, USA, 1977; pp. 121–156.
- Maden, B.; Edward, H. Tetrahydrofolate and tetrahydromethanopterin compared: Functionally distinct carriers in C1 metabolism. Biochem. J. 2000, 350, 609–629.
- Adams, M.W.W.; Mortenson, L.E. Mo reductases: Nitrate reductase and formate dehydrogenase. In Molybdenum Enzymes; Spiro, T.G., Ed.; Wiley Interscience: New York, NY, USA, 1985; pp. 519–593.
- Ferry, J.G. Formate dehydrogenase. FEMS Microbiol. Rev. 1990, 7, 377–382.
- Unden, G.; Bongaerts, J. Alternative respiratory pathways of Escherichia coli: Energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1997, 1320, 217–234.
- Richardson, D.J. Bacterial respiration: A flexible process for a changing environment. Microbiology 2000, 146, 551–571.
- Richardson, D.; Sawers, G. Structural biology—PMF through the redox loop. Science 2002, 295, 1842–1843.
- Vorholt, J.A.; Thauer, R.K. Molybdenum and tungsten enzymes in C1 metabolism. Met. Biol. Sys. 2002, 39, 571–619.
- Sawers, R.G. Formate and its role in hydrogen production in Escherichia coli. Biochem. Soc. Trans. 2005, 33, 42–46.
- Grimaldi, S.; Schoepp-Cothenet, B.; Ceccaldi, P.; Guigliarelli, B.; Magalon, A. The prokaryotic Mo/W-bisPGD enzymes family: A catalytic workhorse in bioenergetic. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 1048–1085.
- Hille, R.; Hall, J.; Basu, P. The Mononuclear Molybdenum Enzymes. Chem. Rev. 2014, 114, 3963–4038.
- Maia, L.B.; Moura, J.J.; Moura, I. Molybdenum and tungsten-dependent formate dehydrogenases. J. Biol. Inorg. Chem. 2015, 20, 287–309.
- Hartmann, T.; Schwanhold, N.; Leimkühler, S. Assembly and catalysis of molybdenum or tungsten-containing formate dehydrogenases from bacteria. Biochim. Biophys. Acta 2015, 1854, 1090–1100.
- Maia, L.B.; Moura, I.; Moura, J.J. Molybdenum and tungsten-containing formate dehydrogenases: Aiming to inspire a catalyst for carbon dioxide utilization. Inorg. Chim. Acta 2017, 455, 350–363.
- Maia, L.B.; Moura, I.; Moura, J.J. Molybdenum and tungsten-containing enzymes: An overview. In Molybdenum and Tungsten Enzymes: Biochemistry; Hille, R., Schulzke, C., Kirk, M., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2017; pp. 1–80.
- Niks, D.; Hille, R. Reductive activation of CO2 by formate dehydrogenases. Methods Enzymol. 2018, 613, 277–295.
- Niks, D.; Hille, R. Molybdenum- and tungsten-containing formate dehydrogenases and formylmethanofuran dehydrogenases: Structure, mechanism, and cofactor insertion. Prot. Sci. 2019, 28, 111–122.
- Nielsen, C.F.; Lange, L.; Meyer, A.S. Classification and enzyme kinetics of formate dehydrogenases for biomanufacturing via CO2 utilization. Biotechnol. Adv. 2019, 37, 107408.
- Maia, L.B.; Moura, I.; Moura, J.J.G. Carbon Dioxide Utilisation—The Formate Route. In Enzymes for Solving Humankind’s Problems; Moura, J.J.G., Moura, I., Maia, L.B., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 29–81.
- Kato, N. Formate dehydrogenase from methylotrophic yeasts. Methods Enzymol. 1990, 188, 459–462.
- Vinals, C.; Depiereux, E.; Feytmans, E. Prediction of structurally conserved regions of D-specific hydroxy acid dehydrogenases by multiple alignment with formate dehydrogenase. Biochem. Biophys. Res. Commun. 1993, 192, 182–188.
- Popov, V.O.; Lamzin, V.S. NAD (+)-dependent formate dehydrogenase. Biochem. J. 1994, 301, 625–643.
- Filippova, E.V.; Polyakov, K.M.; Tikhonova, T.V.; Stekhanova, T.N.; Booeko, K.M.; Popov, V.O. Structure of a new crystal modification of the bacterial NAD-dependent formate dehydrogenase with a resolution of 2.1 Å. Crystallogr. Rep. 2005, 50, 796–800.
- Shabalin, I.G.; Polyakov, K.M.; Tishkov, V.I.; Popov, V.O. Atomic resolution crystal structure of nad+ dependent formate dehydrogenase from bacterium Moraxella sp. C-1. Acta Nat. 2009, 1, 89–93.
- Alekseeva, A.A.; Savin, S.S.; Tishkov, V.I. NAD+-dependent formate dehydrogenase from plants. Acta Nat. 2011, 3, 38–54.
- Hille, R. The mononuclear molybdenum enzymes. Chem. Rev. 1996, 96, 2757–2816.
- Johnson, M.K.; Rees, D.C.; Adams, M.W.W. Tungstoenzymes. Chem. Rev. 1996, 96, 2817–2839.
- Bertram, P.A.; Karrasch, M.; Schmitz, R.A.; Böcher, R.; Albracht, S.P.J.; Thauer, R.K. Formylmethanofuran dehydrogenases from methanogenic Archaea. Substrate specificity, EPR properties and reversible inactivation by cyanide of the molybdenum or tungsten iron-sulfur proteins. Eur. J. Biochem. 1994, 220, 477–484.
- Hochheimer, A.; Hedderich, R.; Thauer, R.K. The formylmethanofuran dehydrogenase isozymes in Methanobacterium wolfeii and Methanobacterium terhmoautotrophicum: Induction of the molybdenum isozyme by molybdate and constitutive synthesis of the tungsten isozyme. Arch. Microbiol. 1998, 170, 389–393.
- Wagner, T.; Ermler, U.; Shima, S. The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 clusters. Science 2016, 354, 114–117.
