Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Feijun Luo | -- | 3869 | 2023-12-17 09:26:54 | | | |
| 2 | Lindsay Dong | -4 word(s) | 3865 | 2023-12-18 06:41:53 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lu, H.; Xie, T.; Wu, Q.; Hu, Z.; Luo, Y.; Luo, F. Alpha-Glucosidase Inhibitory Peptides. Encyclopedia. Available online: https://encyclopedia.pub/entry/52835 (accessed on 07 February 2026).
Lu H, Xie T, Wu Q, Hu Z, Luo Y, Luo F. Alpha-Glucosidase Inhibitory Peptides. Encyclopedia. Available at: https://encyclopedia.pub/entry/52835. Accessed February 07, 2026.
Lu, Han, Tiantian Xie, Qi Wu, Zuomin Hu, Yi Luo, Feijun Luo. "Alpha-Glucosidase Inhibitory Peptides" Encyclopedia, https://encyclopedia.pub/entry/52835 (accessed February 07, 2026).
Lu, H., Xie, T., Wu, Q., Hu, Z., Luo, Y., & Luo, F. (2023, December 17). Alpha-Glucosidase Inhibitory Peptides. In Encyclopedia. https://encyclopedia.pub/entry/52835
Lu, Han, et al. "Alpha-Glucosidase Inhibitory Peptides." Encyclopedia. Web. 17 December, 2023.
Copy Citation
With the change in people’s lifestyle, diabetes has emerged as a chronic disease that poses a serious threat to human health, alongside tumor, cardiovascular, and cerebrovascular diseases. α-glucosidase inhibitors, which are oral drugs, have proven effective in preventing and managing this disease. Studies have suggested that bioactive peptides could serve as a potential source of α-glucosidase inhibitors. These peptides possess certain hypoglycemic activity and can effectively regulate postprandial blood glucose levels by inhibiting α-glucosidase activity, thus intervening and regulating diabetes.
α-glucosidase inhibitory peptide
source
mechanism
bioavailability
1. Introduction
According to the International Diabetes Federation, as of 2021, about 537 million people worldwide have diabetes, with a prevalence of more than 10% [1]. By 2045, the number of diabetes patients in China will reach an unprecedented 174 million, ranking first in the world. Diabetes can lead to severe injury, dysfunction, and failure of multiple organs and tissues, with progressive metabolic complications, such as neuropathy, retinopathy [2][3], kidney disease [4], and cardiovascular disease [5]. There are three types of diabetes: type 1 diabetes, type 2 diabetes, and gestational diabetes. About 80% of diabetes is type 2 diabetes and their pharmacological treatments focus on increasing insulin action or inhibiting carbohydrate digestive enzymes such as glycosidase inhibitors, thereby reducing blood glucose concentration [6][7]. Therefore, the control of blood glucose is a key step in the managements and treatments of diabetes.
Glycosidases are enzymes that play an important role in glucose metabolism. They can hydrolyze glucoside bonds and release monosaccharides. Glucosidase are the most important enzymes in the glycosidase family and their main functions are to hydrolyze glucosidase bonds and release glucose. Glucosidase can be divided into α-glucosidase and β-glucosidase according to the break mode of the glycoside bond. α-Glucosidase (E.C.3.2.1.20) includes maltase, isomaltase, glucoamylase, and sucrose and can bind to the membrane of small intestinal epithelial cells [8]. The function of α-glucosidase is to break the α-glucoside bond and release α-glucose from the non-reducing end of oligosaccharides. The α-amylase secreted by the salivary glands and pancreas can hydrolyze carbohydrates from food into oligosaccharides, which are then broken down into absorbable glucose by α-glucosidase in the small intestine [9]. Glucose is absorbed into the bloodstream by epithelial cells in the upper small intestine, resulting in a postprandial glucose elevation. α-Glucosidase inhibitors can competitively inhibit the activity of small intestinal α-glucosidase and delay or inhibit the absorption of glucose in the small intestine, thereby effectively reducing blood glucose concentrations [10]. Recent studies have found that active peptides have many biological functions [11][12][13][14][15][16]. Some active peptides can obviously inhibit the activity of α-glucosidase; it suggests that α-glucosidase inhibitory peptide has potential application in the prevention and treatment of type 2 diabetes.
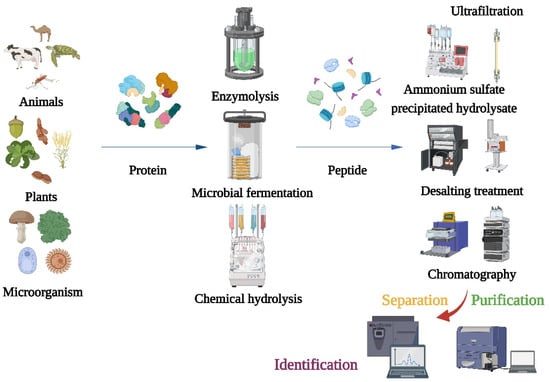
2. Production, Purification, and Characterization of Bioactive Peptides
2.1. Conventional Approach
The main preparation methods of α-glucosidase inhibitory peptides include enzymatic hydrolysis, microbial fermentation, and chemical methods [17][18]. Chemical synthesis can synthesize peptide chains with specific structures and sequences, but the cost is high in mass production and it is easy to cause environmental pollution [19]. The microbial fermentation method utilizes microbial metabolism-produced enzymes to hydrolyze the proteins in the fermented raw materials. It has the advantages of high hydrolytic activity and low cost and does not need to separate and purify the enzyme. The disadvantage is that the reaction process is not easy to control and is susceptible to microbial contamination.
After producing α-glucosidase inhibitory peptides, different separation and purification techniques should be adopted to obtain the purified products (Figure 1). Purifications of bioactive peptides are the prerequisite for identifying the structures of peptides and detecting their activities. The purifications of peptides are mainly based on their molecular size, charge, polarity, solubility, and other physical chemistry properties [20]. Meanwhile, the combination of multidimensional chromatography and various separation methods can obtain better separation results. The first step of purification is usually precipitation of the hydrolysate with ammonium sulfate, followed by desalting. Because α-glucosidase inhibitory peptides are usually small, the hydrolyzed products are typically truncated to 1 kDa, 3 kDa, 5 kDa, and 10 kDa for membrane filtration [21][22].

Figure 1. Flow chart of traditional method for preparation, isolation, purification, and identification of α-glucosidase inhibitory peptide.
