Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Xiaoyue Pan | -- | 2543 | 2023-12-21 16:36:01 | | | |
| 2 | Camila Xu | Meta information modification | 2543 | 2023-12-22 03:45:31 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lee, C.H.; Murrell, C.E.; Chu, A.; Pan, X. Circadian Regulation of Apolipoproteins in the Brain. Encyclopedia. Available online: https://encyclopedia.pub/entry/53041 (accessed on 07 February 2026).
Lee CH, Murrell CE, Chu A, Pan X. Circadian Regulation of Apolipoproteins in the Brain. Encyclopedia. Available at: https://encyclopedia.pub/entry/53041. Accessed February 07, 2026.
Lee, Chaeeun Hannah, Charlotte Ellzabeth Murrell, Alexander Chu, Xiaoyue Pan. "Circadian Regulation of Apolipoproteins in the Brain" Encyclopedia, https://encyclopedia.pub/entry/53041 (accessed February 07, 2026).
Lee, C.H., Murrell, C.E., Chu, A., & Pan, X. (2023, December 21). Circadian Regulation of Apolipoproteins in the Brain. In Encyclopedia. https://encyclopedia.pub/entry/53041
Lee, Chaeeun Hannah, et al. "Circadian Regulation of Apolipoproteins in the Brain." Encyclopedia. Web. 21 December, 2023.
Copy Citation
The circadian rhythm is a 24 h internal clock within the body that regulates various factors, including sleep, body temperature, and hormone secretion. Circadian rhythm disruption is an important risk factor for many diseases including neurodegenerative illnesses. The central and peripheral oscillators’ circadian clock network controls the circadian rhythm in mammals. The clock genes govern the central clock in the suprachiasmatic nucleus (SCN) of the brain.
lipid
apolipoprotein
circadian clock
hormone
neuron
brain
1. Introduction
Sleep is crucial for good health, whereas sleep deprivation affects learning, memory, attention, and physical health. Many factors, including sleep apnea, restless leg syndrome, insomnia, jet lag, shift work, and perturbation of the body’s internal clocks, can cause sleep disorders. Sleep disorders and sleep deficiency are strongly linked to a growing number of health problems, including heart disease, stroke, high blood pressure, type 2 diabetes, obesity, depression, and cancer.
The circadian rhythm is a 24 h cycle in which the body adjusts physically and behaviorally to the light and dark phases throughout the day. When the eye receives light from the surrounding environment, the body controls various factors, including sleep/wake cycles, eating habits, body temperature, hormone release, heart activity, and blood pressure [1][2][3]. However, disruptions to the circadian rhythm commonly occur; examples include clock gene mutations, medication use, stress, aging, irregular work schedules, changes in time zones, and vision impairment [4]. Frequent disruptions to the internal clock are a risk factor for multiple diseases and disorders, including cancer, sleeping disorders (e.g., jet lag), atherosclerosis, obesity, diabetes, viral infections (e.g., COVID-19), and neurodegenerative diseases (e.g., Huntington’s disease and Alzheimer’s disease) [5][6][7][8][9][10][11][12].
Many factors, such as cell phones, microwave radiation, temperature, and food availability have been found to regulate human brain diseases, as recently reviewed elsewhere [13][14][15]. Animal and human studies have indicated that the circadian rhythm system regulates lipid metabolism, including the diurnal rhythm of lipid absorption, storage, and transport [12][16][17][18][19][20][21][22][23][24][25][26][27][28][29]. The mechanisms through which disruption of the circadian rhythm and circadian clock affects the rhythmic expression of lipid metabolism-associated apolipoprotein genes in various brain regions remain unclear.
To understand the association between circadian clock genes and apolipoproteins, this research focuses on the regulation and dysregulation of the circadian clock and apolipoproteins in the brain.
Apolipoproteins, major constituents of lipoproteins, function as lipid transporters by binding lipids and facilitating their secretion into the blood, lymph, and cerebrospinal fluid (CSF). Apolipoprotein synthesis is affected by various factors, such as age and sex, through mechanisms that vary by tissue type. Clinical studies have shown that plasma levels of most apolipoproteins, except apolipoprotein (Apo)H, are higher in women than in men [30]. Regarding age, women show significant negative relationships with plasma levels of ApoB, ApoE, ApoH, and ApoJ from young to mid-life, whereas links at older ages are insignificant or positively related. Plasma levels of apolipoproteins including ApoA1, ApoA2, ApoB, ApoC3, and ApoH are negatively associated with age in middle-aged and old men; however, in men, plasma levels of ApoE and ApoJ show U-shaped curves with the period from young-aged to middle-aged, reacting to higher levels of ApoE and ApoJ in plasma at the oldest ages in men [30].