- Hemmann, J.L.; Wagner, T.; Shima, S.; Vorholt, J.A. Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites. Proc. Natl. Acad. Sci. USA 2019, 116, 25583–25590.
- Niks, D.; Duvvuru, J.; Escalona, M.; Hille, R. Spectroscopic and Kinetic Properties of the Molybdenum-containing, NAD+-dependent Formate Dehydrogenase from Ralstonia eutropha. J. Biol. Chem. 2016, 291, 1162–1174.
- Maia, L.B.; Fonseca, L.; Moura, I.; Moura, J.J. Reduction of carbon dioxide by a molybdenum-containing formate dehydrogenase: A kinetic and mechanistic study. J. Am. Chem. Soc. 2016, 138, 8834–8846.
- Yu, X.; Niks, D.; Mulchandani, A.; Hille, R. Efficient reduction of CO2 by the molybdenum-containing formate dehydrogenase from Cupriavidus necator (Ralstonia eutropha). J. Biol. Chem. 2017, 292, 16872–16879.
- Meneghello, M.; Oliveira, A.R.; Jacq-Bailly, A.; Pereira, I.A.C.; Léger, C.; Fourmond, V. Formate Dehydrogenases Reduce CO2 Rather than HCO3−: An Electrochemical Demonstration. Angew. Chem. 2021, 60, 9964–9967.
- Meneghello, M.; Uzel, A.; Broc, M.; Manuel, R.R.; Magalon, A.; Léger, C.; Pereira, I.A.C.; Walburger, A.; Fourmond, V. Electrochemical Kinetics Support a Second Coordination Sphere Mechanism in Metal-Based Formate Dehydrogenase. Angew. Chem. 2023, 62, e202212224.
- Harmer, J.R.; Hakopian, S.; Niks, D.; Hille, R.; Bernhardt, P.V. Redox Characterization of the Complex Molybdenum Enzyme Formate Dehydrogenase from Cupriavidus necator. J. Am. Chem. Soc. 2023, 145, 25850–25863.
- Leimkühler, S. Metal-Containing Formate Dehydrogenases, a Personal View. Molecules 2023, 28, 5338.
- Stiefel, E.I. Proposed molecular mechanism for the action of molybedenum in enzymes: Coupled proton and electron transfer. Proc. Natl. Acad. Sci. USA 1973, 70, 988–992.
- Stiefel, E.I. The coordination and bioinorganic chemistry of molybdenum. Prog. Inorg. Chem. 1977, 22, 1–223.
- Rajapakshe, A.; Snyder, R.A.; Astashkin, A.V.; Bernardson, P.; Evans, D.J.; Young, C.G.; Evans, D.H.; Enemark, J.H. Insights into the nature of Mo(V) species in solution: Modeling catalytic cycles for molybdenum enzymes. Inorg. Chim. Acta 2009, 362, 4603–4608.
- Maia, L.; Moura, I.; Moura, J.J.G. EPR spectroscopy on mononuclear molybdenum-containing enzymes. In Future Directions in Metalloprotein and Metalloenzyme Research, Biological Magnetic Resonance; Hanson, G., Berliner, L.J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 33, pp. 55–101.
- Kirk, M.L.; Hille, R. Spectroscopic Studies of Mononuclear Molybdenum Enzyme Centers. Molecules 2022, 27, 4802.
- Hille, R.; Niks, D. Application of EPR and related methods to molybdenum-containing enzymes. Methods Enzymol. 2022, 666, 373–412.
- Gladyshev, V.N.; Khangulov, S.V.; Stadtman, T.C. Nicotinic acid hydroxylase from Clostridium barkeri: Electron paramagnetic resonance studies show that selenium is coordinated with molybdenum in the catalytically active selenium-dependent enzyme. Proc. Natl. Acad. Sci. USA 1994, 91, 232–236.
- Gladyshev, V.N.; Khangulov, S.V.; Axley, M.J.; Stadtman, T.C. Coordination of selenium to molybdenum in formate dehydrogenase H from Escherichia coli. Proc. Natl. Acad. Sci. USA 1994, 91, 7708–7711.
- Gladyshev, V.N.; Boyington, J.C.; Khangulov, S.V.; Grahame, D.A.; Stadtman, T.C.; Sun, P.D. Characterization of crystalline formate dehydrogenase H from Escherichia coli: Stabilization, EPR spectroscopy, and preliminary crystallographic analysis. J. Biol. Chem. 1996, 271, 8095–8100.
- Khangulov, S.V.; Gladyshev, V.N.; Dismukes, G.C.; Stadtman, T.C. Selenium-containing formate dehydrogenase H from Escherichia coli: A molybdopterin enzyme that catalyzes formate oxidation without oxygen transfer. Biochemistry 1998, 37, 3518–3528.
- Hanson, G.R.; Wilson, G.L.; Bailey, T.D.; Pilbrow, J.R.; Wedd, A.G. Multifrequency electron spin resonance of molybdenum (V) and tungsten (V) compounds. J. Am. Chem. Soc. 1987, 109, 2609–2616.
- Rivas, M.G.; González, P.J.; Brondino, C.D.; Moura, J.J.; Moura, I. EPR characterization of the molybdenum (V) forms of formate dehydrogenase from Desulfovibrio desulfuricans ATCC 27774 upon formate reduction. J. Inorg. Biochem. 2007, 101, 1617–1622.
- Almendra, M.J.; Brondino, C.D.; Gavel, O.; Pereira, A.S.; Tavares, P.; Bursakov, S.; Moura, I. Purification and characterization of a tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Biochemistry 1999, 38, 16366–16372.
- Raaijmakers, H.; Teixeira, S.; Dias, J.M.; Almendra, M.J.; Brondino, C.D.; Moura, I.; Romão, M.J. Tungsten-containing formate dehydrogenase from Desulfovibrio gigas: Metal identification and preliminary structural data by multi-wavelength crystallography. J. Biol. Inorg. Chem. 2001, 6, 398–404.
- Oliveira, A.R.; Mota, C.; Klymanska, K.; Biaso, F.; Romão, M.J.; Guigliarelli, B.; Pereira, I.C. Spectroscopic and Structural Characterization of Reduced Desulfovibrio vulgaris Hildenborough W-FdhAB Reveals Stable Metal Coordination during Catalysis. ACS Chem. Biol. 2022, 17, 1901–1909.