Traditional methods have many problems in the analytical process: (1) Some peptides have very low levels in the mixture or interact with other components, which make their isolations and purifications very difficult; (2) Some highly active peptides may not be detected using conventional methods, especially those that target novel structures or mechanisms; (3) More experimental steps may be required, which will consume time and resources and, thus, increase the cost of investigation, especially when peptides are isolated and characterized on a large scale; (4) When large numbers of peptides need to be isolated from complex protein mixtures, traditional methods may not be appropriate, and there may be efficiency and scalability issues when purifying peptides on a large scale. To overcome these problems, new technologies and methods have emerged in recent years [23][24][25][26][27].
2.2. Bioinformatics-Assisted Methods
Computer research is also known as “computer methods” or “bioinformatics”. Bioactive peptides are studied using databases and bioinformatics tools. It accelerates the process of discovery and design of α-glucosidase inhibitory peptides by predicting peptide structure, function, and interactions. These predictions are derived and analyzed by computers from existing experimental data. Bioinformatics technology plays an important role in the study of α-glucosidase inhibitory peptides and has great potential to further improve the current state of research. An increasing number of databases are being created and used and bioinformatics tools allow for a range of studies [28]: (1) A large amount of bioactive peptide structural data was collected in the structural database, which can analyze the relationship between structural patterns and functional features of peptides. These analytical results are helpful to understand the similarities and differences among different bioactive peptides, reveal the relationship between structures and activities, and investigate the optimization of their the structures to improve their activities and stabilities. (2) Using structural information from the bioactive peptide database can help researchers design new bioactive peptide molecules using computational simulations. This simulation technique focuses on optimizing peptide design and synthesis strategies by assessing the stabili
 ties or affinities of peptides to their targeting sites. (3) The bioactive peptide database collects a large number of known bioactive peptides and their related data. By utilizing methods such as machine learning and data mining, prediction models can be built to identify and predict potentially bioactive peptide molecules. This prediction can help researchers quickly screen and evaluate the activity of candidate peptides among a large number of peptide samples, thus improving the preparation efficiency and success rate. The application of bioinformatics in the field of peptides provides a great opportunity for the study of α-glucosidase inhibitory peptide.
ties or affinities of peptides to their targeting sites. (3) The bioactive peptide database collects a large number of known bioactive peptides and their related data. By utilizing methods such as machine learning and data mining, prediction models can be built to identify and predict potentially bioactive peptide molecules. This prediction can help researchers quickly screen and evaluate the activity of candidate peptides among a large number of peptide samples, thus improving the preparation efficiency and success rate. The application of bioinformatics in the field of peptides provides a great opportunity for the study of α-glucosidase inhibitory peptide.
3. Source and Structural Characteristics of α-Glucosidase Inhibitory Peptide
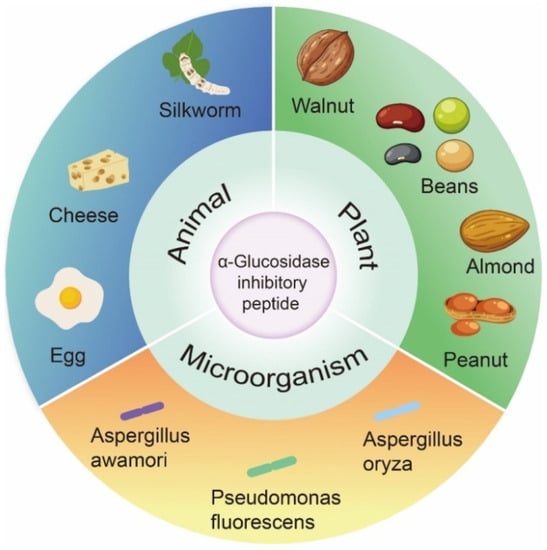
In past studies, the focus of α-glucosidase inhibition peptides has shifted to identification and characterization, which are mainly derived from the partial hydrolysis of proteins from plants, animals, and microorganisms (Figure 2).

Figure 2. Origin of α-glucosidase inhibitory peptides.
There are several steps involved in assessing or predicting the role of α-glucosidase inhibitory peptides using bioinformatics. The first step requires a comparison using data from two databases (protein and peptide sequences), where protein databases include the Protein database, RCSB Protein Data Bank, ExPASy, UniProtKB, etc. The bioactive peptide databases include BIOPEP, Peptide Ranker, Peptidecutter, EnzymePredictor, and so on. Potential sources of bioactive peptides can be obtained by evaluating protein sequences and protein sources of peptides with known sequence peptides. Peptidecutter can perform computer simulations of enzymatic hydrolysis, which predicts where specific substances will cleave a protein sequence under known conditions. Computer simulation of enzymatic hydrolysis can greatly improve the efficiency of active peptide screening, optimize peptide sequence design based on simulation results, and select more promising candidate peptides.
3.1. Animal Origin
The proteins in dairy products have high nutritional value. They also contain a significant amount of animal-derived peptides and are considered to be a high-quality source of bioactive peptides. Milk-derived proteins are a precursor of bioactive peptides with various physiological functions. The main protein in milk is casein, which is around 80% of the total amount. Casein is structurally diverse with many sites exposed so that many proteases break down these proteins to form functional bioactive peptides. Numerous studies have shown that milk hydrolysates such as camel milk, cow milk, and goat milk have significant inhibitory effects on α-glucosidase. Casein hydrolysates extracted from bovine and camel milk were found to have a strong inhibitory effect on α-glucosidase, with IC50 values of 1.04 mg/mL and 0.59 mg/mL, respectively; LPTGWLM, MFE, and GPAHCLL were the most effective α-glucosidase inhibitory peptides [29].
