Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Anriban Mitra | -- | 3434 | 2023-12-21 22:17:50 | | | |
| 2 | Fanny Huang | Meta information modification | 3434 | 2023-12-22 06:52:15 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wang, J.; Ford, J.C.; Mitra, A.K. Metastasis-Initiating Cells. Encyclopedia. Available online: https://encyclopedia.pub/entry/53048 (accessed on 07 February 2026).
Wang J, Ford JC, Mitra AK. Metastasis-Initiating Cells. Encyclopedia. Available at: https://encyclopedia.pub/entry/53048. Accessed February 07, 2026.
Wang, Ji, James C. Ford, Anirban K. Mitra. "Metastasis-Initiating Cells" Encyclopedia, https://encyclopedia.pub/entry/53048 (accessed February 07, 2026).
Wang, J., Ford, J.C., & Mitra, A.K. (2023, December 21). Metastasis-Initiating Cells. In Encyclopedia. https://encyclopedia.pub/entry/53048
Wang, Ji, et al. "Metastasis-Initiating Cells." Encyclopedia. Web. 21 December, 2023.
Copy Citation
Ovarian cancer is the deadliest gynecological malignancy with a high prevalence of transcoelomic metastasis. Metastasis is a multi-step process and only a small percentage of cancer cells, metastasis-initiating cells (MICs), have the capacity to finally establish metastatic lesions. These MICs maintain a certain level of stemness that allows them to differentiate into other cell types with distinct transcriptomic profiles and swiftly adapt to external stresses. Furthermore, they can coordinate with the microenvironment, through reciprocal interactions, to invade and establish metastases. Therefore, identifying, characterizing, and targeting MICs is a promising strategy to counter the spread of ovarian cancer.
ovarian cancer
metastasis
metastasis-initiating cells
microenvironment
1. Introduction
About 120 years ago, the seed and soil hypothesis, formulated by Dr. Stephen Paget, has pointed out the organ-specific secondary growth of cancer cells in breast cancer, instigating extensive research on the factors leading to metastasis in different types of cancer [1]. Two decades later, a hierarchical hematopoietic model was proposed by Dr. Maximow [2], ushering in a new era of stem cell research. Hematopoietic stem cells (HSCs) were identified, and their regenerative characteristic was confirmed by Dr. Becker, Dr. Till, and Dr. McCulloch, indirectly leading to the isolation of AML (acute lymphoid leukemia)-initiating cells and preliminary characterization of them in 1994 [3]. Several decades later, the self-renewal and differentiation features of AML-initiating cells were validated by Dr. Dick, providing the bedrock for the contemporary cancer stem cell (CSC) hierarchical model [4]. Building upon the concept of stemness, Dr. Bapat successfully isolated cancer initiating cells from the ascites of a patient diagnosed with grade IV ovarian serous adenocarcinoma in 2005 [5]. Shortly thereafter, transcoelomic metastasis, a special cancerous dissemination route, was clearly described as a multi-step process for ovarian cancer by Dr. Tan [6]. This delineation spurred subsequent research, placing a heightened focus on understanding the initiation of metastasis in OC. Hosseini et al. established a breast cancer mouse model to recapitulate early lesions of cancer metastasis through expressing the Her2 transgene at puberty of BALB-NeuT mice [7]. MICs in OC, a concept in its relative infancy, chiefly encompasses a group of cells endowed with the capacity to establish clinically significant intraperitoneal metastasis.
2. Origins of MICs
Extensive research has provided compelling evidence for the existence of a specific type of cancer cells, including those in OC, capable of metastasizing to other parts of the body from primary tumors [8][9][10]. Nevertheless, the precise origins of these metastasis-capable cells remain a subject of ongoing debate. At the forefront of this debate is the quest to determine the primary tumor’s origin. Histologically speaking, HGSOC cells bear a resemblance to normal cells lining the fallopian tube (the most anatomically proximal structure), endometrium, and endocervix, leading researchers to hypothesize that OC cells originate from fallopian tube epithelia (FTE). However, some researchers insist that OC cells originate from ovarian surface epithelia (OSE) because of proximity and their epithelial phenotype [11]. Detailed profiling of OSE-derived and FTE-derived OC cells revealed that the latter exhibits a more aggressive phenotype coupled with heightened chemoresistance and a proclivity for invasion [12]. MICs are more likely to originate from FTE-derived OC cells, but whether MICs intrinsically attain their initiation capacity in the beginning or acquire it later remains unknown and more research needs to be done.
3. Characteristics of MICs
To thrive in the dynamic microenvironments and to establish the distant omental metastasis, MICs must possess not only the potential to colonize remote sites but also the remarkable ability to swiftly adapt to external stressors. The defining characteristics of MICs can be comprehensively examined through three distinct perspectives: markers, multi-omic profiles, and the intricacies of their metastatic niches.
3.1. Markers
The isolation of MICs from tumor masses is a pivotal step for researchers to characterize them and to find potential therapeutics targeting them, relying heavily on the selection of markers. Those markers can be categorized into surface markers and intracellular markers. Surface markers are particularly favored over other cellular markers due to their ease of recognition by specialized antibodies without compromising the integrity of the OC cell membranes. This approach facilitates the sorting of a larger population of live cells through flow cytometry techniques, enabling more comprehensive characterizations. Sometimes, some intracellular markers (enzymes and transcription factors) can also be utilized to identify MICs, for they can offer valuable insights into the functional profiles of these cells.
CD24
CD24 is a sialoglycoprotein, originally found to be widely expressed on the surface of B cells to prevent them from terminal differentiating into plasma cells [13]. Because of its intrinsic trait to modulate differentiation, CD24 is also a well-known ovarian cancer surface marker, predicting a poor prognosis in OC patients. High expression of it is found to be positively correlated with an advanced FIGO stage and presence of peritoneal metastasis [14]. In addition, CD24 is a marker for OC CSCs [15][16][17], capable of initiating cancerous lesions in the peritoneal cavity, confirming the metastasis-initiating trait of CD24+ cells. In terms of cancer-initiating capacity, CD24 can increase the phosphorylation of STAT3 via JAK2 to favor the growth of primary tumor spheres and the ability of forming secondary spheres [18], playing an important role in forming spheroids in ascites during metastatic dissemination. Resistance to anoikis (the first step of transcoelomic metastasis) is also closely related to the high expression level of CD24 [6]. In addition, adhesions to the surface of mesothelial cells, a necessary step for invading the peritoneum, are mediated by interactions between CD24 and P-selectins, a mesothelial surface protein [19][20].
CD44
CD44, a glycoprotein ligand, plays a pivotal role in mediating cell-to-cell adhesion through its interactions with a range of molecules, including hyaluronic acids (HAs) [21][22][23], osteopontin [23][24], collagens [25][26] and metalloproteinases (MMPs) [27][28]. There are multiple isoforms of CD44 that are confirmed to be correlated with cancerous phenotypes in various cancers [29][30][31]. Notably, research has highlighted the metastatic initiation potential of CD44 variant 6 (CD44v6) in advanced ovarian cancer (OC) [9][32] with negative correlations observed between good prognoses and the enrichment of CD44v6 positive OC cells [21][33][34]. Furthermore, the mesenchymal isoform of CD44 is able to promote EMT of OC cells and endow them with a stem cell-like phenotype [35]. During the adhesive step of the transcoelomic metastasis of OC, a sandwich model of two CD44 receptors and one stabilized HA/Versican polymer in between them has been proposed to emphasize the supportive role of CD44 during the invasion of the peritoneum [21]. Interestingly, CD44 was found to be transferred from one subpopulation of OC to the others to augment the metastatic potential in low-metastatic cells via exosome-mediated transport [36]. A combination of both CD44 and CD117 has been employed to isolate cells with heightened stemness and increased metastatic capacity [9].
CD133
CD133 is a glycoprotein originally found to be expressed on adult stem cells and widely known to play a critical role in maintenance of stemness [37][38][39]. That stemness-associated characteristic is also applied to OC and previous research has suggested the metastatic-initiation role of CD133+ cells [40]. Furthermore, CD133 is closely associated with EMT mediated by ZEB2 and negatively correlates with good prognoses in OC patients in terms of overall survival (OS) [41]. CD133 can also contribute to the metastasis of OC cells via increasing secretion of MMPs and upregulating the surface markers (PECAM1 and ICAM1) to strengthen adhesions [42]. It can also trigger the transcriptional activity of β-catenin to maintain CSC properties via interacting with integrins in various cancer types. Whether CD133+ cells preserve the CD133-β-catenin pathway remains an intriguing area for exploration in OC [43].