- Oliveira, A.R.; Mota, C.; Mourato, C.; Domingos, R.M.; Santos, M.F.; Gesto, D.; Pereira, I.A. Toward the mechanistic understanding of enzymatic CO2 reduction. ACS Catal. 2020, 10, 3844–3856.
- Graham, J.E.; Niks, D.; Zane, G.M.; Gui, Q.; Hom, K.; Hille, R.; Raman, C.S. How a formate dehydrogenase responds to oxygen: Unexpected O2 insensitivity of an enzyme harboring tungstopterin, selenocysteine, and clusters. ACS Catal. 2022, 12, 10449–10471.
- Raman, C.S. Reply to Comment on ‘How a Formate Dehydrogenase Responds to Oxygen: Unexpected O2 Insensitivity of an Enzyme Harboring Tungstopterin, Selenocysteine, and Clusters’. ACS Catal. 2023, 13, 9629–9632.
- Jollie, D.R.; Lipscomb, J.D. Formate dehydrogenase from Methylosinus trichosporium OB3b. Purification and spectroscopic characterization of the cofactors. J. Biol. Chem. 1991, 266, 21853–21863.
- Cramer, S.P.; Liu, C.L.; Mortenson, L.E.; Spence, J.T.; Liu, S.M.; Yamamoto, I.; Ljungdahl, L.G. Formate dehydrogenase molybdenum and tungsten sites—Observation by EXAFS of structural differences. J. Inorg. Biochem. 1985, 23, 119–124.
- George, G.N.; Colangelo, C.M.; Dong, J.; Scott, R.A.; Khangulov, S.V.; Gladyshev, V.N.; Stadtman, T.C. X-ray absorption spectroscopy of the molybdenum site of Escherichia coli formate dehydrogenase. J. Am. Chem. Soc. 1998, 120, 1267–1273.
- George, G.N.; Costa, C.; Moura, J.J.G.; Moura, I. Observation of ligand-based redox chemistry at the active site of a molybdenum enzyme. J. Am. Chem. Soc. 1999, 121, 2625–2626.
- Schrapers, P.; Hartmann, T.; Kositzki, R.; Dau, H.; Reschke, S.; Schulzke, C.; Haumann, M. Sulfido and cysteine ligation changes at the molybdenum cofactor during substrate conversion by formate dehydrogenase (FDH) from Rhodobacter capsulatus. Inorg. Chem. 2015, 54, 3260–3271.
- Duffus, B.R.; Schrapers, P.; Schuth, N.; Mebs, S.; Dau, H.; Leimkühler, S.; Haumann, M. Anion binding and oxidative modification at the molybdenum cofactor of formate dehydrogenase from Rhodobacter capsulatus studied by X-ray absorption spectroscopy. Inorg. Chem. 2020, 59, 214–225.
- Boyington, J.C.; Gladyshev, V.N.; Khangulov, S.V.; Stadtman, T.C.; Sun, P.D. Crystal structure of formate dehydrogenase H: Catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 1997, 275, 1305–1308.
- Jormakka, M.; Tornroth, S.; Byrne, B.; Iwata, S. Molecular basis of proton motive force generation: Structure of formate dehydrogenase-N. Science 2002, 295, 1863–1868.
- Raaijmakers, H.; Macieira, S.; Dias, J.M.; Teixeira, S.; Bursakov, S.; Huber, R.; Moura, I.; Moura, M.J.; Romão, M.J. Gene sequence and the 1.8 Å crystal structure of the tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Structure 2002, 10, 1261–1272.
- Radon, C.; Mittelstädt, G.; Duffus, B.R.; Burger, J.; Hartmann, T.; Mielke, T.; Teutloff, C.; Leimkuhler, S.; Wendler, P. Cryo-EM structures reveal intricate Fe-S cluster arrangement and charging in Rhodobacter capsulatus formate dehydrogenase. Nat. Commun. 2020, 11, 1912.
- Young, T.; Niks, D.; Hakopian, S.; Tam, T.K.; Yu, X.; Hille, R.; Blaha, G.M. Crystallographic and kinetic analyses of the FdsBG subcomplex of the cytosolic formate dehydrogenase FdsABG from Cupriavidus necator. J. Biol. Chem. 2020, 295, 6570–6585.
- Dietrich, H.M.; Righetto, R.D.; Kumar, A.; Wietrzynski, W.; Trischler, R.; Schuller, S.K.; Schuller, J.M. Membrane-anchored HDCR nanowires drive hydrogen-powered CO2 fixation. Nature 2022, 607, 823–830.
- Yoshikawa, T.; Makino, F.; Miyata, T.; Suzuki, Y.; Tanaka, H.; Namba, K.; Kano, K.; Sowa, K.; Kitazumi, Y.; Shirai, O. Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis. Chem. Commun. 2022, 58, 6478–6481.
- Vilela-Alves, G.; Manuel, R.R.; Oliveira, A.R.; Pereira, I.C.; Romão, M.J.; Mota, C. Tracking W-Formate Dehydrogenase Structural Changes during Catalysis and Enzyme Reoxidation. Int. J. Mol. Sci. 2022, 24, 476.
- Zinoni, F.; Birkmann, A.; Leinfelder, W.; Bock, A. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon. Proc. Natl. Acad. Sci. USA 1987, 84, 3156–3160.
- Axley, M.J.; Böck, A.; Stadtman, T.C. Catalytic properties of an Escherichia coli formate dehydrogenase mutant in which sulfur replaces selenium. Proc. Natl. Acad. Sci. USA 1991, 88, 8450–8454.
- Berry, M.J.; Martin, G.W.; Tujebajeva, R.; Grundner-Culemann, E.; Mansell, J.B.; Morozova, N.; Harney, J.W. Selenocysteine Insertion Sequence Element Characterization and Selenoprotein Expression. Methods Enzymol. 2002, 347, 17–24.