Notably, some peptides composed of amino acids such as Gly, Ser, Glu, Tyr, Arg, Phe, and Pro are considered to be α-glucosidase inhibitors. Lee et al. [30] identified the tripeptides GEY and GYG with α-glucosidase inhibitory activities (IC50 of 2.70 and 1.50 mg/mL). Zhang et al. [31] used the quantitative structure–activity relationship screening method and silkworm protein database to identify α-glucosidase inhibitory peptides. Four peptides that have α-glucosidase inhibition effects were obtained and they were QPGR (IC50 of 65.80 mol/L), SQSPA (IC50 of 20.00 mol/L), QPPT (IC50 of 560.0 mol/L), and NSPR (IC50 of 205.00 mol/L). QPGR could form hydrogen bonds with Lys776 in the active site ofα-glucosidase and SQSPA had potential interactions with Arg520, Lys519, and Asp777 of α-glucosidase. Compared with QPGR, QPPT and NSPR only form a single bond with Lys776 of α-glucosidase. In the three-dimensional structure, Lys776 is located at the edge of the α-glucosidase activity pocket and may be a key target for these peptides to inhibit α-glucosidase activity [31]. SEDSSEVDIDLGN was a new peptide derived from sericin, and it had been found to non-competitively bind α-glucosidase by hydrogen bonding forces or van der Waals forces. The binding might be dominated by the Asn and Ser side chains which were contained in SEDSSEVDIDLGN [32][33]. In addition, CSSV (IC50 = 206.00 μg/mL), SAAP (IC50 = 66.90 μg/mL), PGGP (IC50 = 63.50 μg/mL), LGGGN (IC50 = 42.93 μg/mL), YSFR (IC50 = 162.00 μg/mL) from the protein of Giant Salamander, and GPPGPA from skin collagen hydrolysates of Giant Salamander contained higher contents of Gly and Pro [34][35]. This might be why these peptides have a strong inhibitory effect on α-glucosidase. Peptide sequences were identified from edible insects, KVEGDLK, YETGNGIK, AIGVGAIR, IIAPPER, and FDPFPK exhibited the highest enzyme inhibitory activity [36]. The only peptide containing phenylalanine FDPFPK was identified as the most powerful α-glucosidase inhibitor (IC50 = 5.95 μg/mL). These results suggest that some peptides composed of specific amino acids can be used as α-glucosidase inhibitors.
3.2. Plant Origin
Plants have always been an important source of drug development due to their wide distribution, rich resources, effectiveness, and high safety. Moreover, plant secondary metabolites also can be widely used in pharmaceuticals and other fields. Many researchers have screened α-glucosidase inhibitors from plants and they are a rich source of α-glucosidase inhibitory peptides [37][38][39].
α-Glucosidase inhibitory peptides from various protein-rich plant hydrolysates, such as grains, legumes, and seeds, have been extensively studied. Enzymatic hydrolysis increased the α-glucosidase inhibitory activity of protein hydrolysates [40]. Different proteases have different restriction cutting sites, peptides with different molecular weights, amino acid sequences, and biological activities can be released by cutting peptide bonds at different parts [41]. Commonly used enzymes include alkaline protease, acid protease, pepsin, trypsin, neutral protease, bromelain, flavor protease, etc. Alkaline protease is an endonuclease with a wide range of specificity. It preferentially cleaves the C-terminal peptide bonds of hydrophobic amino acid residues such as Try, Phe, Leu, Ile, Val, Met, etc. This gave it more catalytic sites to cleave peptide bonds in proteins, thus allowing deeper enzymatic cleavage [42]. A longer enzymatic digestion time would result in a higher degree of hydrolysis value of the hydrolyzed product [43]. Trypsin only cleats the C-terminus into Arg and Lys. Flavourzyme are exopeptidases that break the N-terminus of the peptide chain; papain prefers to cleaved peptide bonds between carboxylic acid groups of Lys or Arg and adjacent amino acid residues [44]. Compared with other hydrolysates, the plant protein peptides hydrolyzed by alkaline protease showed the highest α-glucosidase inhibition. Therefore, alkaline proteases has been used to hydrolyze many plant proteins, such as soy protein, Luffa cylindrical (L.) M. Roem seed, and Ginkgo biloba seed [45][46][47]. For better comparison, previous studies used different commercial enzymes for the enzymatic preparation of α-glucosidase inhibitory peptides under the same conditions. Soybean protein peptides prepared using alkaline protease had the highest α-glucosidase inhibitory activity where compared to those prepared using papain and trypsin. Three new α-glucosidase inhibitory peptides LLPLPVLK, SWLRL, and WLRL were found with IC50 values of 237.43 ± 0.52 μmol/Lol/L, 182.05 ± 0.74 μmol/Lol/L, and 165.29 ± 0.74 μmol/Lol/L [45]. Using different proteases to hydrolyze Luffa cylindrical (L.) M. Roem seed, the alkaline protease hydrolysate showed the strongest inhibition of α-glucoside, followed by tryptic hydrolysate, with concentration-dependent inhibition of α-glucosidase (IC50 of 0.48–0.80 mg/mL) [47]. Ginkgo biloba seed was hydrolyzed using alkaline protease and three new peptides with α-glucosidase inhibitory activity were screened: LSMSFPPF, VPKIPPP, and MPGPPSD. LSMSFPPF showed the strongest inhibitory activity (IC50 of 454.33 ± 32.45 μmol/Lol/L), followed by MPGPPSD (IC50 of 943.82 ± 73.10 μmol/L) and VPKIPPP (IC50 of 1446.81 ± 66.98 μmol/Lol/L) [46].
The candidate peptides were further screened based on molecular weight, amino acid composition, and binding energy to α-glucosidase. Low molecular weight peptides have better stability and higher bioavailability, resulting in better biological function in vivo [48][49][50]. Digestion of soy protein with trypsin to obtain hydrolysates with α-glucosidase inhibitory activity (IC50 of 0.27 mg/mL) showed the highest inhibitory activity corresponding to MW < 5 kDa grade fractions and a yield of 76.08% [51]. Two tripeptides GSR (IC50 of 20.4 μmol/L) and EAK (IC50 of 520.2 μmol/L) were obtained, the inhibitory effect of peptides on α-glucosidase is mainly due to the formation of five strong hydrogen bonds between GSR and His674, Asp518, Arg600, Asp616, and Asp282 in α-glucosidase; four hydrogen bonds were formed between EAK and residues Asp282, Asp518, and Asp616. Asp residues are import targets to inhibit the activity of α-glucosidase [51]. In the easy-to-cook and difficult-to-cook bean hydrolysates, the ultrafiltration fractions with MW < 3 kDa showed the highest inhibition of α-glucosidase (34.4–89.2%) [52]. The subunits of gluten hydrolysates were digested using kiwifruit actinidin. The WGLYH (≤1 kDa) group have the highest inhibitory activities on α-amylase and α-glucosidase [53].