CD117
CD117 (c-KIT) is a receptor tyrosine kinase protein and it is normally upregulated in mast cells and certain hematopoietic stem cells, which is closely related to cell differentiation [44][45][46]. Studies have demonstrated that CD117+ OC cells possess the capability not only for self-regeneration but also for generating diverse metastatic variants [47]. Multiple reports have established direct correlations between CD117 expression and various factors such as chemoresistance, advanced clinical stage, malignancy, as well as markers like SOX2, COT4, and NANOG [48][49][50]. Furthermore, the higher expression level of CD117 in extracellular vesicles from OC cells is positively correlated to more aggressive tumor invasion [51]. Given that CD44 also serves as an initiation-associated factor, the combination of CD117 and CD44 has been applied to identify cell subpopulations with relatively higher stemness potential [52][53][54].
ROR1
ROR1 is a receptor tyrosine kinase-like orphan receptor, primarily recognized for its involvement in the modulation of neurite growth, exhibiting higher expression levels in embryonic tissues compared to adult counterparts [55][56][57][58][59]. Some previous reports have indicated that high expression levels of ROR1 have detrimental effects on the prognoses of OC patients in terms of overall survival and progression-free survival rates [60]. Kipps et al. linked stemness to ROR1 and found that more spheroids and a higher expression level of ALDH1 are positively correlated with the enrichment of ROR1+ OC cells [61]. Moreover, the inhibitory effects of silencing ROR1 on invasion and adhesion of peritoneal membranes are validated in vitro, further implicating the potential metastatic initiation capability of ROR1+ cells [62]. Nevertheless, research employing in vivo models and metastatic experiments is warranted to comprehensively elucidate the functions of ROR1+ cells in OC.
ALDH
The family of aldehyde dehydrogenases (ALDH) comprises numerous isoforms responsible for catalyzing the oxidation of aldehydes [63][64]. ALDH1A1, notably identified as a negative prognostic indicator, is a well-established marker for OC CSCs [65][66][67][68] and previous research has shown that ALDH1A1 contributes to the maintenance of stemness via the coordinated regulation of cell cycle checkpoints and DNA repair networks [69]. It has also been observed that ALDH+ cells are enriched in patient ascites-derived spheroids, implicating the resistance to anoikis imparted by ALDH. In addition, those spheroids finally contribute to the formation of metastatic lesions in the peritoneal cavity [70]. The inhibition of conversion from ALDH- OC cells to ALDH+ ones, mediated by DDB2, can decrease peritoneal metastasis, suggesting the metastasis-initiating capability of ALDH [71].
SOX2
The Yamanaka factors, consisting of SOX2, KLF4, OCT4, and MYC, have been pivotal in mediating the induction of pluripotent stem cells from terminally differentiated fibroblasts—an epochal achievement in stem cell research that has reverberated into the study of cancer stem cells [72][73][74][75]. OCT4 and SOX2 were identified to be core transcription factors, whereas KLF4 and MYC to be essential factors in maintaining pluripotency [72]. Most importantly, only a small percentage of cells were confirmed to co-upregulate the expression levels of all 4 of the above factors [76], coinciding with the widely observed phenomenon that cancer-initiating cells account for less than 10% of all cells within the tumor mass. In the context of ovarian cancer (OC), SOX2 assumes a predominant role in the maintenance of cancer stemness, particularly within spheroids, as evidenced by its higher expression levels [50][77]. Notably, SOX2 exhibits a dual role in OC, enhancing metastatic potential while diminishing adhesive capabilities [78], implicating its important role in metastasis initiation. While numerous publications have linked the malignancy of OC to elevated MYC and OCT4 expression levels [79][80][81][82][83], none of them have explicitly indicated the metastasis-initiating capacity of both factors. Interestingly, KLF4 plays a tumor suppressing role during metastasis of OC and previous research has also explicitly indicated its apoptosis-inducing effects and chemo-sensitizing functions [84][85]. Even though the transcriptional activity of SOX2 (the most effective Yamanaka factor for MIC markers) can be applied to identify some MICs, one main limitation of utilizing TFs as markers is that they cannot be used to isolate live cells by FACS.
3.2. Multi-Omic Profiles
In the realm of cancer research, the widespread utilization of genomic, epigenomic, and transcriptomic techniques has empowered the creation of cellular profiles with unprecedented resolution. The potential of comprehensively characterizing MICs by harnessing the capabilities offered by these three aspects—genomic, epigenomic, and transcriptomic are reviewed.
Genomic Profiles
Many studies have implicated the enrichment of metastatic-initiating cells with stemness in spheroids in OC and suggested that CD44+/CD117+ cells possess the potential to initiate metastasis. Comparative genome analysis has been done to compare the CD44+/CD117+ cells with CD44−/CD117− cells isolated from the ascites of six patients and found that there were small genetic differences (rearrangements on chromosome 2) between them in terms of single nucleotide polymorphisms (SNPs) in just one patient [86], indicating potential differentiation from double positive cells to negative cells. Interestingly, hundreds of somatic genomic arrangements between primary and metastatic tumor samples were identified and several somatic breakpoints influenced the cancer-related genes (FANCD2, ERBB4 and ESR1) [87], substantiating genomic instability and variability during metastasis, but under most circumstances, global genomic instability is rarely observed, implicating the selection for MICs (accounting for a small percentage of the whole primary tumor) under external stresses. Recent extensive research involving 250 biopsy pairs from a diverse range of metastatic solid malignancies, including OC, has explored genomic changes in metastases under treatment selection pressure from a pan-cancer perspective. Interestingly, the genomic profiles of metastases remained nearly unchanged during treatment in terms of known actionable genomic points. However, substantial variations emerged in the genomic profiles of metastases during treatment, primarily in the unactionable genomic regions, especially intergenic regions., implicating the influences of epigenomic differences induced by those changes [88]. In summary, relative genomic stability is the major characteristic of MICs regardless of whether they are under treatment or not, and trivial changes of genomic profiles of MICs and non-MICs also confirms that those non-MICs may potentially originate from MICs.
Epigenomic Landscape
Epigenetic regulations of gene expression are key to cellular phenotypical changes [89][90][91][92], and they are more flexible and immediate, compared to genomic changes. Histone modifications [93][94] (such as histone methylation, histone acetylation, and histone phosphorylation), DNA modifications [95][96] (such as DNA methylation and DNA hydroxyl methylation), and non-coding RNA-induced transcriptional changes [97][98][99] mainly account for epigenetic regulatory pathways. During normal differentiation, the global openness of the chromatins of stem cells is decreased and the transcriptional activities of most pluripotent genes will be diminished [100]. Similar to normal adult stem cells, the stemness of MICs is related to global hypomethylation and local hypermethylation of specific genes and that trait imparts great flexibility of manipulating transcriptions of genes to MICs in response to changing external stimuli [101]. In MICs, the variance of the expression levels of all genes is expected to be lower than that of non-MICs and entropy is used to quantitatively assess the stemness of cells [102][103][104]. The higher the entropy value (the quantitative measure for uncertainty) is, the more the number of possible differentiation lines that cells can walk down. Multiple methods have been developed to quantify entropy values. Given that the phenotypes of cancer cells rely heavily on the activation/inhibition of different combinations of various signaling pathways, some researchers calculate the signal entropy values based on the protein–protein interaction (PPI) network and gene expression profiles [105]. Additionally, Dr. Vaidya has developed a method to calculate entropy values based on DNA methylation, where the variation in these values reflects the replication status of stem cells [106].
In addition, PRC2 (polycomb repressive complex 2) is a very important regulator in the epigenetic field and its main job is to close target genomic regions via methylating histone H3 on lysine 27 [107][108][109]. MICs can effectively use EZH2, the histone methyltransferase of PRC2, to mediate the decrease of DAB2IP, a negative regulator of WNT signaling via causing the downregulation of WNT5B, to maintain their stemness. MICs can also leverage DNA methylase and HOTAIR, a non-coding RNA which associates with PRC2 to play a suppressive role in transcription, to achieve the same goal [110].