- Hatfield, D.L.; Carlson, B.A.; Xu, X.M.; Mix, H.; Gladyshev, V.N. Selenocysteine Incorporation Machinery and the Role of Selenoproteins in Development and Health. Prog. Nucleic Acid Res. Mol. Biol. 2006, 81, 97–142.
- Allmang, C.; Wurth, L.; Krol, A. The Selenium to Selenoprotein Pathway in Eukaryotes: More Molecular Partners than Anticipated. Biochim. Biophys. Acta Gen. Subj. 2009, 1790, 1415–1423.
- Yoshizawa, S.; Böck, A. The Many Levels of Control on Bacterial Selenoprotein Synthesis. Biochim. Biophys. Acta Gen. Subj. 2009, 1790, 1404–1414.
- Bulteau, A.-L.; Chavatte, L. Update on Selenoprotein Biosynthesis. Antioxid. Redox Signal. 2015, 23, 775–794.
- Arnér, E.S. Selenoproteins—What unique properties can arise with selenocysteine in place of cysteine? Exp. Cell Res. 2010, 316, 1296–1303.
- Brigelius-Flohe, R. The Evolving Versatility of Selenium in Biology. Antioxid. Redox Signal. 2015, 23, 757–760.
- Bortoli, M.; Torsello, M.; Bickelhaupt, F.M.; Orian, L. Role of the Chalcogen (S, Se, Te) in the Oxidation Mechanism of the Glutathione Peroxidase Active Site. ChemPhysChem 2017, 18, 2990–2998.
- Lubitz, W.; Ogata, H.; Rüdiger, O.; Reijerse, E. Hydrogenases. Chem. Rev. 2014, 114, 4081–4148.
- Ogata, H.; Nishikawa, K.; Lubitz, W. Hydrogens detected by subatomic resolution protein crystallography in a hydrogenase. Nature 2015, 520, 571–574.
- Fauque, G.; Peck, H.D., Jr.; Moura, J.J.G.; Huynh, B.H.; Berlier, Y.; DerVartanian, D.V.; Teixeira, M.; Przybyla, A.E.; Lespinat, P.A.; Moura, I. The three classes of hydrogenases from sulfate-reducing bacteria of the genus Desulfovibrio. FEMS Microbiol. Rev. 1988, 4, 299–344.
- Pereira, A.S.; Tavares, P.; Moura, I.; Moura, J.J.; Huynh, B.H. Mössbauer characterization of the iron-sulfur clusters in Desulfovibrio vulgaris hydrogenase. J. Am. Chem. Soc. 2001, 123, 2771–2782.
- Patil, D.S.; Moura, J.J.; He, S.H.; Teixeira, M.; Prickril, B.C.; DerVartanian, D.V.; Peck, H.D., Jr.; LeGall, J.; Huynh, B.H. EPR-detectable redox centers of the periplasmic hydrogenase from Desulfovibrio vulgaris. J. Biol. Chem. 1988, 263, 18732–18738.
- Moura, J.J.G.; Teixeira, M.; Moura, I. The role of nickel and iron-sulfur centers in the bioproduction of hydrogen. Pure Appl. Chem. 1989, 61, 915–921.
- Wombwell, C.; Caputo, C.A.; Reisner, E. -Hydrogenase Chemistry. Acc. Chem. Res. 2015, 48, 2858–2865.
- Barbosa, T.M.; Baltazar, C.S.A.; Cruz, D.R.; Lousa, D.; Soares, C.M. Studying O2 pathways in - and -hydrogenases. Sci. Rep. 2020, 10, 10540.
- Happe, R.P.; Roseboom, W.; Pierik, A.J.; Albracht, S.P.; Bagley, K.A. Biological Activation of Hydrogen. Nature 1997, 385, 126.
- Bleijlevens, B.; van Broekhuizen, F.A.; De Lacey, A.L.; Roseboom, W.; Fernandez, V.M.; Albracht, S.P. The Activation of the -Hydrogenase from Allochromatium Vinosum. An Infrared Spectro-Electrochemical Study. J. Biol. Inorg. Chem. 2004, 9, 743–752.
- Fichtner, C.; Laurich, C.; Bothe, E.; Lubitz, W. Spectroelectrochemical Characterization of the Hydrogenase of Desulfovibrio vulgaris Miyazaki F. Biochemistry 2006, 5, 9706–9716.
- Frey, M.; Fontecilla-Camps, J.C.; Volbeda, A. Nickel–Iron Hydrogenases. In Handbook of Metalloproteins; Messerschmidt, A., Huber, R., Poulos, T., Wieghardt, K., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2001; p. 880.
- LeGall, J.; Ljungdahl, P.O.; Moura, I.; Peck, H.D., Jr.; Xavier, A.V.; Moura, J.J.G.; Teixera, M.; Huynh, B.H. DerVartanian DV. The presence of redox-sensitive nickel in the periplasmic hydrogenase from Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 1982, 106, 610–616.
- Higuchi, Y.; Yagi, T.; Yasuoka, N. Unusual Ligand Structure in Ni-Fe Active Center and an Additional Mg Site in Hydrogenase Revealed by High Resolution X-Ray Structure Analysis. Structure 1997, 5, 1671–1680.
- Volbeda, A.; Garcin, E.; Piras, C.; deLacey, A.L.; Fernandez, V.M.; Hatchikian, E.C.; Frey, M.; FontecillaCamps, J.C. Structure of the Hydrogenase Active Site: Evidence for Biologically Uncommon Fe Ligands. J. Am. Chem. Soc. 1996, 118, 12989–12996.
- Carepo, M.; Tierney, D.L.; Brondino, C.D.; Yang, T.C.; Pamplona, A.; Telser, J.; Moura, I.; Moura, J.J.; Hoffman, B.M. 17O ENDOR detection of a solvent-derived Ni-(OH(x))-Fe bridge that is lost upon activation of the hydrogenase from Desulfovibrio gigas. J. Am. Chem. Soc. 2002, 124, 281–286.
- Yang, X.; Darensbourg, M.Y. The roles of chalcogenides in O2 protection of H2ase active sites. Chem. Sci. 2020, 11, 9366–9377.