According to previous investigations, the peptides with strong α-glucosidase inhibitory activity are short peptides with relative molecular weights less than 1 kDa; this is because lower-molecular-weight peptides can enter the active site of α-glucosidase and bind to it. [54]. However, VVDLVFFAAAK (MW = 1179.4 Da) from black tea protein also exhibited the best α-glucosidase inhibitory ability compared to peptides with MW < 1 kDa [55]. Therefore, MW < 1 kDa was only used as the first screening condition. On the other hand, amino acid composition also contributes significantly to α-glucosidase inhibitory activity. α-Glucosidase inhibitory peptides are usually accompanied by a high degree of hydrophobicity [29][56]. It suggests that hydrophobic amino acid contents in bioactive peptides are closely related with the inhibition activities of α-glucosidases. Quinoa proteins were subjected to simulated gastrointestinal digestion in vitro. Three peptides were isolated and the peptide IQEGGLT (IC50 of 109.48 μmol/L) containing three hydrophobic residues showed strong inhibitory activity against α-glucosidase. In contrast, at 250 μmol/L, peptides DKDPYPK (22.16 ± 0.61%) and GEHGSDGNV (30.84 ± 0.69%) showed lower inhibition than IQEGGLT (55.85 ± 0.26%), probably due to their higher hydrophilicity [57]. Quinoa proteins contain high amounts of Gln, Glu, Asp, Asn, Arg, Ser, Leu, and Pro [58]. Ujiroghene et al. [59] identified four antidiabetic peptides from germinated quinoa yogurt drink including VAHPVF, LAHMIVAGA, KLTPQMA, and KSFGSSNI. Among these peptides, LAHMIVAGA and VAHPVF showed significant α-glucosidase inhibitory activity with IC50 values of 10.90 mg/mL and 9.00 mg/m.
For the different amino acid composition of peptides, leucine and valine can participate in muscle repair and blood glucose control; proline can improve the hypoglycemic activity of peptide [60]. The peptide GLLGY from rice bran fermentation broth was a non-competitive inhibitor that forms five hydrogen bonds with Asp282, Ser523, Asp616, and His674 of α-glucosidase [61]. GLLGY also maintained excellent α-glucosidase inhibition in the gastrointestinal digestive system [61]. From the structure of α-glucosidase, Asp616 and His674 were the key amino acid residues in the catalytic structural domain, which might be the target of molecular docking of α-glucosidase inhibitors.
The binding energy of plant-derived peptides to α-glucosidase can be predicted by docking. The lower the binding energy required for peptides to bind to α-glucosidase, the easier it is to inhibit the activity of α-glucosidase. Four peptides FYNPAAGR, FFVPPSQQ, PGVLPVAS, and FSYNPQAG were screened from the hydrolysates of hot-pressed peanut meal. Molecular docking indicates that peptides can occupy the active pocket of α-glucosidase through hydrogen bonding, hydrophobic interaction, salt bridges, and π stacking, thus preventing α-glucosidase from forming a complex with substrates. Among these four peptides, PGVLPVAS had the lowest binding energy to α-glucoside, followed by FYNPAAGR, FFVPPSQQ, and FSYNPQAG [62]. Therefore, lower binding energy of peptides than that of acarbose could be used as another screening criterion. The peptide TTGGKGGK (−8.97 kcal/mol) obtained from black bean (Phaseolus vulgaris L.) proteins had a higher α-glucosidase inhibitory potential than acarbose. Black soybean peptides inhibit α-glucosidase through hydrogen bonding, polarity, and hydrophobicity. The main binding sites are Asp34, Thr83, and Asn32 [63]. Among the peptides purified from chickpea (Cicerarietinum L.) protein hydrolyzates, FGKG showed optimal inhibition of α-glucosidase [64]. Since FGKG exhibited the highest inhibition rate and lowest binding energy (−10.047 kcal/mol), the hydrophobic interactions of FGKG with Leu162-Phe1 and Ala198-Phe1 appeared to contribute significantly to the stabilization of the inhibitor–enzyme complex [64].
3.3. Microbial Origin
As mankind’s knowledge of the world deepens, the vast resources contained in microorganisms are being understood. For peptides, microbiota is a high-quality source that is gradually being exploited by mankind. Due to the short life cycle, fast growth, reproduction, and low culture cost of microorganisms, more and more scholars are focusing on obtaining α-glucosidase inhibitory peptides from microorganism.
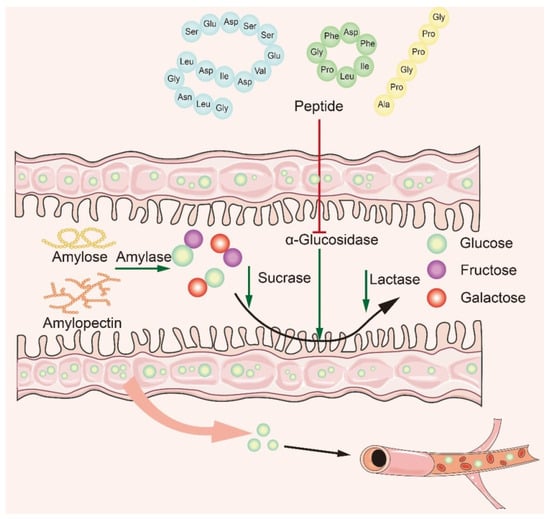
4. Mechanism of α-Glucosidase Inhibitory Peptide
Many studies have shown that naturally active peptides could treat type 2 diabetes by lowering blood sugar, but screening for them is a time-consuming process [65][66]. To discover safer and more effective α-glucosidase inhibitory peptides, it is particularly important to explore the interaction mechanism between α-glucosidase inhibitory peptides and α-glucosidase. Based on recent studies, it is concluded that the main mechanism of action is to inhibit α-glucosidase activity in small intestine mucosa (Figure 3). The bioactive peptide mainly binds to oligosaccharides and enzymes, producing competitive inhibition. The peptides act on the enzyme through the hydrophobic effect and hydrogen bonding van der Waals forces, modulating the amino acids at the catalytic sites of the enzyme to affect its original activity and reduce glucose intake [32][67][68]. In recent years, researchers have summarized the structural features and mechanism of α-glucosidase inhibitory peptides: (1) Low-molecular-weight peptides have better α-glucosidase inhibitory activity [69]; (2) Amino acids contain at least a hydroxyl or basic side chain at the N-terminus, with Pro present in the penultimate position at the C-terminus [8]; (3) The presence of Arg at the C-terminal end improves the stability of the peptide–enzyme binding, ensures strong binding of peptides to key active amino acid residues of the enzyme, and promotes increased inhibitory activity [70]; (4) The higher the content of hydrophobic amino acids, the better the inhibitory activity of α-glucosidase.

Figure 3. Mechanism of α-glucosidase inhibitory peptides.