Many physiological functions and disease pathologies have been ascribed to apolipoproteins. For example, researchers' animal studies have shown that the circadian clock regulates the ApoB-containing lipoprotein pathway, thereby controlling lipid absorption and plasma lipids, in clock circadian regulator (Clock)-mutant mice [31][32]. Global or liver-specific basic helix-loop-helix ARNT like 1 (Bmal1)-deficient mice show elevated assembly and secretion of an ApoB-containing very low-density lipoprotein (VLDL), thus regulating the development of atherosclerosis [26]. In addition, it have been identified that ApoA-IV is targeted primarily by Bmal1, which down-regulates nuclear receptor subfamily 1 group D member 1 (Nr1d1), thereby controlling VLDL lipoprotein assembly and secretion in the liver via cyclic adenosine monophosphate (cAMP)-responsive element-binding protein H [33]. This process directly regulates ApoA4 expression in the liver and small intestine and consequently increases VLDL lipoprotein particle sizes in the blood circulation [33]. ApoA4, a crucial player in the circadian regulation of intestinal lipid absorption, mediates lipoprotein production [34]. A high concentration of total cholesterol in the serum in middle age is a risk factor for Alzheimer’s disease (AD) and other types of dementia in old age. Recently, studies have suggested a strong association between AD and cardiovascular disease (CVD) risk factors such as high-density lipoprotein (HDL) levels, low-density lipoprotein (LDL) levels, hypertension, and atherosclerosis [35]. Studies have shown that cholesterol-lowering agents such as statins decrease the incidence of AD [36][37], although the mechanism remains unknown.
In mammalian peripheral tissues, many genes associated with lipid biosynthesis and metabolism are rhythmically controlled by circadian clock genes [6][38][39][40], thereby suggesting that dysfunction in clock genes may be associated with abnormal lipid metabolism and impaired lipid absorption. For example, Nocturne (gene name Ccrn4l) plays a critical role in dealing with dietary lipids in the small intestinal enterocytes and, by extension, efficient absorption of lipids [19]; Nocturne−/− mice have diminished chylomicron/VLDL triglyceride and cholesterol levels [19]. Several studies have indicated that circadian clock regulation is necessary for normal physiology and pathology mechanisms.
The circadian clock may significantly regulate cerebral lipid metabolism and help prevent apolipoprotein-associated neurodegenerative diseases, such as AD [18][41][42][43][44][45]. Several apolipoproteins have been directly implicated in the etiopathology of AD, including APOE, clusterin (CLU or APOJ), APOC1, and APOB [46][47][48][49].
2. Function of SCN
2.1. SCN
The brain SCN is a minor area within the brain’s hypothalamus. One unilateral SCN of the brain contains approximately 10,000 neurons in two anatomic subdivisions. The SCN is the primary input and output location for light, as well as neuronal and hormonal activities [50][51][52]. The SCN includes many cell types, and many neurons contain multiple neuropeptides, neurotransmitters, and input or output connections. Wen et al. have identified eight primary cell types, each with a specific mode of circadian gene expression, according to single-cell RNA-sequencing [53]. Interestingly, several studies have shown that astrocytes execute the circadian rhythm in the SCN [54][55][56][57]. Studies have suggested that the SCN is the most critical region for sleep, mood, control of circadian timing, and non-neural circadian gene expression and regulation. Moreover, in mammals, removal of the SCN ablates the circadian rhythm [58][59][60], thus suggesting that the SCN is the primary center for circadian regulation. In addition, individual neurons of the SCN produce individualistic circadian oscillations of circadian clock gene expression and nerve reflex on the basis of ex vivo culture.
2.2. Neurons and Hormones in the SCN
Several peptide hormones, such as vasopressin (AVP), vasoactive intestinal peptide (VIP), peptide histidine-isoleucine, and neurotransmitters, are found in the SCN [61]. In the mouse, VIP is found in 10% of all SCN cells, whereas AVP neurons are found in 20% of all SCN cells [62]. AVP regulates blood pressure [63][64], whereas VIP governs the release of various bodily chemicals, such as gastric acid, during digestion [65]. Peptide histidine-isoleucine regulates food consumption behavior [66]. These hormones are essential in controlling energy and glucose homeostasis. The circadian rhythms of secretion of AVP and VIP have been found to be affected by gap junction (cell–cell junction) blockers in rat SCN slice cultures [67]. Because the removal of these gap junction blockers from cultures results in the restoration of the circadian rhythms of AVP and VIP, these hormones may be regulated by cell–cell interactions in the SCN, such as neuron-astrocyte cross-talk [67]. The central circadian clock may regulate physiological and pathological functions in peripheral tissues through hormones and peripheral tissue cross-talk.
Sex hormone receptors, such as estrogen receptor-α (ERα), estrogen receptor-β (ERβ), progesterone receptors, and androgen receptor (AR), are expressed in the SCN [68]. The SCN regulates the circadian rhythms of sex hormone release; conversely, sex hormone feedback influences SCN function. Several groups have reported that sex hormone receptors are associated with SCN function and the circadian clock in ERα-deficient and circadian clock gene knockout mice [69][70]. The difference in the SCN function between females and males has been suggested to be due to ERα expression, which is statistically significantly higher in the SCN in female than male animals [69]. Pre-astroglia (astrocytes and ependymal glia) have a sex-specific role in the organization of the SCN but not the sexually dimorphic area, and neuroglia have also been found to have a sex-specific role in the mature SCN in Mongolian gerbils [71].
Glucagon-like peptide 1 (GLP-1) is produced primarily in the intestine but is also found in the brain, particularly in the nucleus tractus solitarius, which is situated in the brainstem [72]. In addition, gene expression of GLP-1 is found in hypothalamus neurons, particularly in the paraventricular and arcuate nucleus [73]. The GLP-1 receptor is also expressed in the central and peripheral nervous systems. The FDA has recently approved synthetic GLP-1 receptor agonists for treating type 2 diabetes mellitus and weight loss [74][75]. Several studies in animal models have shown that GLP-1 receptor agonists may ameliorate motor and cognitive symptoms in Parkinson’s disease (PD) or AD [61], thus slowing the development of neurodegeneration.