Previous research on OC also suggests that global histone acetylation and hypomethylation can augment the stemness of OC cells to generate more MICs [111]. Furthermore, histone lysine demethylases are responsible for upregulating SOX2 to elevate the initiation potentials of MICs via modifying the chromatin landscape in OC [112].
Transcriptomic Landscape
The transcriptomic characteristics of MICs can be summarized into two categories: plasticity and stemness. Transcriptomic plasticity is mainly imparted by flexible epigenomic regulations, and it can endow cells with the ability to switch between EMT and MET (mesenchymal epithelial transition) [113][114][115][116]. EMT, a well-studied biological process, is required for cancer cells to acquire a mesenchymal phenotype to invade into stromal tissues via breaking the mesothelial layer and pave the way for metastatic colonization [117]. There are multiple signaling pathways (Wnt/β-catenin signaling pathway, PI3K/Akt/mTOR signaling pathway, TGF β signaling pathway) which can activate EMT [118][119][120]. The Wnt/β-catenin signaling pathway includes canonical and non-canonical pathways, the former of which is studied heavily and requires the destruction of the β-catenin complex to mediate the transcription of downstream genes [119]. Cysteine-rich intestinal protein 1 (CRIP1) and BRMS1 = like transcriptional repressor (BRMS1L) have been identified to be associated with EMT via augmenting or diminishing the expression of Wnt/β-catenin-mediated genes (TCF, MYC and CCND1) in OC cancer cells, respectively [121][122]. In addition, previous research has found that PCNP can promote the EMT of OC cells via increasing the accumulation of β-catenin in the nucleus. Interestingly, the simultaneous acquisition of stemness during the EMT process has been reported in many cancer types including OC, implicating the importance of EMT in MICs [123]. In contrast to EMT, MET can restore OC cells back to an epithelial phenotype with more proliferative potentials, which is necessary for cancer cells to spread throughout the greater omentum, but there is a very limited number of publications about MET in OC cancer cells. Previous research has found that SMAD7 is a negative regulator of the TGF-β signaling pathway and the SMAD7-mediated-MET phenomenon was observed in OC stem cells [124]. Importantly, the process of transitioning between epithelial and mesenchymal phenotypes is not dichotomous but rather comprises a spectrum of intermediate cellular states known as hybrid EMT cells [123][125]. However, whether MICs are hybrid EMT cells remains to be explored.
The stemness trait lies at the heart of cancer-initiating cells, engaging MICs in the potential differentiation lines towards other cell types with different functions, and the lower proliferative capacity of this trait makes MICs less susceptible to standard chemotherapeutic drugs. Except for Yamanaka factors, epigenetic regulators should be implementors of this program, flexibly and constantly switching chromatic regions on or off to upregulate or downregulate specific downstream genes [71][126][127][128][129][130]. Previous research has found that KDM4C can upregulate OCT4 expression to increase the stemness of OC cells via the trimethylation of lysine 9 of histone 3 at the promoter region of OCT4 [129]. In addition, DDB2, responsible for the trimethylation of lysine 27 of histone 3, was discovered to bind to the ALDH1A1 gene promoter to interfere with the dedifferentiation process in OC, implicating its role in the differentiation of MICs [71].
3.3. Metastatic Niche
Successful metastatic initiation hinges on the intricate coordination of multiple cell types to build a microenvironment (niche) suitable for MICs to proliferate, differentiate, and maintain (Figure 1). However, the composition and architectural features of these niches can vary significantly depending on the target organ and cancer type. The greater omentum, which consists of a great number of adipocytes, is the most preferable metastatic site for OC cells, where it is lined by a double layer of mesothelial cells, a first-line defense for cancerous invasion [6][131]. In addition, milky spots (specialized secondary lymphoid organs), aggregates of immune cells (T cells, B cells, granulocytes, and macrophages), stromal cells (fibroblasts, the high endothelium of veins, and mesothelial cells), and extracellular matrices (ECMs), are widely distributed across the greater omentum. Under normal circumstances, milky spots mediate immunity against foreign bodies in the peritoneal cavity whereas they often initiate insufficient anti-tumor immunity and finally become accomplices of metastatic cancer cells to augment the progression of metastatic initiation [132][133]. Furthermore, tumors are classified into hot, variable, and cold based on the infiltration of immune cells and their pro-/anti-inflammatory traits, informing therapeutic targeting of immune checkpoints [134]. OC tumors enriched in pro-inflammatory immune cells manifest better prognoses compared to ones having regulatory T cells, which can also be used for predicting responses to immunotherapies [135][136]. The formation of the immune landscape of OC is a dynamic process mediated by the elimination and escape phases of immunoediting. Throughout the process, cytotoxic functions, elicited by CD8+ T cells, NK T cells, NK cells, and IFN production, facilitate the annihilation of tumor cells with recognizable antigens. However, the failure of the induction of killing them finally leads to the survival of tumor cells which upregulate multiple ligands to evade normal cancer immunosurveillance [137].
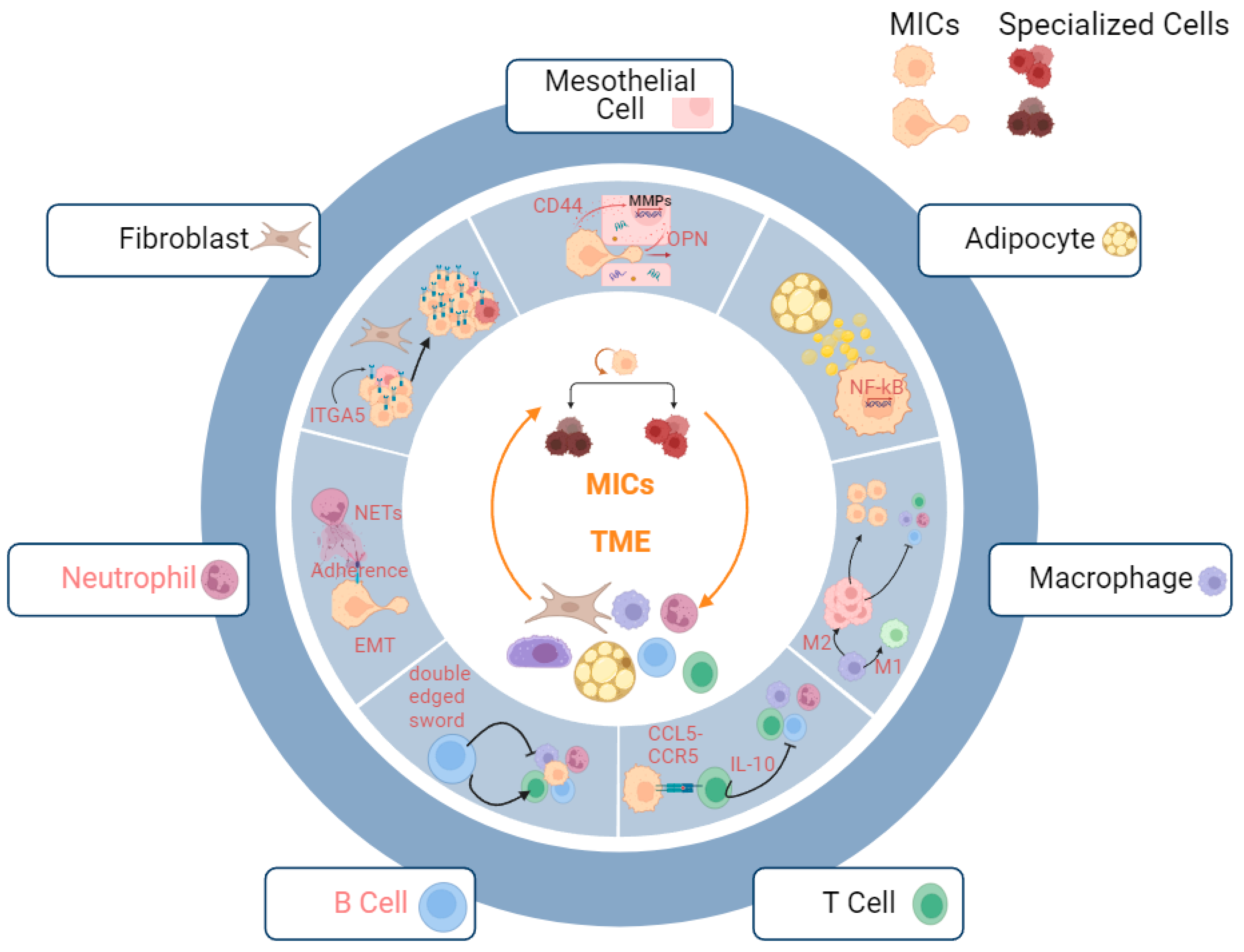
Figure 1. Metastatic niche of MICs. Productive interactions with multiple stromal and immune cells help MICs in establishing metastases. Mesothelial cells: OPN secreted by mesothelial cells facilitate the invasion of MICs; adipocytes: lipid droplets secreted by adipocyte amplify the stemness of MICs via upregulating NF-kB signaling pathways; macrophages: M2 macrophages assist immune escape of MICs by mediating suppression of pro-inflammatory immune microenvironment; T Cells: binding of CCR5 and CCL5 suppresses pro-inflammatory immune microenvironment; B Cells: B cells serve as both pro-inflammatory and anti-inflammatory mediators for cancer immune microenvironment; neutrophils: neutrophils increase adhesions of MICs to omentum via NETs; fibroblasts: fibroblasts facilitate the adaptation of MICs to microenvironment via upregulating ITGA5 and provide a supportive niche. The interactions between B cells/ neutrophils (red highlight) and the MICs remain to be explored.