Hydrogen bond and hydrophobic interaction play an important role in α-glucosidase inhibition. Qiu et al. [71] used papain to hydrolyze soft-shelled turtle (Pelodiscus sinensis) egg to obtain potential anti-diabetic peptides. Furthermore, three oligopeptides, HNKPEVEVR, ARDASVLK, and SGTLLHK, were synthesized, which had strong inhibitory effects on α-glucosidase activity (IC50 = 56,195 and 289 μmol/Lol/L, respectively). Li et al. [72] investigated the action mechanism of ARDASVLK and HNKPEVEVR on α-glucosidase using synchronous fluorescence, circular dichroism spectroscopy, and molecular modeling. The amino acid residues that ARDASVLK binds to α-glucosidase mainly include Pro1159, Arg1250, Tyr1251, Gln1254, Glu1258, Trp1355, Trp1369, Gln1372, Arg1377, Asp1526, Phe1559, Ile1587, Gly1588, and Arg1591. HNKPEVEVR interacted with amino acid residues of α-glucosidase as follows: Arg1156, Tyr1251, Trp1355, Leu1367, Asp1368, Trp1369, Asp1370, Phe1427, Asn1429, Val1432, Ser1452, Arg1455, Ser1459, Lys1460, and Ile1587 [72]. Molecular docking and thermodynamic results demonstrated that α-glucosidase interactions with ARDASVLK and HNKPEVEVR were driven by hydrogen bonding and hydrophobic action [72]. The detection by circular dichroism spectroscopy showed that the content of some secondary structures (α-helix and β-sheet) of HNKPEVEVR were altered. Although the cause of these secondary structure changes was unknown, it was clear for minor conformational changes that blocking active center or preventing substrate binding would inactivate the enzyme. Therefore, ARDASVLLK and HNKPEVEVR, which were obtained from turtle eggs, could burst α-glucosidase by forming α-glucosidase-peptide complexes. In this case, the active vacancy of the enzyme was occupied and the conformation was changed, eventually the enzyme activity was diminished.
The presence of van der Waals forces also affects the activity of α-glucosidase. Three new peptides including LSMSFPPF, VPKIPPPHE, and MPGPPS which had α-glucosidase inhibitory activity were selected from the protein isolates of Ginkgo biloba seed cake. There were 12 effector sites of van der Waals force between LSMSFPPF and α-glucosidase [46]. For these sites, Ala454, Phe455, Ala514, Asp440, Thr517, Thr519, Leu525, Asn447, and Arg520 were on α-glucosidase A chain and Leu93, His91, and Lys89 were on the chain B. In addition, VPKIPPP and α-glucosidase also had 12 effector sites of van der Waals force, including Glu377, Thr339, Leu227, Leu300, Pru602, Gly228, Arg400, Tyr389, Glu396, Asn233, Ala229, and Glu231 on chain A.
For the α-glucosidase inhibitory peptide, it can form hydrogen bonds with the active site of the α-glucosidase which makes the binding more stable. At the same time, the inhibitory peptide interacts with the hydrophobic part of the enzyme to enhance the affinity between the inhibitory peptide and enzyme; the stability of the binding site has also been enhanced. This ultimately prevents the enzyme from binding to oligosaccharides, thereby effectively inhibiting changes in blood glucose. Van der Waals force interactions between α-glucosidase inhibitory peptides and enzymes can affect the binding and dissociation kinetics of inhibitory peptides, thus regulating enzyme activity, and some inhibitory peptides can improve the affinity and selectivity of inhibitory peptides by enhancing van der Waals force interactions with enzymes.
5. Bioavailability of α-Glucosidase Inhibitory Peptides
Systemic delivery of functional peptides via the oral route has been a daunting task. Low bioavailability of peptide was due to denaturation in the digestive tract, susceptibility in enzymatic reaction, and poor permeability in the cell membrane [73]. The bioavailability of peptide components was usually assessed via simulating gastrointestinal digestion. The stability of peptide bioactivity depends not only on structural maintenance but also on other factors [74]. Because the behavior of peptides that mimic the digestive species in the gastrointestinal tract can provide important information about their possible behavior in the digestive system. α-Glucosidase inhibiting peptides could exert hypoglycemic effects in vivo after entering the bloodstream in their active form. Therefore, peptides must withstand hydrolysis by gastrointestinal enzymes and continue to exert biological activity through the intestinal wall after oral administration [57]. Whether the peptide is consumed as a functional food ingredient or as a nutritional product, the ability to resist gastrointestinal digestion or interpretation is very important.
6. Conclusions
Foods not only provide nutrition, but also provide many functional ingredients that are beneficial to health. Food-derived α-glucosidase inhibitory peptides are non-toxic and can inhibit the increase in blood glucose concentration, which shows great potential in the management and treatment of diabetes. It will be helpful to screen α-glucosidase inhibitory peptides more efficiently. Meanwhile, the discoveries of α-glucosidase inhibitory peptides can be accelerated by bioinformatics analysis. As dietary active peptides, it is also necessary to consider whether they will be degraded by digestive enzymes; thus, increasing the stability and bioavailability of α-glucosidase inhibitory peptides are important issues. Especially, will these bioactive peptides affect the gut microbiota and provide additional mechanisms for lowering blood glucose? These issues require further study.
References
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119.
- Dehdashtian, E.; Mehrzadi, S.; Yousefi, B.; Hosseinzadeh, A.; Reiter, R.J.; Safa, M.; Ghaznavi, H.; Naseripour, M. Diabetic retinopathy pathogenesis and the ameliorating effects of melatonin; involvement of autophagy, inflammation and oxidative stress. Life Sci. 2018, 193, 20–33.
- Simó-Servat, O.; Hernández, C.; Simó, R. Diabetic retinopathy in the context of patients with diabetes. Ophthalmic Res. 2019, 62, 211–217.
- Sandholm, N.; Groop, P.H. Genetic basis of diabetic kidney disease and other diabetic complications. Curr. Opin. Genet. Dev. 2018, 50, 17–24.
- Harding, J.L.; Pavkov, M.E.; Magliano, D.J.; Shaw, J.E.; Gregg, E.W. Global trends in diabetes complications: A review of current evidence. Diabetologia 2019, 62, 3–16.
- Gloster, T.M.; Davies, G.J. Glycosidase inhibition: Assessing mimicry of the transition state. Org. Biomol. Chem. 2010, 8, 305–320.
- Chiarelli, F.; Marzio, D.D. Peroxisome proliferator-activated receptor-γ agonists and diabetes: Current evidence and future perspectives. Vasc. Health Risk Man. 2008, 4, 297–304.
- Ibrahim, M.A.; Bester, M.J.; Neitz, A.W.H.; Gaspar, A.R.M. Structural properties of bioactive peptides with α-glucosidase inhibitory activity. Chem. Biol. Drug Des. 2018, 91, 370–379.