2.3. Immune Factors in the SCN
The SCN has immense biological importance, functioning as the brain’s internal clock and guiding sleep patterns and ovulation cycles, as well as other regulatory functions. In the SCN, glial fibrillary acidic protein (GFAP), a specific marker for astrocytes, is present, and the levels of blood GFAP can be used to predict early-stage AD [76]. The glial cells of the SCN regulate the signals entering the circadian system by receiving them from the immune system via NF-kappaB signaling, according to studies in animal models [76][77]. Interleukin-1β (IL-1β) and interleukin-1 type 1 receptor (IL-1R1) are expressed in the SCN in young and old mice: IL-1β is more highly expressed in younger mice, whereas IL-1R1 shows rhythmic expression in only younger mice. The rhythmic phrases of both IL-1β and IL-1R1 in the paraventricular nucleus of the brain are affected by age in mice [78]. After treatment of mice with lipopolysaccharide (LPS; 5 mg/kg) to stimulate an immune response, only the expression of IL-1R1 increases after 6 or 24 h [78]. These data suggest that the levels of IL-1β and IL-1R1 in the mouse SCN are dependent on the diurnal clock, age, and the immune system. In animal models, LPS induces interferon-γ and tumor necrosis factor-α (TNF-α), thereby decreasing excitatory SCN activity [79]. Immune system dysfunction disrupts SCN function and can result in destructive sleep patterns. Thus, alterations in the synaptic mechanisms of SCN neurons might have integral roles in sleep disorders, and proinflammatory cytokines participate in these changes.
3. Circadian Rhythm of Circadian Clock Genes in the SCN
The SCN controls the central circadian clock, whereas peripheral clocks are located in every tissue and organ system of the body. In the brain, the two nuclei are located near the hypothalamus, and approximately 10,000 neurons hold a spectrum of changes occurring within the body activity throughout the day. Numerous circadian clock genes control the circadian rhythm, including Clock, Bmal1, period circadian regulators (Per1, Per2, and Per3), and cryptochrome circadian regulators (Cry1 and Cry2). The Bmal1 and Clock genes encode proteins that form a heterodimer. The heterodimer of Clock and Bmal1 binds sites called E-box enhancers and subsequently increases transcription upstream of the Per1, Per2, Per3, Cry1, and Cry2 genes. After these genes are transcribed, the Per and Cry proteins heterodimerize, thereby preventing their self-transcription through interaction with the Bmal1:Clock/neuronal PAS domain protein 2 (Npas2) complex [80].
Several studies have reported that the circadian rhythm in humans is associated with neurodegenerative diseases; however, the mechanisms are unknown [81][82]. Kress et al. have shown that mice with peripheral Bmal1 deficiency have elevated ApoE in the brain parenchyma, which regulates the fibrillar plaque deposition [83]. Recently, Lee et al. have determined that Rev-erbα deficiency in microglia enhances lipid accumulation in the microglia and influences inflammatory signaling in male mice [28][84], thus suggesting that the circadian clock gene Rev-erbα might serve as a treatment target for AD.
Using the SCNseq database, it was found that circadian rhythm-associated circadian clock genes are expressed in the SCN in male mice. Figure 1 shows a similar expression of all seven circadian clock genes, but the highest expression for Cry2. All identified circadian clock-associated genes show rhythmic expression over a 24 h cycle. The shaded parts of the graph represent the dark phases of the 24 h cycle. Similar patterns are shown in Figure 2. Bmal1 (Arntl), Per1, Per2, Per3, and Cry2 all peak during the daytime and have the lowest expression in the nighttime. Furthermore, because expression levels were measured in mice, nocturnal behavior must also be considered: mice have an inactive phase during the daytime and an active phase during the nighttime.
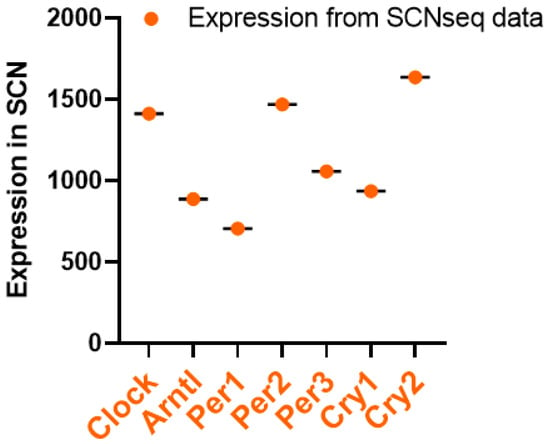
Figure 1. Circadian clock pathway gene expression levels in the SCN; data retrieved from SCNseq. Gene information, including expression levels, was obtained through the SCNseq database. The expression levels in the SCN were organized, and highly or weakly expressed genes were analyzed in GraphPad Prism 9.