- Moura, J.J.G.; Moura, I.; Huynh, B.H.; Krüger, H.J.; Teixeira, M.; DuVarney, R.C.; DerVartanian, D.V.; Xavier, A.V.; Peck, H.D., Jr.; LeGall, J. Unambiguous identification of the nickel EPR signal in 61Ni-enriched Desulfovibrio gigas hydrogenase. J. Biochem. Biophys. Res. Commun. 1982, 108, 1388–1393.
- Gutierrez-Sanz, O.; Marques, M.C.; Baltazar, C.S.; Fernandez, V.M.; Soares, C.M.; Pereira, I.A.; De Lacey, A.L. Influence of the Protein Structure Surrounding the Active Site on the Catalytic Activity of Hydrogenases. J. Biol. Inorg. Chem. 2013, 18, 419–427.
- He, S.H.; Teixeira, M.; LeGall, J.; Patil, D.S.; Moura, I.; Moura, J.J.; DerVartanian, D.V.; Huynh, B.H.; Peck, H.D., Jr. EPR studies with 77Se-enriched (NiFeSe) hydrogenase of Desulfovibrio baculatus. Evidence for a selenium ligand to the active site nickel. J. Biol. Chem. 1989, 264, 2678–2682.
- Eidsness, M.K.; Scott, R.A.; Prickril, B.C.; DerVartanian, D.V.; LeGall, J.; Moura, I.; Moura, J.J.; Peck, H.D., Jr. Evidence for selenocysteine coordination to the active site nickel in the hydrogenases from Desulfovibrio baculatus. Proc. Natl. Acad. Sci. USA 1989, 86, 147–151.
- Teixeira, M.; Fauque, G.; Moura, I.; Lespinat, P.A.; Berlier, Y.; Prickril, B.; Peck, H.D., Jr.; Xavier, A.V.; LeGall, J.; Moura, J.J.G. Nickel--selenium-containing hydrogenases from Desulfovibrio baculatus (DSM 1743). Redox centers and catalytic properties. Eur. J. Biochem. 1987, 167, 47–58.
- Teixeira, M.; Moura, I.; Fauque, G.; Czechowski, M.; Berlier, Y.; Lespinat, P.A.; Le Gall, J.; Xavier, A.V.; Moura, J.J.G. Redox properties and activity studies on a nickel-containing hydrogenase isolated from a halophilic sulfate reducer Desulfovibrio salexigens. Biochimie 1986, 68, 75–84.
- Moura, I.; Cordas, C.; Moura, J.J.G. Direct electrochemistry study of the multiple redox centers of hydrogenase from Desulfovibrio gigas. Bioelectrochemistry 2008, 74, 83–89.
- Zacarias, S.; Vélez, M.; Pita, M.; De Lacey, A.L.; Matias, P.M.; Pereira, I.A.C. Characterization of the hydrogenase from Desulfovibrio vulgaris Hildenborough. Methods Enzymol. 2018, 613, 169–201.
- Marques, M.C.; Tapia, C.; Gutierrez-Sanz, O.; Ramos, A.R.; Keller, K.L.; Wall, J.D.; De Lacey, A.L.; Matias, P.M.; Pereira, I.A.C. The Direct Role of Selenocysteine in Hydrogenase Maturation and Catalysis. Nat. Chem. Biol. 2017, 13, 544–550.
- Stolwijk, J.M.; Garje, R.; Sieren, J.C.; Buettner, G.R.; Zakharia, Y. Understanding the Redox Biology of Selenium in the Search of Targeted Cancer Therapies. Antioxidants 2020, 9, 420.
- Weaver, K.; Skouta, R. The Selenoprotein Glutathione Peroxidase 4: From Molecular Mechanisms to Novel Therapeutic Opportunities. Biomedicines 2022, 10, 891.
- Mills, G.C. Hemoglobin catabolism. I. Glutathione peroxidase, an erythrocyte enzyme which protects hemoglobin from oxidative breakdown. J. Biol. Chem. 1957, 229, 189–197.
- Little, C.; Olinescu, R.; Reid, K.G.; O’Brien, P.J. Properties and Regulation of Glutathione peroxidase. J. Biol. Chem. 1970, 245, 3632–3636.
- Flohe, L.; Günzler, W.A.; Schock, H.H. Glutathione peroxidase: A selenoenzyme. FEBS Lett. 1973, 32, 132–134.
- Rotruck, J.T.; Pope, A.L.; Ganther, H.E.; Swanson, A.B.; Hafeman, D.G.; Hoekstra, W.G. Selenium: Biochemical role as a component of glutathione peroxidase. Science 1973, 179, 588–590.
- Forstrom, J.W.; Zakowski, J.J.; Tappel, A.L. Identification of the catalytic site of rat liver glutathione peroxidase as selenocysteine. Biochemistry 1978, 17, 2639–2644.
- Trenz, T.S.; Delaix, C.L.; Turchetto-Zolet, A.C.; Zamocky, M.; Lazzarotto, F.; Margis-Pinheiro, M. Going Forward and Back: The Complex Evolutionary History of the GPx. Biology 2021, 10, 1165.
- Brigelius-Flohé, R.; Flohé, L. Regulatory Phenomena in the Glutathione Peroxidase Superfamily. Antioxid. Redox Signal. 2020, 33, 498–516.
- Labrecque, C.L.; Fuglestad, B. Electrostatic Drivers of GPx4 Interactions with Membrane, Lipids, and DNA. Biochemistry 2021, 60, 2761–2772.
- Kraus, R.J.; Foster, S.J.; Ganther, H.E. Identification of selenocysteine in glutathione peroxidase by mass spectroscopy. Biochemistry 1983, 22, 5853–5858.
- Gladyshev, V.N.; Factor, V.M.; Housseau, F.; Hatfield, D.L. Contrasting patterns of regulation of the antioxidant selenoproteins, thioredoxin reductase, and glutathione peroxidase, in cancer cells. Biochem. Biophys. Res. Commun. 1998, 251, 488–493.