- Zhang, B.W.; Li, X.; Sun, W.L.; Xing, Y.; Xiu, Z.L.; Zhuang, C.L.; Dong, Y.S. Dietary flavonoids and acarbose synergistically inhibit α-glucosidase and lower postprandial blood glucose. J. Agric. Food Chem. 2017, 65, 8319–8330.
- Ha, T.J.; Song, S.B.; Ko, J.; Park, C.H.; Ko, J.M.; Choe, M.E.; Kwak, D.Y.; Lee, J.H. Isolation and identification of α-glucosidase inhibitory constituents from the seeds of vigna nakashimae: Enzyme kinetic study with active phytochemical. Food Chem. 2018, 266, 483–489.
- Aslam, M.Z.; Aslam, M.S.; Firdos, S.; Ghous, G.; Firdos, G.; Zhao, H.F.; Zhang, B.L. Role of bioactive peptides in reducing the severity of hypertension with the inhibition of ACE. Int. J. Pept. Res. Ther. 2019, 25, 1639–1649.
- Yan, J.; Zhao, J.G.; Yang, R.J.; Zhao, W. Bioactive peptides with antidiabetic properties: A review. Int. J. Food Sci. Technol. 2019, 54, 1909–1919.
- Wu, Q.; Luo, F.; Wang, X.L.; Lin, Q.; Liu, G.Q. Angiotensin I-converting enzyme inhibitory peptide: An emerging candidate for vascular dysfunction therapy. Crit. Rev. Biotechnol. 2022, 42, 736–755.
- Liu, J.; Wu, Q.; Yang, T.; Yang, F.; Guo, T.; Zhou, Y.; Han, S.; Luo, Y.; Guo, T.; Luo, F.; et al. Bioactive peptide F2d isolated from rice residue exerts antioxidant effects via Nrf2 signaling pathway. Oxid. Med. Cell Longev. 2021, 2021, 2637577.
- Fernández-Tomé, S.; Sanchón, J.; Recio, I.; Hernandez-Ledesma, B. Transepithelial transport of lunasin and derived peptides: Inhibitory effects on the gastrointestinal cancer cells viability. J. Food Compos. Anal. 2018, 68, 101–110.
- Sun, S.; Niu, H.; Yang, T.; Lin, Q.; Luo, F.; Ma, M. Antioxidant and anti-fatigue activities of egg white peptides prepared by pepsin digestion. J. Sci. Food Agric. 2014, 94, 3195–3200.
- Ma, X.M.; Fan, X.; Wang, D.P.; Li, X.A.; Wang, X.Y.; Yang, J.Y.; Qiu, C.G.; Liu, X.L.; Pang, G.X.; Abra, R.; et al. Study on preparation of chickpea peptide and its effect on blood glucose. Front. Nutr. 2022, 9, 988628.
- Zhang, Y.; Chen, Y.; Liu, X.Y.; Wang, W.; Wang, J.H.; Li, X.; Sun, S.L. Preparation and Identification of Peptides with α-Glucosidase Inhibitory Activity from Shiitake Mushroom (Lentinus edodes) Protein. Foods 2023, 12, 2534.
- Espitia, P.J.P.; Soares, N.D.F.; Coimbra, J.S.D.; de Andrade, N.J.; Cruz, R.S.; Medeiros, E.A.A. Bioactive Peptides: Synthesis, Properties, and Applications in the Packaging and Preservation of Food. Compr. Rev. Food Sci. Food Saf. 2012, 11, 187–204.
- Arumugam, V.; Venkatesan, M.; Ramachandran, S.; Sundaresan, U. Bioactive peptides from marine ascidians and future drug development—A review. Int. J. Pept. Res. Ther. 2018, 24, 13–18.
- Huang, F.J.; Wu, T. Purification and characterization of a new peptide (s-8300) from shark liver. J. Food Biochem. 2010, 34, 962–970.
- Olagunju, A.I.; Omoba, O.S.; Enujiugha, V.N.; Alashi, A.M.; Aluko, R.E. Thermoase-hydrolysed pigeon pea protein and its membrane fractions possess in vitro bioactive properties (antioxidative, antihypertensive, and antidiabetic). J. Food Biochem. 2021, 45, e13429.
- Cox, J. Prediction of peptide mass spectral libraries with machine learning. Nat. Biotechnol. 2023, 41, 33–43.
- Neagu, A.N.; Jayathirtha, M.; Baxter, E.; Donnelly, M.; Petre, B.A.; Darie, C.C. Applications of tandem mass spectrometry (MS/MS) in protein analysis for biomedical research. Molecules 2022, 27, 2411.
- Qiu, X.I.; Ruterbories, K.J.; Ji, Q.C.; Jenkins, G.J. Signature peptide selection workflow for biomarker quantification using LC–MS-based targeted proteomics. Bioanalysis 2023, 15, 295–300.
- Yan, Y.Q.; Wang, L.; Wang, H. Functional peptides from one-bead one-compound high-throughput screening technique. Chem. Res. Chin. Univ. 2023, 39, 83–91.
- Guo, H.; Hao, Y.; Yang, X.; Ren, G.; Richel, A. Exploration on bioactive properties of quinoa protein hydrolysate and peptides: A review. Crit. Rev. Food Sci. 2023, 63, 2896–2909.
- Zhang, S.L.; Liu, L.L.; Gao, L.; Zhao, Y.H.; Shi, N.; Wang, H.B.; Xu, H.Y. Bioinformatics and computer simulation approaches to the discovery and analysis of bioactive peptides. Curr. Pharm. Biotechnol. 2022, 23, 1541–1555.
- Mudgil, P.; Kamal, H.; Kilari, B.P.; Salim, M.A.S.M.; Gan, C.Y.; Maqsood, S. Simulated gastrointestinal digestion of camel and bovine casein hydrolysates: Identification and characterization of novel anti-diabetic bioactive peptides. Food Chem. 2021, 353, 129374.
- Lee, H.J.; Lee, H.S.; Choi, J.W.; Ra, K.S.; Kim, J.M.; Suh, H.J. Novel tripeptides with α-glucosidase inhibitory activity isolated from silk cocoon hydrolysate. J. Agric. Food Chem. 2011, 59, 11522–11525.
- Zhang, Y.; Wang, N.; Wang, W.; Wang, J.H.; Zhu, Z.Y.; Lia, X. Molecular mechanisms of novel peptides from silkworm pupae that inhibit α-glucosidase. Peptides 2016, 76, 45–50.