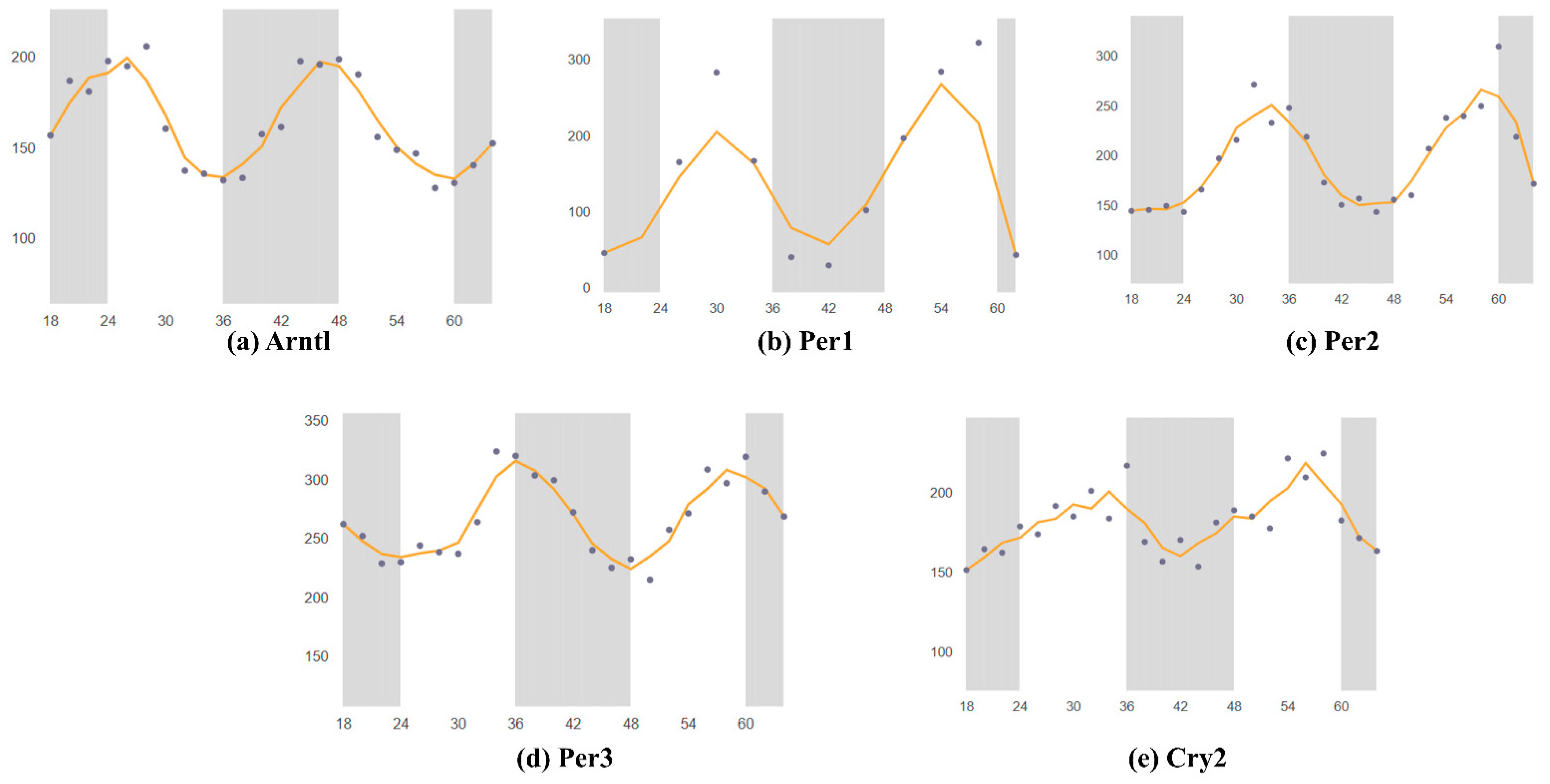
Figure 2. Mouse gene expression levels in the brain over two 24 h periods; data retrieved from CircaDB. The gray bar is dark period, dots is shown the different time gene expression through 66 h. Horizontal is over two 24 h periods 0–66 h. the wave pattern shown the circadian rhythm of the different time point data. (a) Arntl (Bmal1) expression in the brain stem; Mouse 1. OST Brain Stem (Affymetrix). (b) Per1 expression in the SCN; Mouse SCN MAS4 Panda 2002 (Affymetrix). (c) Per2 expression in the SCN; Mouse 1. OST SCN 2014 (Affymetrix). (d) Per3 expression in the brain stem; Mouse 1. OST (Affymetrix). (e) Cry2 expression in the SCN; Mouse 1. OST SCN 2014 (Affymetrix). Through the same database as in Figure 1, which determined whether each gene is rhythmically expressed over 24 h. The graphs are divided into two sections: light and dark, representing daytime and nighttime, respectively. The graphs were compared to determine when the genes were most highly expressed throughout the day. Similar patterns were identified in circadian rhythms. Past studies on Mus musculus brain tissue were collected through the Circadian Expression Profiles Database (CircaDB), compared with the graphs from SCNseq, and verified for consistency. The graphs from CircaDB also show the available data on the circadian rhythmic expression of the genes. Similar to the graphs from SCNseq, light and night phases are shown in the CircaDB graphs. High and low peaks were identified. All collected data from Affymetrix assays were searched, thus enabling analysis of gene expression levels.
As described above, the circadian cock regulates the immune system, which in turn plays a major role in the expression of circadian clock genes in the SCN. Adults with prenatal LPS exposure have elevated anxiety-like behavior along with altered rhythms of circadian clock genes in the SCN [85]: the most significant effect is on Nr1d1 expression, whereas Per2 is the least affected. However, the developing SCN also shows adaptive flexibility to immune challenges in the early stages of development. In the same study, whereas Nr1d1 expression in the LPS-treated animal group was arrhythmic at postnatal day 3, it showed improvements, on the basis of a higher amplitude, by postnatal day 20 [85], thus suggesting the circadian clock may assist the immune system in dealing with challenges later in life.