- Masuda, R.; Kimura, R.; Karasaki, T.; Sase, S.; Goto, K. Modeling the Catalytic Cycle of Glutathione Peroxidase by Nuclear Magnetic Resonance Spectroscopic Analysis of Selenocysteine Selenenic Acids. J. Am. Chem. Soc. 2021, 143, 6345–6350.
- Epp, O.; Ladenstein, R.; Wendel, A. The refined structure of the selenoenzyme glutathione peroxidase at 0.2-nm resolution. Eur. J. Biochem. 1983, 133, 51–69.
- Tosatto, S.C.E.; Bosello, V.; Fogolari, F.; Mauri, P.; Roveri, A.; Toppo, S.; Flohe, L.; Ursini, F.; Maiorino, M. The Catalytic Site of Glutathione Peroxidases. Antioxid. Redox Signal. 2008, 10, 1515–1526.
- Mustacich, D.; Powis, G. Thioredoxin reductase. Biochem. J. 2000, 346, 1–8.
- Williams, C.H., Jr. Chemistry and Biochemistry of Flavoenzymes; Müller, F., Ed.; CRC: Boca Raton, FL, USA, 1992; Volume III, pp. 121–211.
- Arscott, L.D.; Gromer, S.; Schirmer, R.H.; Williams, C.H., Jr. The mechanism of thioredoxin reductase from human placenta is similar to the mechanisms of lipoamide dehydrogenase and glutathione reductase and is distinct from the mechanism of thioredoxin reductase from Escherichia coli. Proc. Natl. Acad. Sci. USA 1997, 94, 3621–3626.
- Williams, C.H.; Arscott, L.D.; Müller, S.; Lennon, B.W.; Ludwig, M.L.; Wang, P.F.; Veine, D.M.; Becker, K.; Schirmer, R.H. Thioredoxin reductase two modes of catalysis have evolved. Eur. J. Biochem. 2000, 267, 6110–6117.
- Williams, C.H., Jr. Mechanism and structure of thioredoxin reductase from Escherichia coli. FASEB J. 1995, 9, 1267–1276.
- Zhong, L.; Arnér, E.S.; Ljung, J.; Aslund, F.; Holmgren, A. Rat and calf thioredoxin reductase are homologous to glutathione reductase with a carboxyl-terminal elongation containing a conserved catalytically active penultimate selenocysteine residue. J. Biol. Chem. 1998, 273, 8581–8591.
- Gladyshev, V.N.; Jeang, K.T.; Stadtman, T.C. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc. Natl. Acad. Sci. USA 1996, 93, 6146–6151.
- Miranda-Vizuete, A.M.; Damdimopoulos, A.E.; Pedrajas, J.R.; Gustafsson, J.-Å.; Spyrou, G. Human mitochondrial thioredoxin reductase. Eur. J. Biochem. 1999, 261, 405–412.
- Lee, S.R.; Bar-Noy, S.; Kwon, J.; Levine, R.L.; Stadtman, T.C.; Rhee, S.G. Mammalian thioredoxin reductase: Oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc. Natl Acad. Sci. USA 2000, 97, 2521–2526.
- Holmgren, A.; Björnstedt, M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995, 252, 199–208.
- Lee, S.R.; Kim, J.R.; Kwon, K.S.; Yoon, H.W.; Levine, R.L.; Ginsburg, A.; Rhee, S.G. Molecular cloning and characterization of a mitochondrial selenocysteine-containing thioredoxin reductase from rat liver. J. Biol. Chem. 1999, 274, 4722–4734.
- Turanov, A.A.; Su, D.; Gladyshev, V.N. Characterization of alternative cytosolic forms and cellular targets of mouse mitochondrial thioredoxin reductase. J. Biol. Chem. 2006, 281, 22953–22963.
- Arnér, E.S.J. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim. Biophys. Acta 2009, 1790, 495–526.
- Sandalova, T.; Zhong, L.; Lindqvist, Y.; Holmgren, A.; Schneider, G. Three-dimensional structure of a mammalian thioredoxin reductase: Implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc. Natl Acad. Sci. USA 2001, 98, 9533–9538.
- Biterova, E.I.; Turanov, A.A.; Gladyshev, V.N.; Barycki, J.J. Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism. Proc. Natl. Acad. Sci. USA 2005, 102, 15018–15023.
- Karplus, P.A.; Schulz, G.E. Refined structure of glutathione reductase at 1.54 A resolution. J. Mol. Biol. 1987, 195, 701–729.
- Fritz-Wolf, K.; Urig, S.; Becker, K. The structure of human thioredoxin reductase 1 provides insights into C-terminal rearrangements during catalysis. J. Mol. Biol. 2007, 370, 116–127.
- Eckenroth, B.; Harris, K.; Turanov, A.A.; Gladyshev, V.N.; Raines, R.T.; Hondal, R.J. Semisynthesis and characterization of mammalian thioredoxin reductase. Biochemistry 2006, 45, 5158–5170.
- Fritz-Wolf, K.; Kehr, S.; Stumpf, M.; Rahlfs, S.; Becker, K. Crystal structure of the human thioredoxin reductase-thioredoxin complex. Nat. Commun. 2011, 2, 383.
- Marsan, E.S.; Bayse, C.A. A Halogen Bonding Perspective on Iodothyronine Deiodinase Activity. Molecules 2020, 25, 1328.
- Behne, D.; Kyriakopoulos, A.; Meinhold, H.; Köhrle, J. Identification of type I iodothyronine 5′-deiodinase as a selenoenzyme. Biochem. Biophys. Res. Commun. 1990, 173, 1143–1149.
- Berry, M.J.; Banu, L.; Larsen, P.R. Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature 1991, 349, 438–440.
- Larsen, P.R.; Berry, M.J. Nutritional and hormonal regulation of thyroid hormone deiodinases. Annu. Rev. Nutr. 1995, 15, 323–352.
- Bianco, A.C.; Salvatore, D.; Gereben, B.; Berry, M.J.; Larsen, P.R. Biochemistry, Cellular and Molecular Biology, and Physiological Roles of the Iodothyronine Selenodeiodinases. Endocr. Rev. 2002, 23, 38–89.
- Köhrle, J. Iodothyronine deiodinases. Methods Enzymol. 2002, 347, 125–167.