- Xie, F.; Wang, S.Y.; Zhang, L.; Wu, J.H.; Wang, Z.W. Investigating inhibitory activity of novel synthetic sericin peptide on α-D-glucosidase: Kinetics and interaction mechanism study using a docking simulation. J. Sci. Food Agric. 2018, 98, 1502–1510.
- Fang, Y.W.; Wang, S.Y.; Wu, J.H.; Zhang, L.; Wang, Z.W.; Gan, L.; He, J.J.; Shi, H.M.; Hou, J.L. The kinetics and mechanism of α-glucosidase inhibition by F5-SP, a novel compound derived from sericin peptides. Food Funct. 2017, 8, 323–332.
- Ramadhan, A.H.; Nawas, T.; Zhang, X.W.; Pembe, W.M.; Xia, W.S.; Xu, Y.S. Purification and identification of a novel antidiabetic peptide from Chinese giant salamander (Andrias davidianus) protein hydrolysate against α-amylase and α-glucosidase. Int. J. Food Prop. 2017, 20, S3360–S3372.
- Zhou, M.; Ren, G.Y.; Zhang, B.; Ma, F.L.; Fan, J.L.; Qiu, Z.J. Screening and identification of a novel antidiabetic peptide from collagen hydrolysates of Chinese giant salamander skin: Network pharmacology, inhibition kinetics and protection of IR-HepG2 cells. Food Funct. 2022, 13, 3329–3342.
- Zielińska, E.; Karaś, M.; Baraniak, B.; Jakubczyk, A. Evaluation of ACE, α-glucosidase, and lipase inhibitory activities of peptides obtained by in vitro digestion of selected species of edible insects. Eur. Food Res. Technol. 2020, 246, 1361–1369.
- Le, T.K.D.; Danova, A.; Aree, T.; Duong, T.H.; Koketsu, M.; Ninomiya, M.; Sawada, Y.; Kamsri, P.; Pungpo, P.; Chavasiri, W. α-Glucosidase inhibitors from the stems of knema globularia. J. Nat. Prod. 2022, 85, 776–786.
- Dirir, A.M.; Daou, M.; Yousef, A.F.; Yousef, L.F. A review of alpha-glucosidase inhibitors from plants as potential candidates for the treatment of type-2 diabetes. Phytochem. Rev. 2022, 21, 1049–1079.
- Teng, H.; Chen, L. α-Glucosidase and α-amylase inhibitors from seed oil: A review of liposoluble substance to treat diabetes. Crit. Rev. Food Sci. 2017, 57, 3438–3448.
- Famuwagun, A.A.; Alashi, A.M.; Gbadamosi, O.S.; Taiwo, K.A.; Oyedele, D.; Adebooye, O.C.; Aluko, R.E. Antioxidant and enzymes inhibitory properties of amaranth leaf protein hydrolyzates and ultrafiltration peptide fractions. J. Food Biochem. 2021, 45, e13396.
- Chen, N.; Yang, H.; Sun, Y.; Niu, J.; Liu, S.Y. Purification and identification of antioxidant peptides from walnut (Juglans regia L.) protein hydrolysates. Peptides 2012, 38, 344–349.
- Dong, Z.Y.; Li, M.Y.; Tian, G.; Zhang, T.H.; Ren, H.; Quek, S.Y. Effects of ultrasonic pretreatment on the structure and functionality of chicken bone protein prepared by enzymatic method. Food Chem. 2019, 299, 125103.
- Nourmohammadi, E.; SadeghiMahoonak, A.; Alami, M.; Ghorbani, M. Amino acid composition and antioxidative properties of hydrolysed pumpkin (Cucurbita pepo L.) oil cake protein. Int. J. Food Prop. 2017, 20, 3244–3255.
- Ambigaipalan, P.; Al-Khalifa, A.S.; Shahidi, F. Antioxidant and angiotensin I converting enzyme (ACE) inhibitory activities of date seed protein hydrolysates prepared using alcalase, flavourzyme and thermolysin. J. Funct. Foods 2015, 18, 1125–1137.
- Wang, R.C.; Zhao, H.X.; Pan, X.X.; Orfila, C.; Lu, W.H.; Ma, Y. Preparation of bioactive peptides with antidiabetic, antihypertensive, and antioxidant activities and identification of α-glucosidase inhibitory peptides from soy protein. Food Sci. Nutr. 2019, 7, 1848–1856.
- Wang, X.; Deng, Y.J.; Xie, P.J.; Liu, L.J.; Zhang, C.H.; Cheng, J.; Zhang, Y.; Liu, Y.; Huang, L.X.; Jiang, J.X. Novel bioactive peptides from ginkgo biloba seed protein and evaluation of their α-glucosidase inhibition activity. Food Chem. 2023, 404, 134481.
- Arise, R.O.; Idi, J.J.; Mic-Braimoh, I.M.; Korode, E.; Ahmed, R.N.; Osemwegie, O. In vitro Angiotesin-1-converting enzyme, α-amylase and α-glucosidase inhibitory and antioxidant activities of Luffa cylindrical (L.) M. Roem seed protein hydrolysate. Heliyon 2019, 5, e01634.
- Hong, H.; Fan, H.B.; Chalamaiah, M.; Wu, J.P. Preparation of low-molecular-w4eight, collagen hydrolysates (peptides): Current progress, challenges, and future perspectives. Food Chem. 2019, 301, 125222.
- Ngoh, Y.Y.; Gan, C.Y. Enzyme-assisted extraction and identification of antioxidative and α-amylase inhibitory peptides from Pinto beans (Phaseolus vulgaris cv. Pinto). Food Chem. 2016, 190, 331–337.
- Oseguera-Toledo, M.E.; de Mejia, E.G.; Amaya-Llano, S.L. Hard-to-cook bean (Phaseolus vulgaris L.) proteins hydrolyzed by alcalase and bromelain produced bioactive peptide fractions that inhibit targets of type-2 diabetes and oxidative stress. Food Res. Int. 2015, 76, 839–851.
- Jiang, M.Z.; Yan, H.; He, R.H.; Ma, Y.K. Purification and a molecular docking study of α-glucosidase-inhibitory peptides from a soybean protein hydrolysate with ultrasonic pretreatment. Eur. Food Res. Technol. 2018, 244, 1995–2005.
- Valencia-Mejía, E.; Batista, K.A.; Fernández, J.J.A.; Fernandes, K.F. Antihyperglycemic and hypoglycemic activity of naturally occurring peptides and protein hydrolysates from easy-to-cook and hard-to-cook beans (Phaseolus vulgaris L.). Food Res. Int. 2019, 121, 238–246.