Deletion of Bmal1 in the SCN of the mouse brain leads to amyloid-β buildup within the brain. Moreover, disruption of the circadian rhythm of Bmal1 locally in the brain parenchyma increases the expression of ApoE, and fibrillar plaques begin to accumulate [86][87][88], thus suggesting that the circadian clock has an essential role in regulating amyloid-β and ApoE dynamics and pathology. AD is characterized by plaque accumulation in the brain, including amyloid-β buildup, and changes in the sleep cycle are symptoms of AD.
References
- Roenneberg, T.; Merrow, M. The Circadian Clock and Human Health. Curr. Biol. 2016, 26, R432–R443.
- Green, C.B.; Takahashi, J.S.; Bass, J. The meter of metabolism. Cell 2008, 134, 728–742.
- Bass, J.T. The circadian clock system’s influence in health and disease. Genome Med. 2017, 9, 94.
- Lananna, B.V.; Musiek, E.S. The wrinkling of time: Aging, inflammation, oxidative stress, and the circadian clock in neurodegeneration. Neurobiol. Dis. 2020, 139, 104832.
- Prasai, M.J.; George, J.T.; Scott, E.M. Molecular clocks, type 2 diabetes and cardiovascular disease. Diab Vasc. Dis. Res. 2008, 5, 89–95.
- Maury, E.; Ramsey, K.M.; Bass, J. Circadian rhythms and metabolic syndrome: From experimental genetics to human disease. Circ. Res. 2010, 106, 447–462.
- Firsov, D.; Bonny, O. Circadian rhythms and the kidney. Nat. Rev. Nephrol. 2018, 14, 626–635.
- Pan, X.; Mota, S.; Zhang, B. Circadian Clock Regulation on Lipid Metabolism and Metabolic Diseases. Adv. Exp. Med. Biol. 2020, 1276, 53–66.
- Nassan, M.; Videnovic, A. Circadian rhythms in neurodegenerative disorders. Nat. Rev. Neurol. 2022, 18, 7–24.
- Zhang, D.; Pollock, D.M. Circadian regulation of kidney function: Finding a role for Bmal1. Am. J. Physiol. Renal Physiol. 2018, 314, F675–F678.
- Levi, F.; Schibler, U. Circadian rhythms: Mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 593–628.
- Gooley, J.J. Circadian regulation of lipid metabolism. Proc. Nutr. Soc. 2016, 75, 440–450.
- Mumtaz, S.; Rana, J.N.; Choi, E.H.; Han, I. Microwave Radiation and the Brain: Mechanisms, Current Status, and Future Prospects. Int. J. Mol. Sci. 2022, 23, 9288.
- Wardzinski, E.K.; Jauch-Chara, K.; Haars, S.; Melchert, U.H.; Scholand-Engler, H.G.; Oltmanns, K.M. Mobile Phone Radiation Deflects Brain Energy Homeostasis and Prompts Human Food Ingestion. Nutrients 2022, 14, 339.
- Inskip, P.D.; Hoover, R.N.; Devesa, S.S. Brain cancer incidence trends in relation to cellular telephone use in the United States. Neuro Oncol. 2010, 12, 1147–1151.
- Shimba, S.; Ishii, N.; Ohta, Y.; Ohno, T.; Watabe, Y.; Hayashi, M.; Wada, T.; Aoyagi, T.; Tezuka, M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 12071–12076.
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045.
- Yang, G.; Zhang, J.; Jiang, T.; Monslow, J.; Tang, S.Y.; Todd, L.; Pure, E.; Chen, L.; FitzGerald, G.A. Bmal1 Deletion in Myeloid Cells Attenuates Atherosclerotic Lesion Development and Restrains Abdominal Aortic Aneurysm Formation in Hyperlipidemic Mice. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 1523–1532.
- Douris, N.; Kojima, S.; Pan, X.; Lerch-Gaggl, A.F.; Duong, S.Q.; Hussain, M.M.; Green, C.B. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr. Biol. 2011, 21, 1347–1355.
- Pan, X.; Hussain, M.M. Clock is important for food and circadian regulation of macronutrient absorption in mice. J. Lipid Res. 2009, 50, 1800–1813.
- Gomez-Santos, C.; Gomez-Abellan, P.; Madrid, J.A.; Hernandez-Morante, J.J.; Lujan, J.A.; Ordovas, J.M.; Garaulet, M. Circadian rhythm of clock genes in human adipose explants. Obesity 2009, 17, 1481–1485.
- Pan, X.; Hussain, M.M. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J. Biol. Chem. 2007, 282, 24707–24719.
- Chua, E.C.; Shui, G.; Lee, I.T.; Lau, P.; Tan, L.C.; Yeo, S.C.; Lam, B.D.; Bulchand, S.; Summers, S.A.; Puvanendran, K.; et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 14468–14473.
- Brown, S.A. Circadian Metabolism: From Mechanisms to Metabolomics and Medicine. Trends Endocrinol. Metab. 2016, 27, 415–426.