References
- Paget, S. The distribution of secondary growths in cancer of the breast. Lancet 1889, 133, 571–573.
- Alexander, M. Untersuchungen Über Blut Und Bindegewebe. Arch. Für Mikrosk. Anat. 1922, 73, 444–561.
- Becker, A.J.; Mcculloch, E.A.; Till, J.E. Cytological Demonstration of the Clonal Nature of Spleen Colonies Derived from Transplanted Mouse Marrow Cells. Nature 1963, 197, 452–454.
- Bonnet, D.; Dick, J.E. Human Acute Myeloid Leukemia Is Organized as a Hierarchy That Originates from a Primitive Hematopoietic Cell. Nat. Med. 1997, 3, 730–737.
- Bapat, S.A.; Mali, A.M.; Koppikar, C.B.; Kurrey, N.K. Stem and Progenitor-Like Cells Contribute to the Aggressive Behavior of Human Epithelial Ovarian Cancer. Cancer Res. 2005, 65, 3025–3029.
- Tan, D.S.; Agarwal, R.; Kaye, S.B. Mechanisms of Transcoelomic Metastasis in Ovarian Cancer. Lancet Oncol. 2006, 7, 925–934.
- Hosseini, H.; Obradović, M.M.S.; Hoffmann, M.; Harper, K.L.; Sosa, M.S.; Werner-Klein, M.; Nanduri, L.K.; Werno, C.; Ehrl, C.; Maneck, M.; et al. Early Dissemination Seeds Metastasis in Breast Cancer. Nature 2016, 540, 552–558.
- Motohara, T.; Masuda, K.; Morotti, M.; Zheng, Y.; El-Sahhar, S.; Chong, K.Y.; Wietek, N.; Alsaadi, A.; Karaminejadranjbar, M.; Hu, Z.; et al. An Evolving Story of the Metastatic Voyage of Ovarian Cancer Cells: Cellular and Molecular Orchestration of the Adipose-Rich Metastatic Microenvironment. Oncogene 2019, 38, 2885–2898.
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.-C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.-M.; Nephew, K.P. Identification and Characterization of Ovarian Cancer-Initiating Cells from Primary Human Tumors. Cancer Res. 2008, 68, 4311–4320.
- Massagué, J.; Ganesh, K. Metastasis-Initiating Cells and Ecosystems. Cancer Discov. 2021, 11, 971–994.
- Riso, P.L.; Villa, C.E.; Gasparoni, G.; Vingiani, A.; Luongo, R.; Manfredi, A.; Jungmann, A.; Bertolotti, A.; Borgo, F.; Garbi, A.; et al. A Cell-of-Origin Epigenetic Tracer Reveals Clinically Distinct Subtypes of High-Grade Serous Ovarian Cancer. Genome Med. 2020, 12, 94.
- Zhang, S.; Dolgalev, I.; Zhang, T.; Ran, H.; Levine, D.A.; Neel, B.G. Both Fallopian Tube and Ovarian Surface Epithelium Are Cells-of-Origin for High-Grade Serous Ovarian Carcinoma. Nat. Commun. 2019, 10, 5367.
- Fischer, G.F.; Majdic, O.; Gadd, S.; Knapp, W. Signal Transduction in Lymphocytic and Myeloid Cells via CD24, a New Member of Phosphoinositol-Anchored Membrane Molecules. J. Immunol. 1990, 144, 638–641.
- Nakamura, K.; Terai, Y.; Tanabe, A.; Ono, Y.J.; Hayashi, M.; Maeda, K.; Fujiwara, S.; Ashihara, K.; Nakamura, M.; Tanaka, Y.; et al. CD24 Expression Is a Marker for Predicting Clinical Outcome and Regulates the Epithelial-Mesenchymal Transition in Ovarian Cancer via Both the Akt and ERK Pathways. Oncol. Rep. 2017, 37, 3189–3200.
- Gao, M.-Q.; Choi, Y.-P.; Kang, S.; Youn, J.H.; Cho, N.-H. CD24+ Cells from Hierarchically Organized Ovarian Cancer Are Enriched in Cancer Stem Cells. Oncogene 2010, 29, 2672–2680.
- Kang, K.S.; Choi, Y.P.; Gao, M.-Q.; Kang, S.; Kim, B.G.; Lee, J.H.; Kwon, M.J.; Shin, Y.K.; Cho, N.H. CD24+ Ovary Cancer Cells Exhibit an Invasive Mesenchymal Phenotype. Biochem. Biophys. Res. Commun. 2013, 432, 333–338.
- Koh, J.; Lee, S.; Park, H.; Lee, H.J.; Cho, N.H.; Kim, J. Susceptibility of CD24+ Ovarian Cancer Cells to Anti-Cancer Drugs and Natural Killer Cells. Biochem. Biophys. Res. Commun. 2012, 427, 373–378.
- Burgos-Ojeda, D.; Wu, R.; McLean, K.; Chen, Y.-C.; Talpaz, M.; Yoon, E.; Cho, K.R.; Buckanovich, R.J. CD24+ Ovarian Cancer Cells Are Enriched for Cancer-Initiating Cells and Dependent on JAK2 Signaling for Growth and Metastasis. Mol. Cancer Ther. 2015, 14, 1717–1727.
- Carroll, M.J.; Fogg, K.C.; Patel, H.A.; Krause, H.B.; Mancha, A.-S.; Patankar, M.S.; Weisman, P.S.; Barroilhet, L.; Kreeger, P.K. Alternatively Activated Macrophages Upregulate Mesothelial Expression of P-Selectin to Enhance Adhesion of Ovarian Cancer Cells. Cancer Res. 2018, 78, 3560–3573.
- Li, Y.; Sun, X.; Wang, H. Role of CD24 in Anoikis Resistance of Ovarian Cancer Cells. J. Huazhong Univ. Sci. Technol. Med. Sci. 2015, 35, 390–396.
- Ween, M.P.; Oehler, M.K.; Ricciardelli, C. Role of Versican, Hyaluronan and CD44 in Ovarian Cancer Metastasis. Int. J. Mol. Sci. 2011, 12, 1009–1029.
- Herrera-Gayol, A.; Jothy, S. The impact of hyaluronan on the in vitro invasive properties of human breast cancer cell lines with high CD44 expression. Hyaluronan 2002, 1, 443–446.
- Sano, N.; Kitazawa, K.; Sugisaki, T. Localization and Roles of CD44, Hyaluronic Acid and Osteopontin in IgA Nephropathy. Nephron 2001, 89, 416–421.
- Weber, G.F.; Ashkar, S.; Glimcher, M.J.; Cantor, H. Receptor-Ligand Interaction Between CD44 and Osteopontin (Eta-1). Science 1996, 271, 509–512.
- Ishii, S.; Ford, R.; Thomas, P.; Nachman, A.; Steele, G.; Jessup, J.M. CD44 Participates in the Adhesion of Human Colorectal Carcinoma Cells to Laminin and Type IV Collagen. Surg. Oncol. 1993, 2, 255–264.