- Koehrle, J.; Auf’mkolk, M.; Rokos, H.; Hesch, R.D.; Cody, V. Rat liver iodothyronine monodeiodinase. Evaluation of the iodothyronine ligand-binding site. J. Biol. Chem. 1986, 261, 11613–11622.
- Kuiper, G.G.J.M.; Kester, M.H.A.; Peeters, R.P.; Visser, T.J. Biochemical mechanisms of thyroid hormone deiodination. Thyroid 2005, 15, 787–798.
- Visser, T.J.; Schoenmakers, C.H. Characteristics of type III iodothyronine deiodinase. Acta Med. Austriaca 1992, 19, 18–21.
- Köhrle, J. Local activation and inactivation of thyroid hormones: The deiodinase family. Mol. Cell Endocrinol. 1999, 151, 103–119.
- Köhrle, J.; Jakob, F.; Contempré, B.; Dumont, J.E. Selenium, the thyroid, and the endocrine system. Endocr. Rev. 2005, 26, 944–984.
- Handy, D.E.; Loscalzo, J. The role of glutathione peroxidase-1 in health and disease. Free Radic. Biol. Med. 2022, 188, 146–161.
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione Peroxidase-1 in Health and Disease: From Molecular Mechanisms to Therapeutic Opportunities. Antioxid. Redox Signal. 2011, 15, 1957–1997.
- Zhang, M.-L.; Wu, H.-T.; Chen, W.-J.; Xu, Y.; Ye, Q.-Q.; Shen, J.-X.; Liu, J. Involvement of glutathione peroxidases in the occurrence and development of breast cancers. J. Transl. Med. 2020, 18, 247.
- Baliga, M.S.; Wang, H.; Zhuo, P.; Schwartz, J.L.; Diamond, A.M. Selenium and GPx-1 overexpression protect mammalian cells against UV-induced DNA damage. Biol. Trace. Elem. Res. 2007, 115, 227–242.
- Liu, J.; Du, J.; Zhang, Y.; Sun, W.; Smith, B.J.; Oberley, L.W.; Cullen, J.J. Suppression of the malignant phenotype in pancreatic cancer by overexpression of phospholipid hydroperoxide glutathione peroxidase. Hum. Gene Ther. 2006, 17, 105–116.
- Metere, A.; Frezzotti, F.; Graves, C.E.; Vergine, M.; De Luca, A.; Pietraforte, D.P.; Giacomelli, L. A possible role for selenoprotein glutathione peroxidase (GPx1) and thioredoxin reductases (TrxR1) in thyroid cancer: Our experience in thyroid surgery. Cancer Cell Int. 2018, 18, 7.
- Min, S.Y.; Kim, H.S.; Jung, E.J.; Jung, E.J.; Jee, C.D.; Kim, W.H. Prognostic significance of glutathione peroxidase 1 (GPX1) down-regulation and correlation with aberrant promoter methylation in human gastric cancer. Anticancer Res. 2012, 32, 3169–3175.
- Nalkiran, I.; Turan, S.; Arikan, S.; Kahraman, Ö.T.; Acar, L.; Yaylim, I.; Ergen, A. Determination of Gene Expression and Serum Levels of MnSOD and GPX1 in Colorectal Cancer. Anticancer Res. 2015, 35, 255–259.
- Cheng, Y.; Xu, T.; Li, S.; Ruan, H. GPX1, a biomarker for the diagnosis and prognosis of kidney cancer, promotes the progression of kidney cancer. Aging 2019, 11, 12165–12176.
- Meng, Q.; Xu, J.; Liang, C.; Liu, J.; Hua, J.; Zhang, Y.; Ni, Q.; Shi, S.; Yu, X. GPx1 is involved in the induction of protective autophagy in pancreatic cancer cells in response to glucose deprivation. Cell Death Dis. 2018, 9, 1187.
- Al-Taie, O.H.; Uceyler, N.; Eubner, U.; Jakob, F.; Mork, H.; Scheurlen, M.; Brigelius-Flohe, R.; Schottker, K.; Abel, J.; Thalheimer, A.; et al. Expression Profiling and Genetic Alterations of the Selenoproteins GI-GPx and SePP in Colorectal Carcinogenesis. Nutr. Cancer 2004, 48, 6–14.
- Woenckhaus, M.; Klein-Hitpass, L.; Grepmeier, U.; Merk, J.; Pfeifer, M.; Wild, P.; Bettstetter, M.; Wuensch, P.; Blaszyk, H.; Hartmann, A.; et al. Smoking and cancer-related gene expression in bronchial epithelium and non-small-cell lung cancers. J. Pathol. 2006, 210, 192–204.
- Banning, A.; Kipp, A.; Schmitmeier, S.; Löwinger, M.; Florian, S.; Krehl, S.; Thalmann, S.; Thierbach, R.; Steinberg, P.; Brigelius-Flohé, R. Glutathione Peroxidase 2 Inhibits Cyclooxygenase-2–Mediated Migration and Invasion of HT-29 Adenocarcinoma Cells but Supports Their Growth as Tumors in Nude Mice. Cancer Res. 2008, 68, 9746–9753.
- Imyanitov, E.N.; Togo, A.V.; Hanson, K.P. Searching for cancer-associated gene polymorphisms: Promises and obstacles. Cancer Lett. 2004, 204, 3–14.
- Jablonska, E.; Gromadzinska, J.; Peplonska, B.; Fendler, W.; Reszka, E.; Krol, M.B.; Wieczorek, E.; Bukowska, A.; Gresner, P.; Galicki, M.; et al. Lipid peroxidation and glutathione peroxidase activity relationship in breast cancer depends on functional polymorphism of GPX1. BMC Cancer 2015, 15, 657.
- Hu, J.; Zhou, G.-W.; Wang, N.; Wang, Y.-J. GPX1 Pro198Leu polymorphism and breast cancer risk: A meta-analysis. Breast Cancer Res. Treat. 2010, 124, 425–431.