- Mousavi, B.; Azizi, M.H.; Abbasi, S. Antidiabetic bio-peptides of soft and hard wheat glutens. Food Chem. Mol. Sci. 2022, 4, 100104.
- Wei, R.T.; Lin, L.K.; Li, T.T.; Li, C.; Chen, B.; Shen, Y.H. Separation, identification, and design of α-glucosidase inhibitory peptides based on the molecular mechanism from Paeonia ostii ‘Feng Dan’seed protein. J. Food Sci. 2022, 87, 4892–4904.
- Zhao, B.L.; Su, K.Y.; Mao, X.L.; Zhang, X.W. Separation and identification of enzyme inhibition peptides from dark tea protein. Bioorg. Chem. 2020, 99, 103772.
- Zhao, Q.; Wei, G.Q.; Li, K.L.; Duan, S.H.; Ye, R.; Huang, A.X. Identification and molecular docking of novel α-glucosidase inhibitory peptides from hydrolysates of Binglangjiang buffalo casein. LWT-Food Sci. Technol. 2022, 156, 113062.
- Vilcacundo, R.; Martínez-Villaluenga, C.; Hernández-Ledesma, B. Release of dipeptidyl peptidase IV, α-amylase and α-glucosidase inhibitory peptides from quinoa (Chenopodium quinoa Willd.) during in vitro simulated gastrointestinal digestion. J. Funct. Foods 2017, 35, 531–539.
- Dakhili, S.; Abdolalizadeh, L.; Hosseini, S.M.; Shojaee-Aliabadi, S.; Mirmoghtadaie, L. Quinoa protein: Composition, structure and functional properties. Food Chem. 2019, 299, 125161.
- Ujiroghene, O.J.; Liu, L.; Zhang, S.W.; Lu, J.; Pang, X.Y.; Lv, J.P. α-Glucosidase and ACE dual inhibitory protein hydrolysates and peptide fractions of sprouted quinoa yoghurt beverages inoculated with Lactobacillus casei. Food Chem. 2019, 299, 124985.
- Sadri, H.; Larki, N.N.; Kolahian, S. Hypoglycemic and hypolipidemic effects of leucine, zinc, and chromium, alone and in combination, in rats with type 2 diabetes. Biol. Trace Elem. Res. 2017, 180, 246–254.
- Hu, J.F.; Lai, X.H.; Wu, X.D.; Wang, H.Y.; Weng, N.H.; Lu, J.; Lyu, M.; Wang, S.J. Isolation of a novel anti-diabetic α-glucosidase oligo-peptide inhibitor from fermented rice bran. Foods 2023, 12, 183.
- Yang, X.Y.; Wang, D.; Dai, Y.Y.; Zhao, L.P.; Wang, W.T.; Ding, X.Z. Identification and molecular binding mechanism of novel α-glucosidase inhibitory peptides from hot-pressed peanut meal protein hydrolysates. Foods 2023, 12, 663.
- Mojica, L.; De Mejía, E.G. Optimization of enzymatic production of anti-diabetic peptides from black bean (Phaseolus vulgaris L.) proteins, their characterization and biological potential. Food Funct. 2016, 7, 713–727.
- Quintero-Soto, M.F.; Chávez-Ontiveros, J.; Garzón-Tiznado, J.A.; Salazar-Salas, N.Y.; Pineda-Hidalgo, K.V.; Delgado-Vargas, F.; Lopez-Valenzuela, J.A. Characterization of peptides with antioxidant activity and antidiabetic potential obtained from chickpea (Cicer arietinum L.) protein hydrolyzates. J. Food Sci. 2021, 86, 2962–2977.
- Patil, P.; Mandal, S.; Tomar, S.K.; Anand, S. Food protein-derived bioactive peptides in management of type 2 diabetes. Eur. J. Nutr. 2015, 54, 863–880.
- Rivero-Pino, F.; Guadix, A.; Guadix, E.M. Identification of novel dipeptidyl peptidase IV and α-glucosidase inhibitory peptides from Tenebrio molitor. Food Funct. 2021, 12, 873–880.
- Karimi, A.; Azizi, M.H.; Gavlighi, H.A. Fractionation of hydrolysate from corn germ protein by ultrafiltration: In vitro antidiabetic and antioxidant activity. Food Sci. Nutr. 2020, 8, 2395–2405.
- Park, J.H.; Kim, Y.N.; Kim, J.K.; Park, H.Y.; Song, B.S. Viscothionin purified from mistletoe (Viscum album var. coloratum Ohwi) induces insulin secretion from pancreatic beta cells. J. Ethnopharmacol. 2019, 234, 172–179.
- Sun, W.J.; Wu, C.E.; Fan, G.J.; Hao, G.; Shi, H.J.; Zhang, C. Preparation of a functional beverage with α-glucosidase inhibitory peptides obtained from ginkgo seeds. J. Food Sci. Technol. 2021, 58, 4495–4503.
- Liu, W.; Li, H.; Wen, Y.; Liu, Y.; Wang, J.; Sun, B. Molecular mechanism for the α-glucosidase inhibitory effect of wheat germ peptides. J. Agric. Food Chem. 2021, 69, 15231–15239.
- Qiu, L.Y.; Deng, Z.Y.; Zhao, C.D.; Xiao, T.; Weng, C.; Li, J.; Zheng, L.F. Nutritional composition and proteomic analysis of soft-shelled turtle (Pelodiscus sinensis) egg and identification of oligopeptides with alpha-glucosidase inhibitory activity. Food Res. Int. 2021, 145, 110414.
- Li, N.; Qiu, L.Y.; Song, H.L.; Xiao, T.; Luo, T.; Deng, Z.Y.; Zheng, L.F. Inhibition mechanism of oligopeptides from soft-shelled turtle egg against α-glucosidase and their gastrointestinal digestive properties. J. Food Biochem. 2022, 46, e14328.
- Tyagi, P.; Pechenov, S.; Subramony, J.A. Oral peptide delivery: Translational challenges due to physiological effects. J. Control Release 2018, 287, 167–176.
- Megías, C.; del Mar Yust, M.; Pedroche, J.; Lquari, H.; Girón-Calle, J.; Alaiz, M.; Millán, F.; Vioque, J. Purification of an ACE inhibitory peptide after hydrolysis of sunflower (Helianthus annuus L.) protein isolates. J. Agric. Food Chem. 2004, 52, 1928–1932.
More
Information
Subjects:
Food Science & Technology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
2 times
(View History)
Update Date:
18 Dec 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