- Knutson, J.R.; Iida, J.; Fields, G.B.; McCarthy, J.B. CD44/Chondroitin Sulfate Proteoglycan and Alpha 2 Beta 1 Integrin Mediate Human Melanoma Cell Migration on Type IV Collagen and Invasion of Basement Membranes. Mol. Biol. Cell 1996, 7, 383–396.
- Yu, Q.; Stamenkovic, I. Localization of Matrix Metalloproteinase 9 to the Cell Surface Provides a Mechanism for CD44-Mediated Tumor Invasion. Gene Dev. 1999, 13, 35–48.
- Chellaiah, M.A.; Ma, T. Membrane Localization of Membrane Type 1 Matrix Metalloproteinase by CD44 Regulates the Activation of Pro-Matrix Metalloproteinase 9 in Osteoclasts. Biomed. Res. Int. 2013, 2013, 302392.
- Zhang, H.; Brown, R.L.; Wei, Y.; Zhao, P.; Liu, S.; Liu, X.; Deng, Y.; Hu, X.; Zhang, J.; Gao, X.D.; et al. CD44 Splice Isoform Switching Determines Breast Cancer Stem Cell State. Gene Dev. 2019, 33, 166–179.
- Tanabe, K.K.; Nishi, T.; Saya, H. Novel Variants of CD44 Arising from Alternative Splicing: Changes in the CD44 Alternative Splicing Pattern of MCF-7 Breast Carcinoma Cells Treated with Hyaluronidase. Mol. Carcinog. 1993, 7, 212–220.
- Wang, S.J.; Wreesmann, V.B.; Bourguignon, L.Y.W. Association of CD44 V3-containing Isoforms with Tumor Cell Growth, Migration, Matrix Metalloproteinase Expression, and Lymph Node Metastasis in Head and Neck Cancer. Head Neck 2007, 29, 550–558.
- Tjhay, F.; Motohara, T.; Tayama, S.; Narantuya, D.; Fujimoto, K.; Guo, J.; Sakaguchi, I.; Honda, R.; Tashiro, H.; Katabuchi, H. CD44 Variant 6 Is Correlated with Peritoneal Dissemination and Poor Prognosis in Patients with Advanced Epithelial Ovarian Cancer. Cancer Sci. 2015, 106, 1421–1428.
- Zhao, L.; Gu, C.; Huang, K.; Zhang, Z.; Ye, M.; Fan, W.; Han, W.; Meng, Y. The Prognostic Value and Clinicopathological Significance of CD44 Expression in Ovarian Cancer: A Meta-Analysis. Arch. Gynecol. Obstet. 2016, 294, 1019–1029.
- Zhang, H.-F.; Hu, P.; Fang, S.-Q. Understanding the Role of CD44V6 in Ovarian Cancer. Oncol. Lett. 2017, 14, 1989–1992.
- Bhattacharya, R.; Mitra, T.; Chaudhuri, S.R.; Roy, S.S. Mesenchymal Splice Isoform of CD44 (CD44s) Promotes EMT/Invasion and Imparts Stem-like Properties to Ovarian Cancer Cells. J. Cell Biochem. 2018, 119, 3373–3383.
- Shen, X.; Wang, C.; Zhu, H.; Wang, Y.; Wang, X.; Cheng, X.; Ge, W.; Lu, W. Exosome-Mediated Transfer of CD44 from High-Metastatic Ovarian Cancer Cells Promotes Migration and Invasion of Low-Metastatic Ovarian Cancer Cells. J. Ovarian Res. 2021, 14, 38.
- Coskun, V.; Wu, H.; Blanchi, B.; Tsao, S.; Kim, K.; Zhao, J.; Biancotti, J.C.; Hutnick, L.; Krueger, R.C.; Fan, G.; et al. CD133+ Neural Stem Cells in the Ependyma of Mammalian Postnatal Forebrain. Proc. Natl. Acad. Sci. USA 2008, 105, 1026–1031.
- Böhmer, R.M. Erythropoiesis from Adult but Not Fetal Blood-Derived CD133+ Stem Cells Depends Strongly on Interleukin-3. Growth Factors 2009, 22, 45–50.
- Handgretinger, R.; Gordon, P.R.; Leimig, T.; Chen, X.; Bühring, H.; Niethammer, D.; Kuçi, S. Biology and Plasticity of CD133+ Hematopoietic Stem Cells. Ann. N. Y. Acad. Sci. 2003, 996, 141–151.
- Li, Y.; Fei, H.; Lin, Q.; Liang, F.; You, Y.; Li, M.; Wu, M.; Qu, Y.; Li, P.; Yuan, Y.; et al. ZEB2 Facilitates Peritoneal Metastasis by Regulating the Invasiveness and Tumorigenesis of Cancer Stem-like Cells in High-Grade Serous Ovarian Cancers. Oncogene 2021, 40, 5131–5141.
- Zhou, Q.; Chen, A.; Song, H.; Tao, J.; Yang, H.; Zuo, M. Prognostic Value of Cancer Stem Cell Marker CD133 in Ovarian Cancer: A Meta-Analysis. Int. J. Clin. Exp. Med. 2014, 8, 3080–3088.
- Roy, L.; Bobbs, A.; Sattler, R.; Kurkewich, J.L.; Dausinas, P.B.; Nallathamby, P.; Dahl, K.D.C. CD133 Promotes Adhesion to the Ovarian Cancer Metastatic Niche. Cancer Growth Metastasis 2018, 11, 1179064418767882.
- Su, Y.-J.; Lin, W.-H.; Chang, Y.-W.; Wei, K.-C.; Liang, C.-L.; Chen, S.-C.; Lee, J.-L. Polarized Cell Migration Induces Cancer Type-Specific CD133/Integrin/Src/Akt/GSK3β/β-Catenin Signaling Required for Maintenance of Cancer Stem Cell Properties. Oncotarget 2015, 6, 38029–38045.
- Massa, S.; Balciunaite, G.; Ceredig, R.; Rolink, A.G. Critical Role for C-kit (CD117) in T Cell Lineage Commitment and Early Thymocyte Development in vitro. Eur. J. Immunol. 2006, 36, 526–532.
- Matsuda, R.; Takahashi, T.; Nakamura, S.; Sekido, Y.; Nishida, K.; Seto, M.; Seito, T.; Sugiura, T.; Ariyoshi, Y.; Takahashi, T. Expression of the C-Kit Protein in Human Solid Tumors and in Corresponding Fetal and Adult Normal Tissues. Am. J. Pathol. 1993, 142, 339–346.
- Natali, P.G.; Nicotra, M.R.; Sures, I.; Santoro, E.; Bigotti, A.; Ullrich, A. Expression of C-Kit Receptor in Normal and Transformed Human Nonlymphoid Tissues. Cancer Res. 1992, 52, 6139–6143.
- Luo, L.; Zeng, J.; Liang, B.; Zhao, Z.; Sun, L.; Cao, D.; Yang, J.; Shen, K. Ovarian Cancer Cells with the CD117 Phenotype Are Highly Tumorigenic and Are Related to Chemotherapy Outcome. Exp. Mol. Pathol. 2011, 91, 596–602.
- Al-Shami, S.A.; Al-Kaabi, M.M.; Mahdi, A.K.; Al-Attar, Z. Immunohistochemical Expression of CD117 in Borderline, Low- and High-Grade Ovarian Surface Epithelial Tumours: A Clinicopathological Study. Malays. J. Pathol. 2023, 45, 229–236.
- Conic, I.; Stanojevic, Z.; Velickovic, L.J.; Stojnev, S.; Petrovic, A.R.; Krstic, M.; Stanojevic, M.; Bogdanović, D.; Stefanovic, V. Epithelial Ovarian Cancer with CD117 Phenotype Is Highly Aggressive and Resistant to Chemotherapy. J. Obstet. Gynaecol. Res. 2015, 41, 1630–1637.
- Robinson, M.; Gilbert, S.F.; Waters, J.A.; Lujano-Olazaba, O.; Lara, J.; Alexander, L.J.; Green, S.E.; Burkeen, G.A.; Patrus, O.; Sarwar, Z.; et al. Characterization of SOX2, OCT4 and NANOG in Ovarian Cancer Tumor-Initiating Cells. Cancers 2021, 13, 262.