- Arsova-Sarafinovska, Z.; Matevska, N.; Eken, A.; Petrovski, D.; Banev, S.; Dzikova, S.; Georgiev, V.; Sikole, A.; Erdem, O.; Sayal, A.; et al. Glutathione peroxidase 1 (GPX1) genetic polymorphism, erythrocyte GPX activity, and prostate cancer risk. Int. Urol. Nephrol. 2009, 41, 63–70.
- Raaschou-Nielsen, O.; Sørensen, M.; Hansen, R.D.; Frederiksen, K.; Tjønneland, A.; Overvad, K.; Vogel, U. GPX1 Pro198Leu polymorphism, interactions with smoking and alcohol consumption, and risk for lung cancer. Cancer Lett. 2007, 247, 293–300.
- Men, T.; Zhang, X.; Yang, J.; Shen, B.; Li, X.; Chen, D.; Wang, J. The rs1050450 C > T polymorphism of GPX1 is associated with the risk of bladder but not prostate cancer: Evidence from a meta-analysis. Tumor Biol. 2014, 35, 269–275.
- Bănescu, C.; Trifa, A.P.; Voidăzan, S.; Moldovan, V.G.; Macarie, I.; Lazar, E.B.; Dima, D.; Duicu, C.; Dobreanu, M. CAT, GPX1, MnSOD, GSTM1, GSTT1, and GSTP1 Genetic Polymorphisms in Chronic Myeloid Leukemia: A Case-Control Study. Oxid. Med. Cell. Longev. 2014, 2014, 875861.
- Hansen, R.; Saebø, M.; Skjelbred, C.F.; Nexø, B.A.; Hagen, P.C.; Bock, G.; Lothe, I.M.B.; Johnson, E.; Aase, S.; Hansteen, I.-L.; et al. GPX Pro198Leu and OGG1 Ser326Cys polymorphisms and risk of development of colorectal adenomas and colorectal cancer. Cancer Lett. 2005, 229, 85–91.
- Zachary Bloomgarden, Z. Evolution of type 2 diabetes mellitus treatment approaches: 2. J. Diabetes Res. 2019, 11, 4–6.
- Jetton, T.L.; Lausier, J.; LaRock, K.; Trotman, W.E.; Larmie, B.; Habibovic, A.; Peshavaria, M.; Leahy, J.L. Mechanisms of compensatory beta-cell growth in insulin-resistant rats: Roles of Akt kinase. Diabetes 2005, 54, 2294–2304.
- Italiani, P.; Boraschi, D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 2014, 4, 514.
- Evren Okur, M.E.; Karantas, I.D.; Siafaka, P.I. Diabetes Mellitus: A Review on Pathophysiology, Current Status of Oral Pathophysiology, Current Status of Oral Medications and Future Perspectives. ACTA Pharm. Sci. 2017, 55, 61–82.
- Chatterjee, S.; Khunti, K.; Davies, M.J. Type 2 diabetes. Lancet 2017, 389, 2239–2251.
- Burgos-Morón, E.; Abad-Jiménez, Z.; de Marañón, A.M.; Iannantuoni, F.; Escribano-López, I.; López-Domènech, S.; Salom, C.; Jover, A.; Mora, V.; Roldan, I.; et al. Relationship Between Oxidative Stress, ER Stress, and Inflammation in Type 2 Diabetes: The Battle Continues. J. Clin. Med. 2019, 8, 1385.
- Karunakaran, U.; Park, K.-G. A systematic review of oxidative stress and safety of antioxidants in diabetes: Focus on islets and their defense. Diabetes Metab. J. 2013, 37, 106–112.
- Molteni, C.G.; Principi, N.; Esposito, S. Reactive oxygen and nitrogen species during viral infections. Free Radic. Res. 2014, 48, 1163–1169.
- Khomich, O.A.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Redox Biology of Respiratory Viral Infections. Viruses 2018, 10, 392.
- Seet, R.C.S.; Lee, C.-Y.J.; Lim, E.C.H.; Quek, A.M.L.; Yeo, L.L.L.; Huang, S.-H.; Halliwell, B. Oxidative damage in dengue fever. Free Radic. Biol. Med. 2009, 47, 375–380.
- Korenaga, M.; Wang, T.; Li, Y.; Showalter, L.A.; Chan, T.; Sun, J.; Weinman, S.A. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J. Biol. Chem. 2005, 280, 37481–37488.
- Hogan, C.; Perkins, A.V. Selenoproteins in the Human Placenta: How Essential Is Selenium to a Healthy Start to Life? Nutrients 2022, 14, 628.
- Hussain, T.; Murtaza, G.; Metwally, E.; Kalhoro, D.H.; Kalhoro, M.S.; Rahu, B.A.; Sahito, R.G.A.; Yin, Y.; Yang, H.; Chughtai, M.I.; et al. The Role of Oxidative Stress and Antioxidant Balance in Pregnancy. Mediat. Inflamm. 2021, 2021, 9962860.
- Chiarello, D.I.; Abad, C.; Rojas, D.; Toledo, F.; Vázquez, C.M.; Mate, A.; Sobrevia, L.; Marín, R. Oxidative stress: Normal pregnancy versus preeclampsia. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2020, 1866, 165354.
- Scaife, P.J.; Simpson, A.; Kurlak, L.O.; Briggs, L.V.; Gardner, D.S.; Broughton Pipkin, F.; Jones, C.J.P.; Mistry, H.D. Increased Placental Cell Senescence and Oxidative Stress in Women with Pre-Eclampsia and Normotensive Post-Term Pregnancies. Int. J. Mol. Sci. 2021, 22, 7295.
- Farzin, L.; Sajadi, F. Comparison of serum trace element levels in patients with or without pre-eclampsia. J. Res. Med. Sci. 2012, 17, 938–941.
- Khera, A.; Vanderlelie, J.J.; Perkins, A.V. Selenium supplementation protects trophoblast cells from mitochondrial oxidative stress. Placenta 2013, 34, 594–598.
- Biswas, K.; McLay, J.; Campbell, F.M. Selenium Supplementation in Pregnancy-Maternal and Newborn Outcomes. J. Nutr. Metab. 2022, 2022, 4715965.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
585
Revisions:
3 times
(View History)
Update Date:
29 Dec 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