- Hussain, M.M.; Pan, X. Circadian regulators of intestinal lipid absorption. J. Lipid Res. 2015, 56, 761–770.
- Pan, X.; Bradfield, C.A.; Hussain, M.M. Global and hepatocyte-specific ablation of Bmal1 induces hyperlipidaemia and enhances atherosclerosis. Nat. Commun. 2016, 7, 13011.
- Cheng, B.; Anea, C.B.; Yao, L.; Chen, F.; Patel, V.; Merloiu, A.; Pati, P.; Caldwell, R.W.; Fulton, D.J.; Rudic, R.D. Tissue-intrinsic dysfunction of circadian clock confers transplant arteriosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 17147–17152.
- Lee, J.; Dimitry, J.M.; Song, J.H.; Son, M.; Sheehan, P.W.; King, M.W.; Travis Tabor, G.; Goo, Y.A.; Lazar, M.A.; Petrucelli, L.; et al. Microglial REV-ERBalpha regulates inflammation and lipid droplet formation to drive tauopathy in male mice. Nat. Commun. 2023, 14, 5197, Correction to Nat. Commun. 2023, 14, 7760.
- Bass, J.; Lazar, M.A. Circadian time signatures of fitness and disease. Science 2016, 354, 994–999.
- Muenchhoff, J.; Song, F.; Poljak, A.; Crawford, J.D.; Mather, K.A.; Kochan, N.A.; Yang, Z.; Trollor, J.N.; Reppermund, S.; Maston, K.; et al. Plasma apolipoproteins and physical and cognitive health in very old individuals. Neurobiol. Aging 2017, 55, 49–60.
- Pan, X.; Zhang, Y.; Wang, L.; Hussain, M.M. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010, 12, 174–186.
- Pan, X.; Jiang, X.C.; Hussain, M.M. Impaired cholesterol metabolism and enhanced atherosclerosis in clock mutant mice. Circulation 2013, 128, 1758–1769.
- Pan, X.; Hussain, M.M. Bmal1 regulates production of larger lipoproteins by modulating cAMP-responsive element-binding protein H and apolipoprotein AIV. Hepatology 2022, 76, 78–93.
- Pan, X.; Munshi, M.K.; Iqbal, J.; Queiroz, J.; Sirwi, A.A.; Shah, S.; Younus, A.; Hussain, M.M. Circadian regulation of intestinal lipid absorption by apolipoprotein AIV involves forkhead transcription factors A2 and O1 and microsomal triglyceride transfer protein. J. Biol. Chem. 2013, 288, 20464–20476.
- Martins, I.J.; Hone, E.; Foster, J.K.; Sunram-Lea, S.I.; Gnjec, A.; Fuller, S.J.; Nolan, D.; Gandy, S.E.; Martins, R.N. Apolipoprotein E, cholesterol metabolism, diabetes, and the convergence of risk factors for Alzheimer’s disease and cardiovascular disease. Mol. Psychiatry 2006, 11, 721–736.
- Dufouil, C.; Richard, F.; Fievet, N.; Dartigues, J.F.; Ritchie, K.; Tzourio, C.; Amouyel, P.; Alperovitch, A. APOE genotype, cholesterol level, lipid-lowering treatment, and dementia: The Three-City Study. Neurology 2005, 64, 1531–1538.
- Sparks, D.L.; Sabbagh, M.N.; Connor, D.J.; Lopez, J.; Launer, L.J.; Browne, P.; Wasser, D.; Johnson-Traver, S.; Lochhead, J.; Ziolwolski, C. Atorvastatin for the treatment of mild to moderate Alzheimer disease: Preliminary results. Arch. Neurol. 2005, 62, 753–757.
- Reppert, S.M.; Weaver, D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001, 63, 647–676.
- Hussain, M.M.; Pan, X. Clock genes, intestinal transport and plasma lipid homeostasis. Trends Endocrinol. Metab. 2009, 20, 177–185.
- Froy, O. The circadian clock and metabolism. Clin. Sci. 2011, 120, 65–72.
- Schnell, A.; Chappuis, S.; Schmutz, I.; Brai, E.; Ripperger, J.A.; Schaad, O.; Welzl, H.; Descombes, P.; Alberi, L.; Albrecht, U. The nuclear receptor REV-ERBalpha regulates Fabp7 and modulates adult hippocampal neurogenesis. PLoS ONE 2014, 9, e99883.
- Landgraf, D.; Long, J.E.; Proulx, C.D.; Barandas, R.; Malinow, R.; Welsh, D.K. Genetic Disruption of Circadian Rhythms in the Suprachiasmatic Nucleus Causes Helplessness, Behavioral Despair, and Anxiety-like Behavior in Mice. Biol. Psychiatry 2016, 80, 827–835.
- Wolff, S.E.C.; Wang, X.L.; Jiao, H.; Sun, J.; Kalsbeek, A.; Yi, C.X.; Gao, Y. The Effect of Rev-erbalpha Agonist SR9011 on the Immune Response and Cell Metabolism of Microglia. Front. Immunol. 2020, 11, 550145.
- McKee, C.A.; Lee, J.; Cai, Y.; Saito, T.; Saido, T.; Musiek, E.S. Astrocytes deficient in circadian clock gene Bmal1 show enhanced activation responses to amyloid-beta pathology without changing plaque burden. Sci. Rep. 2022, 12, 1796.