- Shnaider, P.V.; Petrushanko, I.Y.; Aleshikova, O.I.; Babaeva, N.A.; Ashrafyan, L.A.; Borovkova, E.I.; Dobrokhotova, J.E.; Borovkov, I.M.; Shender, V.O.; Khomyakova, E. Expression Level of CD117 (KIT) on Ovarian Cancer Extracellular Vesicles Correlates with Tumor Aggressiveness. Front. Cell Dev. Biol. 2023, 11, 1057484.
- Gao, Z.; Ye, X.; Bordeaux, A.; Hettich, S.; Lin, S.; Han, F.; Jia, Y. MiR-26b Regulates Cell Proliferation and Apoptosis of CD117+CD44+ Ovarian Cancer Stem Cells by Targeting PTEN. Eur. J. Histochem. 2021, 65, 3186.
- Fan, Y.; Cheng, H.; Liu, Y.; Liu, S.; Lowe, S.; Li, Y.; Bentley, R.; King, B.; Tuason, J.P.W.; Zhou, Q.; et al. Metformin Anticancer: Reverses Tumor Hypoxia Induced by Bevacizumab and Reduces the Expression of Cancer Stem Cell Markers CD44/CD117 in Human Ovarian Cancer SKOV3 Cells. Front. Pharmacol. 2022, 13, 955984.
- Chen, M.; Su, J.; Feng, C.; Liu, Y.; Zhao, L.; Tian, Y. Chemokine CCL20 Promotes the Paclitaxel Resistance of CD44+CD117+ Cells via the Notch1 Signaling Pathway in Ovarian Cancer. Mol. Med. Rep. 2021, 24, 635.
- Paganoni, S.; Ferreira, A. Neurite Extension in Central Neurons: A Novel Role for the Receptor Tyrosine Kinases Ror1 and Ror2. J. Cell Sci. 2005, 118, 433–446.
- Tseng, H.-C.; Lyu, P.-C.; Lin, W. Nuclear Localization of Orphan Receptor Protein Kinase (Ror1) Is Mediated through the Juxtamembrane Domain. BMC Cell Biol. 2010, 11, 48.
- Widhopf, G.F.; Cui, B.; Ghia, E.M.; Chen, L.; Messer, K.; Shen, Z.; Briggs, S.P.; Croce, C.M.; Kipps, T.J. ROR1 Can Interact with TCL1 and Enhance Leukemogenesis in Eµ-TCL1 Transgenic Mice. Proc. Natl. Acad. Sci. USA 2014, 111, 793–798.
- Borcherding, N.; Kusner, D.; Liu, G.-H.; Zhang, W. ROR1, an Embryonic Protein with an Emerging Role in Cancer Biology. Protein Cell 2014, 5, 496–502.
- Paganoni, S.; Bernstein, J.; Ferreira, A. Ror1-Ror2 Complexes Modulate Synapse Formation in Hippocampal Neurons. Neuroscience 2010, 165, 1261–1274.
- Zhang, H.; Qiu, J.; Ye, C.; Yang, D.; Gao, L.; Su, Y.; Tang, X.; Xu, N.; Zhang, D.; Xiong, L.; et al. ROR1 Expression Correlated with Poor Clinical Outcome in Human Ovarian Cancer. Sci. Rep. 2014, 4, 5811.
- Zhang, S.; Cui, B.; Lai, H.; Liu, G.; Ghia, E.M.; Widhopf, G.F.; Zhang, Z.; Wu, C.C.N.; Chen, L.; Wu, R.; et al. Ovarian Cancer Stem Cells Express ROR1, Which Can Be Targeted for Anti–Cancer-Stem-Cell Therapy. Proc. Natl. Acad. Sci. USA 2014, 111, 17266–17271.
- Henry, C.; Hacker, N.; Ford, C. Silencing ROR1 and ROR2 Inhibits Invasion and Adhesion in an Organotypic Model of Ovarian Cancer Metastasis. Oncotarget 2017, 8, 112727–112738.
- Rodriguez-Torres, M.; Allan, A.L. Aldehyde Dehydrogenase as a Marker and Functional Mediator of Metastasis in Solid Tumors. Clin. Exp. Metastas 2016, 33, 97–113.
- Marcato, P.; Dean, C.A.; Giacomantonio, C.A.; Lee, P.W.K. Aldehyde Dehydrogenase: Its Role as a Cancer Stem Cell Marker Comes down to the Specific Isoform. Cell Cycle 2011, 10, 1378–1384.
- Muralikrishnan, V.; Fang, F.; Given, T.C.; Podicheti, R.; Chchterbinine, M.; Sriramkumar, S.; O’Hagan, H.M.; Hurley, T.D.; Nephew, K.P. A Novel ALDH1A1 Inhibitor Blocks Platinum-Induced Senescence and Stemness in Ovarian Cancer. Biorxiv 2022, 14, 3437.
- Sharbatoghli, M.; Shamshiripour, P.; Fattahi, F.; Kalantari, E.; Shams, Z.H.; Panahi, M.; Totonchi, M.; Asadi-Lari, Z.; Madjd, Z.; Zanjani, L.S. Co-Expression of Cancer Stem Cell Markers, SALL4/ALDH1A1, Is Associated with Tumor Aggressiveness and Poor Survival in Patients with Serous Ovarian Carcinoma. J. Ovarian Res. 2022, 15, 17.
- Sterzyńska, K.; Klejewski, A.; Wojtowicz, K.; Świerczewska, M.; Nowacka, M.; Kaźmierczak, D.; Andrzejewska, M.; Rusek, D.; Brązert, M.; Brązert, J.; et al. Mutual Expression of ALDH1A1, LOX, and Collagens in Ovarian Cancer Cell Lines as Combined CSCs- and ECM-Related Models of Drug Resistance Development. Int. J. Mol. Sci. 2018, 20, 54.
- Landen, C.N.; Goodman, B.; Katre, A.A.; Steg, A.D.; Nick, A.M.; Stone, R.L.; Miller, L.D.; Mejia, P.V.; Jennings, N.B.; Gershenson, D.M.; et al. Targeting Aldehyde Dehydrogenase Cancer Stem Cells in Ovarian Cancer. Mol. Cancer Ther. 2010, 9, 3186–3199.
- Meng, E.; Mitra, A.; Tripathi, K.; Finan, M.A.; Scalici, J.; McClellan, S.; da Silva, L.M.; Reed, E.; Shevde, L.A.; Palle, K.; et al. ALDH1A1 Maintains Ovarian Cancer Stem Cell-Like Properties by Altered Regulation of Cell Cycle Checkpoint and DNA Repair Network Signaling. PLoS ONE 2014, 9, e107142.
- Jiang, Y.-X.; Siu, M.K.-Y.; Wang, J.-J.; Mo, X.-T.; Leung, T.H.-Y.; Chan, D.W.; Cheung, A.N.-Y.; Ngan, H.Y.-S.; Chan, K.K.-L. Ascites-Derived ALDH+CD44+ Tumour Cell Subsets Endow Stemness, Metastasis and Metabolic Switch via PDK4-Mediated STAT3/AKT/NF-ΚB/IL-8 Signalling in Ovarian Cancer. Br. J. Cancer 2020, 123, 275–287.
- Cui, T.; Srivastava, A.K.; Han, C.; Wu, D.; Wani, N.; Liu, L.; Gao, Z.; Qu, M.; Zou, N.; Zhang, X.; et al. DDB2 Represses Ovarian Cancer Cell Dedifferentiation by Suppressing ALDH1A1. Cell Death Dis. 2018, 9, 561.
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676.
- Huang, J.; Chen, T.; Liu, X.; Jiang, J.; Li, J.; Li, D.; Liu, X.S.; Li, W.; Kang, J.; Pei, G. More Synergetic Cooperation of Yamanaka Factors in Induced Pluripotent Stem Cells than in Embryonic Stem Cells. Cell Res. 2009, 19, 1127–1138.
- Wang, H.F.; He, H.X. Regulation of Yamanaka Factors during H5N1 Virus Infection in A549 Cells and HEK293T Cells. Biotechnol. Biotechnol. Equip. 2018, 32, 1548–1557.
- Liu, X.; Huang, J.; Chen, T.; Wang, Y.; Xin, S.; Li, J.; Pei, G.; Kang, J. Yamanaka Factors Critically Regulate the Developmental Signaling Network in Mouse Embryonic Stem Cells. Cell Res. 2008, 18, 1177–1189.
- Plath, K.; Lowry, W.E. Progress in Understanding Reprogramming to the Induced Pluripotent State. Nat. Rev. Genet. 2011, 12, 253–265.
- Wen, Y.; Hou, Y.; Huang, Z.; Cai, J.; Wang, Z. SOX2 Is Required to Maintain Cancer Stem Cells in Ovarian Cancer. Cancer Sci. 2017, 108, 719–731.
- Wang, X.; Ji, X.; Chen, J.; Yan, D.; Zhang, Z.; Wang, Q.; Xi, X.; Feng, Y. SOX2 Enhances the Migration and Invasion of Ovarian Cancer Cells via Src Kinase. PLoS ONE 2014, 9, e99594.
- Wang, J.; Ma, X.; Jones, H.M.; Chan, L.L.-Y.; Song, F.; Zhang, W.; Bae-Jump, V.L.; Zhou, C. Evaluation of the Antitumor Effects of C-Myc-Max Heterodimerization Inhibitor 100258-F4 in Ovarian Cancer Cells. J. Transl. Med. 2014, 12, 226.
- Islam, S.; Dutta, P.; Sahay, O.; Gopalakrishnan, K.; Muhury, S.R.; Parameshwar, P.; Shetty, P.; Santra, M.K. Feedback-regulated Transcriptional Repression of FBXO31 by C-Myc Triggers Ovarian Cancer Tumorigenesis. Int. J. Cancer 2022, 150, 1512–1524.
- Dimova, I.; Raitcheva, S.; Dimitrov, R.; Doganov, N.; Toncheva, D. Correlations between C-Myc Gene Copy-Number and Clinicopathological Parameters of Ovarian Tumours. Eur. J. Cancer 2006, 42, 674–679.
- Liu, L.; Zhang, J.; Fang, C.; Zhang, Z.; Feng, Y.; Xi, X. OCT4 Mediates FSH-Induced Epithelial–Mesenchymal Transition and Invasion through the ERK1/2 Signaling Pathway in Epithelial Ovarian Cancer. Biochem. Biophys. Res. Commun. 2015, 461, 525–532.
- Wu, D.; Xie, W.; Wang, H.; Chen, W.; Chen, X.; Sun, H. OCT4 Promotes Ovarian Cancer Cell Metastasis and Angiogenesis via Modulating VEGFR2/LRPPRC Pathway. Preprint 2021.
- Chen, Z.; Wang, Y.; Liu, W.; Zhao, G.; Lee, S.; Balogh, A.; Zou, Y.; Guo, Y.; Zhang, Z.; Gu, W.; et al. Doxycycline Inducible Kruppel-Like Factor 4 Lentiviral Vector Mediates Mesenchymal to Epithelial Transition in Ovarian Cancer Cells. PLoS ONE 2014, 9, e105331.
- Wang, B.; Shen, A.; Ouyang, X.; Zhao, G.; Du, Z.; Huo, W.; Zhang, T.; Wang, Y.; Yang, C.; Dong, P.; et al. KLF4 Expression Enhances the Efficacy of Chemotherapy Drugs in Ovarian Cancer Cells. Biochem. Biophys. Res. Commun. 2017, 484, 486–492.
- Mazzoldi, E.L.; Pastò, A.; Pilotto, G.; Minuzzo, S.; Piga, I.; Palumbo, P.; Carella, M.; Frezzini, S.; Nicoletto, M.O.; Amadori, A.; et al. Comparison of the Genomic Profile of Cancer Stem Cells and Their Non-Stem Counterpart: The Case of Ovarian Cancer. J. Clin. Med. 2020, 9, 368.
- Hoogstraat, M.; de Pagter, M.S.; Cirkel, G.A.; van Roosmalen, M.J.; Harkins, T.T.; Duran, K.; Kreeftmeijer, J.; Renkens, I.; Witteveen, P.O.; Lee, C.C.; et al. Genomic and Transcriptomic Plasticity in Treatment-Naïve Ovarian Cancer. Genome Res. 2014, 24, 200–211.
- van de Haar, J.; Hoes, L.R.; Roepman, P.; Lolkema, M.P.; Verheul, H.M.W.; Gelderblom, H.; de Langen, A.J.; Smit, E.F.; Cuppen, E.; Wessels, L.F.A.; et al. Limited Evolution of the Actionable Metastatic Cancer Genome under Therapeutic Pressure. Nat. Med. 2021, 27, 1553–1563.
- Zucconi, F. Epigenetic regulation in plants. Isr. J. Plant Sci. 1988, 37, 131–144.
- Göndör, A. Epigenetic Regulation. Br. J. Surg. 2008, 95, 939–940.
- Klein, C.J.; Benarroch, E.E. Epigenetic Regulation. Neurology 2014, 82, 1833–1840.
- Wu, H.; Sun, Y.E. Epigenetic Regulation of Stem Cell Differentiation. Pediatr. Res. 2006, 59, 21R–25R.
- Seto, E. Histone Modifications. Methods 2003, 31, 1–2.
- Peterson, C.L.; Laniel, M.-A. Histones and Histone Modifications. Curr. Biol. 2004, 14, R546–R551.
- Sang, F. DNA Modifications. Methods Mol. Biol. 2020, 2198, 441–450.
- Tang, F.; Yuan, B.-F. DNA Modification Detection Methods. In DNA Modification Detection Methods; Springer Protocols Handbooks; Humana: New York, NY, USA, 2021; pp. 181–194.
- Lucia, F.D.; Dean, C. Long Non-Coding RNAs and Chromatin Regulation. Curr. Opin. Plant Biol. 2011, 14, 168–173.
- Saxena, A.; Carninci, P. Long Non-coding RNA Modifies Chromatin. Bioessays 2011, 33, 830–839.
- Yamamoto, T.; Saitoh, N. Non-Coding RNAs and Chromatin Domains. Curr. Opin. Cell Biol. 2019, 58, 26–33.
- Efroni, S.; Duttagupta, R.; Cheng, J.; Dehghani, H.; Hoeppner, D.J.; Dash, C.; Bazett-Jones, D.P.; Grice, S.L.; McKay, R.D.G.; Buetow, K.H.; et al. Global Transcription in Pluripotent Embryonic Stem Cells. Cell Stem Cell 2008, 2, 437–447.
- Berry, N.B.; Bapat, S.A. Ovarian Cancer Plasticity and Epigenomics in the Acquisition of a Stem-like Phenotype. J. Ovarian Res. 2008, 1, 8.
- Teschendorff, A.E.; Enver, T. Single-Cell Entropy for Accurate Estimation of Differentiation Potency from a Cell’s Transcriptome. Nat. Commun. 2017, 8, 15599.
- Kannan, S.; Farid, M.; Lin, B.L.; Miyamoto, M.; Kwon, C. Transcriptomic Entropy Benchmarks Stem Cell-Derived Cardiomyocyte Maturation against Endogenous Tissue at Single Cell Level. Biorxiv 2020, 17, e1009305.
- Banerji, C.R.S.; Severini, S.; Caldas, C.; Teschendorff, A.E. Intra-Tumour Signalling Entropy Determines Clinical Outcome in Breast and Lung Cancer. PLoS Comput. Biol. 2015, 11, e1004115.
- Feng, L.; Sun, Y.-D.; Li, C.; Li, Y.-X.; Chen, L.-N.; Zeng, R. Pan-Cancer Network Disorders Revealed by Overall and Local Signaling Entropy. J. Mol. Cell Biol. 2021, 13, 622–635.
- Vaidya, H.; Jeong, H.S.; Keith, K.; Maegawa, S.; Calendo, G.; Madzo, J.; Jelinek, J.; Issa, J.-P.J. DNA Methylation Entropy as a Measure of Stem Cell Replication and Aging. Genome Biol. 2023, 24, 27.
- Son, J.; Shen, S.S.; Margueron, R.; Reinberg, D. Nucleosome-Binding Activities within JARID2 and EZH1 Regulate the Function of PRC2 on Chromatin. Gene Dev. 2013, 27, 2663–2677.
- Liu, N.; Zhu, B. Chapter 10—Regulation of PRC2 Activity. In Polycomb Group Proteins; Pirrotta, V., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 225–258. ISBN 978-0-12-809737-3.