- Lee, J.; Kim, D.E.; Griffin, P.; Sheehan, P.W.; Kim, D.H.; Musiek, E.S.; Yoon, S.Y. Inhibition of REV-ERBs stimulates microglial amyloid-beta clearance and reduces amyloid plaque deposition in the 5XFAD mouse model of Alzheimer’s disease. Aging Cell 2020, 19, e13078.
- Hofman, A.; Ott, A.; Breteler, M.M.; Bots, M.L.; Slooter, A.J.; van Harskamp, F.; van Duijn, C.N.; Van Broeckhoven, C.; Grobbee, D.E. Atherosclerosis, apolipoprotein E, and prevalence of dementia and Alzheimer’s disease in the Rotterdam Study. Lancet 1997, 349, 151–154.
- Wingo, T.S.; Cutler, D.J.; Wingo, A.P.; Le, N.A.; Rabinovici, G.D.; Miller, B.L.; Lah, J.J.; Levey, A.I. Association of Early-Onset Alzheimer Disease With Elevated Low-Density Lipoprotein Cholesterol Levels and Rare Genetic Coding Variants of APOB. JAMA Neurol. 2019, 76, 809–817.
- Marchant, N.L.; Reed, B.R.; Sanossian, N.; Madison, C.M.; Kriger, S.; Dhada, R.; Mack, W.J.; DeCarli, C.; Weiner, M.W.; Mungas, D.M.; et al. The aging brain and cognition: Contribution of vascular injury and abeta to mild cognitive dysfunction. JAMA Neurol. 2013, 70, 488–495.
- Power, M.C.; Rawlings, A.; Sharrett, A.R.; Bandeen-Roche, K.; Coresh, J.; Ballantyne, C.M.; Pokharel, Y.; Michos, E.D.; Penman, A.; Alonso, A.; et al. Association of midlife lipids with 20-year cognitive change: A cohort study. Alzheimers Dement. 2018, 14, 167–177.
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179.
- Earnest, D.J.; Olschowka, J.A. Circadian regulation of c-fos expression in the suprachiasmatic pacemaker by light. J. Biol. Rhythms 1993, 8, S65–S71.
- Brancaccio, M.; Enoki, R.; Mazuski, C.N.; Jones, J.; Evans, J.A.; Azzi, A. Network-mediated encoding of circadian time: The suprachiasmatic nucleus (SCN) from genes to neurons to circuits, and back. J. Neurosci. 2014, 34, 15192–15199.
- Wen, S.; Ma, D.; Zhao, M.; Xie, L.; Wu, Q.; Gou, L.; Zhu, C.; Fan, Y.; Wang, H.; Yan, J. Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat. Neurosci. 2020, 23, 456–467.
- Barca-Mayo, O.; Pons-Espinal, M.; Follert, P.; Armirotti, A.; Berdondini, L.; De Pietri Tonelli, D. Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat. Commun. 2017, 8, 14336.
- Tso, C.F.; Simon, T.; Greenlaw, A.C.; Puri, T.; Mieda, M.; Herzog, E.D. Astrocytes Regulate Daily Rhythms in the Suprachiasmatic Nucleus and Behavior. Curr. Biol. 2017, 27, 1055–1061.
- Brancaccio, M.; Patton, A.P.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron 2017, 93, 1420–1435.e1425.
- Brancaccio, M.; Edwards, M.D.; Patton, A.P.; Smyllie, N.J.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 2019, 363, 187–192.
- Granados-Fuentes, D.; Prolo, L.M.; Abraham, U.; Herzog, E.D. The suprachiasmatic nucleus entrains, but does not sustain, circadian rhythmicity in the olfactory bulb. J. Neurosci. 2004, 24, 615–619.
- Ralph, M.R.; Foster, R.G.; Davis, F.C.; Menaker, M. Transplanted suprachiasmatic nucleus determines circadian period. Science 1990, 247, 975–978.
- Lehman, M.N.; Silver, R.; Gladstone, W.R.; Kahn, R.M.; Gibson, M.; Bittman, E.L. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J. Neurosci. 1987, 7, 1626–1638.
- Harmar, A.J.; Marston, H.M.; Shen, S.; Spratt, C.; West, K.M.; Sheward, W.J.; Morrison, C.F.; Dorin, J.R.; Piggins, H.D.; Reubi, J.C.; et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 2002, 109, 497–508.
- Welsh, D.K.; Takahashi, J.S.; Kay, S.A. Suprachiasmatic nucleus: Cell autonomy and network properties. Annu. Rev. Physiol. 2010, 72, 551–577.
- Pittman, Q.J. Vasopressin and central control of the cardiovascular system: A 40-year retrospective. J. Neuroendocrinol. 2021, 33, e13011.
- Smith, M.C.; Dunn, M.J. The role of prostaglandins in human hypertension. Am. J. Kidney Dis. 1985, 5, A32–A39.
- Reghunandanan, V. Vasopressin in circadian function of SCN. J. Biosci. 2020, 45, 140.
- Olszewski, P.K.; Wirth, M.M.; Shaw, T.J.; Grace, M.K.; Levine, A.S. Peptides that regulate food intake: Effect of peptide histidine isoleucine on consummatory behavior in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 284, R1445–R1453.
- Shinohara, K.; Funabashi, T.; Mitushima, D.; Kimura, F. Effects of gap junction blocker on vasopressin and vasoactive intestinal polypeptide rhythms in the rat suprachiasmatic nucleus in vitro. Neurosci. Res. 2000, 38, 43–47.