- Wang, W.; Cho, H.; Kim, D.; Park, Y.; Moon, J.H.; Lim, S.J.; Yoon, S.M.; McCane, M.; Aicher, S.A.; Kim, S.; et al. PRC2 Acts as a Critical Timer That Drives Oligodendrocyte Fate over Astrocyte Identity by Repressing the Notch Pathway. Cell Rep. 2020, 32, 108147.
- Zong, X.; Wang, W.; Ozes, A.; Fang, F.; Sandusky, G.E.; Nephew, K.P. EZH2-Mediated Downregulation of the Tumor Suppressor DAB2IP Maintains Ovarian Cancer Stem Cells. Cancer Res. 2020, 80, 4371–4385.
- Baba, T.; Convery, P.A.; Matsumura, N.; Whitaker, R.S.; Kondoh, E.; Perry, T.; Huang, Z.; Bentley, R.C.; Mori, S.; Fujii, S.; et al. Epigenetic Regulation of CD133 and Tumorigenicity of CD133+ Ovarian Cancer Cells. Oncogene 2009, 28, 209–218.
- Ramadoss, S.; Sen, S.; Ramachandran, I.; Roy, S.; Chaudhuri, G.; Farias-Eisner, R. Lysine-Specific Demethylase KDM3A Regulates Ovarian Cancer Stemness and Chemoresistance. Oncogene 2017, 36, 1537–1545.
- Lamouille, S.; Subramanyam, D.; Blelloch, R.; Derynck, R. Regulation of Epithelial–Mesenchymal and Mesenchymal–Epithelial Transitions by MicroRNAs. Curr. Opin. Cell Biol. 2013, 25, 200–207.
- Banyard, J.; Bielenberg, D.R. The Role of EMT and MET in Cancer Dissemination. Connect. Tissue Res. 2015, 56, 403–413.
- Hei, B.; Wang, J.; Wu, G.; Ouyang, J.; Liu, R. Verbascoside Suppresses the Migration and Invasion of Human Glioblastoma Cells via Targeting C-Met-Mediated Epithelial-Mesenchymal Transition. Biochem. Biophys. Res. Commun. 2019, 514, 1270–1277.
- Tang, H.; Massi, D.; Hemmings, B.A.; Mandalà, M.; Hu, Z.; Wicki, A.; Xue, G. AKT-Ions with a TWIST between EMT and MET. Oncotarget 2016, 7, 62767–62777.
- Loret, N.; Denys, H.; Tummers, P.; Berx, G. The Role of Epithelial-to-Mesenchymal Plasticity in Ovarian Cancer Progression and Therapy Resistance. Cancers 2019, 11, 838.
- Cao, L.; Shao, M.; Schilder, J.; Guise, T.; Mohammad, K.S.; Matei, D. Tissue Transglutaminase Links TGF-β, Epithelial to Mesenchymal Transition and a Stem Cell Phenotype in Ovarian Cancer. Oncogene 2012, 31, 2521–2534.
- Teeuwssen, M.; Fodde, R. Wnt Signaling in Ovarian Cancer Stemness, EMT, and Therapy Resistance. J. Clin. Med. 2019, 8, 1658.
- Deng, J.; Bai, X.; Feng, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Inhibition of PI3K/Akt/MTOR Signaling Pathway Alleviates Ovarian Cancer Chemoresistance through Reversing Epithelial-Mesenchymal Transition and Decreasing Cancer Stem Cell Marker Expression. BMC Cancer 2019, 19, 618.
- Cao, P.; Zhao, S.; Sun, Z.; Jiang, N.; Shang, Y.; Wang, Y.; Gu, J.; Li, S. BRMS1L Suppresses Ovarian Cancer Metastasis via Inhibition of the β-Catenin-Wnt Pathway. Exp. Cell Res. 2018, 371, 214–221.
- Liu, Y.; Li, W.; Luo, J.; Wu, Y.; Xu, Y.; Chen, T.; Zhang, W.; Fu, F. Cysteine-Rich Intestinal Protein 1 Served as an Epithelial Ovarian Cancer Marker via Promoting Wnt/β-Catenin-Mediated EMT and Tumour Metastasis. Dis. Markers 2021, 2021, 3566749.
- Bocci, F.; Zhou, P.; Nie, Q. Single-Cell RNA-Seq Analysis Reveals the Acquisition of Cancer Stem Cell Traits and Increase of Cell–Cell Signaling during EMT Progression. Cancers 2021, 13, 5726.
- Li, Y.; Gong, W.; Ma, X.; Sun, X.; Jiang, H.; Chen, T. Smad7 Maintains Epithelial Phenotype of Ovarian Cancer Stem-like Cells and Supports Tumor Colonization by Mesenchymal-Epithelial Transition. Mol. Med. Rep. 2015, 11, 309–316.
- Jia, D.; George, J.T.; Tripathi, S.C.; Kundnani, D.L.; Lu, M.; Hanash, S.M.; Onuchic, J.N.; Jolly, M.K.; Levine, H. Testing the Gene Expression Classification of the EMT Spectrum. Biorxiv 2018, 16, 452508.
- Loh, K.M.; Lim, B. Actors in the Cell Reprogramming Drama. Nature 2012, 488, 599–600.
- Lin, S.L. Mechanism of MicroRNA-Mediated Global DNA Demethylation in Human iPS Cells. In Advances in Stem Cell Research. Stem Cell Biology and Regenerative Medicine; Baharvand, H., Aghdami, N., Eds.; Humana Press: Totowa, NJ, USA, 2012.
- Niimi, P.; Levine, M.; Meer, M. Epigenetic Trajectories of Aging and Reprogramming. Innov. Aging 2021, 5, 664–665.
- Chen, G.-Q.; Ye, P.; Ling, R.-S.; Zeng, F.; Zhu, X.-S.; Chen, L.; Huang, Y.; Xu, L.; Xie, X.-Y. Histone Demethylase KDM4C Is Required for Ovarian Cancer Stem Cell Maintenance. Stem Cells Int. 2020, 2020, 8860185.
- Fatma, H.; Siddique, H.R. Pluripotency Inducing Yamanaka Factors: Role in Stemness and Chemoresistance of Liver Cancer. Expert. Rev. Anticancer 2021, 21, 853–864.
- Yeung, T.-L.; Leung, C.S.; Yip, K.-P.; Yeung, C.L.A.; Wong, S.T.C.; Mok, S.C. Cellular and Molecular Processes in Ovarian Cancer Metastasis. A Review in the Theme: Cell and Molecular Processes in Cancer Metastasis. Am. J. Physiol.-Cell Physiol. 2015, 309, C444–C456.
- Psaila, B.; Lyden, D. The Metastatic Niche: Adapting the Foreign Soil. Nat. Rev. Cancer 2009, 9, 285–293.
- Liu, J.; Geng, X.; Li, Y. Milky Spots: Omental Functional Units and Hotbeds for Peritoneal Cancer Metastasis. Tumor Biol. 2016, 37, 5715–5726.
- Galon, J.; Mlecnik, B.; Bindea, G.; Angell, H.K.; Berger, A.; Lagorce, C.; Lugli, A.; Zlobec, I.; Hartmann, A.; Bifulco, C.; et al. Towards the Introduction of the ‘Immunoscore’ in the Classification of Malignant Tumours. J. Pathol. 2014, 232, 199–209.
- Zhang, M.; Shi, M.; Yu, Y.; Sang, J.; Wang, H.; Shi, J.; Duan, P.; Ge, R. The Immune Subtypes and Landscape of Advanced-Stage Ovarian Cancer. Vaccines 2022, 10, 1451.
- Sato, E.; Olson, S.H.; Ahn, J.; Bundy, B.; Nishikawa, H.; Qian, F.; Jungbluth, A.A.; Frosina, D.; Gnjatic, S.; Ambrosone, C.; et al. Intraepithelial CD8+ Tumor-Infiltrating Lymphocytes and a High CD8+/Regulatory T Cell Ratio Are Associated with Favorable Prognosis in Ovarian Cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 18538–18543.
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The Immunobiology of Cancer Immunosurveillance and Immunoediting. Immunity 2004, 21, 137–148.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
638
Revisions:
2 times
(View History)
Update Date:
22 Dec 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