- Kruijver, F.P.; Swaab, D.F. Sex hormone receptors are present in the human suprachiasmatic nucleus. Neuroendocrinology 2002, 75, 296–305.
- Bailey, M.; Silver, R. Sex differences in circadian timing systems: Implications for disease. Front. Neuroendocrinol. 2014, 35, 111–139.
- Alvord, V.M.; Kantra, E.J.; Pendergast, J.S. Estrogens and the circadian system. Semin. Cell Dev. Biol. 2022, 126, 56–65.
- O’Brien, E.R.; Howarth, C.; Sibson, N.R. The role of astrocytes in CNS tumors: Pre-clinical models and novel imaging approaches. Front. Cell. Neurosci. 2013, 7, 40.
- Brierley, D.I.; Holt, M.K.; Singh, A.; de Araujo, A.; McDougle, M.; Vergara, M.; Afaghani, M.H.; Lee, S.J.; Scott, K.; Maske, C.; et al. Central and peripheral GLP-1 systems independently suppress eating. Nat. Metab. 2021, 3, 258–273.
- Rowlands, J.; Heng, J.; Newsholme, P.; Carlessi, R. Pleiotropic Effects of GLP-1 and Analogs on Cell Signaling, Metabolism, and Function. Front. Endocrinol. 2018, 9, 672.
- Nauck, M.A.; Meier, J.J. GLP-1 receptor agonists and SGLT2 inhibitors: A couple at last? Lancet Diabetes Endocrinol. 2016, 4, 963–964.
- Hussein, H.; Zaccardi, F.; Khunti, K.; Davies, M.J.; Patsko, E.; Dhalwani, N.N.; Kloecker, D.E.; Ioannidou, E.; Gray, L.J. Efficacy and tolerability of sodium-glucose co-transporter-2 inhibitors and glucagon-like peptide-1 receptor agonists: A systematic review and network meta-analysis. Diabetes Obes. Metab. 2020, 22, 1035–1046.
- Leone, M.J.; Marpegan, L.; Bekinschtein, T.A.; Costas, M.A.; Golombek, D.A. Suprachiasmatic astrocytes as an interface for immune-circadian signalling. J. Neurosci. Res. 2006, 84, 1521–1527.
- Hainich, E.C.; Pizzio, G.A.; Golombek, D.A. Constitutive activation of the ERK-MAPK pathway in the suprachiasmatic nuclei inhibits circadian resetting. FEBS Lett. 2006, 580, 6665–6668.
- Liu, X.; Quan, N. Microglia and CNS Interleukin-1: Beyond Immunological Concepts. Front. Neurol. 2018, 9, 8.
- Lundkvist, G.B.; Hill, R.H.; Kristensson, K. Disruption of circadian rhythms in synaptic activity of the suprachiasmatic nuclei by African trypanosomes and cytokines. Neurobiol. Dis. 2002, 11, 20–27.
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998, 280, 1564–1569.
- Leng, Y.; Musiek, E.S.; Hu, K.; Cappuccio, F.P.; Yaffe, K. Association between circadian rhythms and neurodegenerative diseases. Lancet Neurol. 2019, 18, 307–318.
- Musiek, E.S.; Holtzman, D.M. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science 2016, 354, 1004–1008.
- Kress, G.J.; Liao, F.; Dimitry, J.; Cedeno, M.R.; FitzGerald, G.A.; Holtzman, D.M.; Musiek, E.S. Regulation of amyloid-beta dynamics and pathology by the circadian clock. J. Exp. Med. 2018, 215, 1059–1068.
- Sheehan, P.W.; Nadarajah, C.J.; Kanan, M.F.; Patterson, J.N.; Novotny, B.; Lawrence, J.H.; King, M.W.; Brase, L.; Inman, C.E.; Yuede, C.M.; et al. An astrocyte BMAL1-BAG3 axis protects against alpha-synuclein and tau pathology. Neuron 2023, 111, 2383–2398.e7.
- Spisska, V.; Pacesova, D.; Mikova, H.; Pohanova, P.; Telensky, P.; Novotny, J.; Bendova, Z. Prenatal exposure to lipopolysaccharide induces changes in the circadian clock in the SCN and AA-NAT activity in the pineal gland. Brain Res. 2020, 1743, 146952.
- Huo, M.; Huang, Y.; Qu, D.; Zhang, H.; Wong, W.T.; Chawla, A.; Huang, Y.; Tian, X.Y. Myeloid Bmal1 deletion increases monocyte recruitment and worsens atherosclerosis. FASEB J. 2017, 31, 1097–1106.
- Ma, Z.; Jiang, W.; Zhang, E.E. Orexin signaling regulates both the hippocampal clock and the circadian oscillation of Alzheimer’s disease-risk genes. Sci. Rep. 2016, 6, 36035.
- Chen, Q.; Peng, X.D.; Huang, C.Q.; Hu, X.Y.; Zhang, X.M. Association between ARNTL (BMAL1) rs2278749 polymorphism T >C and susceptibility to Alzheimer disease in a Chinese population. Genet. Mol. Res. 2015, 14, 18515–18522.
More
Information
Subjects:
Physics, Fluids & Plasmas
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
543
Revisions:
2 times
(View History)
Update Date:
22 Dec 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




