Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dong-Wei Di | -- | 2309 | 2023-12-13 07:17:21 | | | |
| 2 | Lindsay Dong | Meta information modification | 2309 | 2023-12-18 03:03:26 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Luo, P.; Li, T.; Shi, W.; Ma, Q.; Di, D. GRETCHEN HAGEN3 (GH3)-Dependent Auxin Conjugation in Plant. Encyclopedia. Available online: https://encyclopedia.pub/entry/52647 (accessed on 07 February 2026).
Luo P, Li T, Shi W, Ma Q, Di D. GRETCHEN HAGEN3 (GH3)-Dependent Auxin Conjugation in Plant. Encyclopedia. Available at: https://encyclopedia.pub/entry/52647. Accessed February 07, 2026.
Luo, Pan, Ting-Ting Li, Wei-Ming Shi, Qi Ma, Dong-Wei Di. "GRETCHEN HAGEN3 (GH3)-Dependent Auxin Conjugation in Plant" Encyclopedia, https://encyclopedia.pub/entry/52647 (accessed February 07, 2026).
Luo, P., Li, T., Shi, W., Ma, Q., & Di, D. (2023, December 13). GRETCHEN HAGEN3 (GH3)-Dependent Auxin Conjugation in Plant. In Encyclopedia. https://encyclopedia.pub/entry/52647
Luo, Pan, et al. "GRETCHEN HAGEN3 (GH3)-Dependent Auxin Conjugation in Plant." Encyclopedia. Web. 13 December, 2023.
Copy Citation
The precise control of free auxin (indole-3-acetic acid, IAA) gradient, which is orchestrated by biosynthesis, conjugation, degradation, hydrolyzation, and transport, is critical for all aspects of plant growth and development. Of these, the GRETCHEN HAGEN 3 (GH3) acyl acid amido synthetase family, pivotal in conjugating IAA with amino acids, has garnered significant interest.
auxin
IAA
GH3
chemical inhibitor
transcriptional regulation
1. Introduction
Auxins, a group of phytohormones, are integral to the regulation of plant development and stress responses [1][2]. Previous research has identified three naturally occurring auxins: indole-3-acetic acid (IAA), phenylacetic acid (PAA), and 4-chloro-indole-3-acetic acid (4-Cl-IAA), with IAA being the most prevalent and significant in plants [1]. The meticulous modulation of IAA levels, governed by biosynthesis, transport, and inactivation, is essential for normal plant growth, development, and adaptation to both biotic and abiotic environmental stresses [1][3][4].
Four primary pathways have been documented for IAA inactivation in plants: (i) IAA CARBOXYL METHYLTRANSFERASE1 (IAMT1) converts IAA to methyl IAA; (ii) UDP-glucosyltransferase (UGTs) generate ester-linked IAA conjugates; (iii) GRETCHEN HAGEN 3 (GH3) acyl amido synthetases facilitate the formation of amide-linked IAA conjugates; and (iv) IAA oxidation carried out by DIOXYGENASE FOR AUXIN OXIDATION (DAO) [5][6][7][8][9][10]. Methyl IAA and ester-linked IAA, both subject to reconversion into IAA by specific hydrolases, are predominantly regarded as forms of IAA storage [11]. Contrastingly, the reversibility of amide-linked conjugates varies based on the amino acid involved. Most, such as IAA-alanine (IAA-Ala), IAA-phenylalanine (IAA-Phe), and IAA-leucine (IAA-Leu), revert to free IAA, while others like IAA-glutamate (IAA-Glu) and IAA-aspartate (IAA-Asp) are directly degraded [5][11][12][13].
The first GH3 gene was identified from Glycine max as a rapid early auxin-responsive gene. Subsequent research has established the widespread distribution of the GH3 gene family across the plant kingdom, encompassing species from Arabidopsis thaliana to Oryza sativa, Zea mays, Triticum aestivum, and even non-vascular plants like Physcomitrella patens and Marchantia polymorpha [8][11][14][15][16][17][18]. Based on sequence homology and substrate specificity, the GH3 family in Arabidopsis is categorized into three distinct groups: Group I, II, and III. Group I GH3 genes are known to encode enzymes that synthesize amides from jasmonic acid (JA) or salicylic acid (SA). Group II GH3 enzymes function as IAA-amido synthetases, and Group III has been shown to catalyze the conjugation between amino acids and 4-substituted benzoates or indole-3-butyric acid (IBA) [8][19].
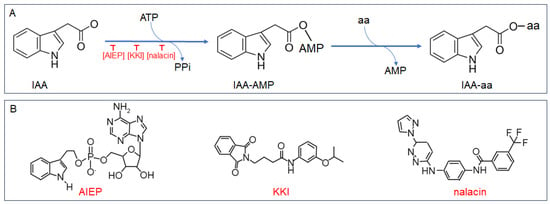
2. The Catalytic Mechanisms and Substrate Specificity of GH3 Acyl Acid Amido Synthetase Enzyme
Chen et al. firstly utilized a combination of initial velocity and product inhibition analyses, alongside mass spectrometry, to delineate the kinetic and chemical mechanisms governing OsGH3.8 activity [20]. They discovered that the conjugation of IAA with Asp operates via a ‘Bi Uni Uni Bi Ping Pong’ mechanism, as depicted in Figure 1A. The process initiates with the binding of IAA and ATP, in the presence of Mg2+, to the unoccupied enzyme. This interaction results in the formation of an adenylated IAA intermediate (IAA-AMP) and the concurrent release of pyrophosphate (PPi). Following this, Asp attaches to the enzyme•IAA•AMP complex, leading to the displacement of AMP and the establishment of an amide linkage between IAA and Asp. The final reaction products, IAA-Asp and AMP, are then released from the OsGH3.8 enzyme’s active site [20].

Figure 1. Catalytic reaction and inhibitor structures of Group II GH3 amido synthetase. (A) Schematic representation of the total reaction mediated by Group II GH3 amido synthetases. (B) Chemical structures of inhibitors targeting Group II GH3 amido synthetases: AIEP (adenosine-59-[2-(1H-indol-3-yl) ethyl] phosphate), KKI (kakeimide), and nalacin (N-[4-[[6-(1H-pyrazol-1-yl)-3-pyridazinyl] amino] phenyl]-3-(trifluoromethyl)benzamide).
Structural analyses of the GH3 enzyme in Arabidopsis, grape, and rice have illuminated that both monocotyledons and dicotyledons employ a similar mechanism for AMP and IAA binding [21]. The GH3 enzyme exhibits distinct acyl acid binding preferences, with specific residues within its active site conferring selectivity for particular substrates [21]. In OsGH3.8, the amino acids Arg130 and Leu137 play a crucial role in substrate specificity. The mutation of Arg130 to Leu (Arg130-Leu) shifts the enzyme’s substrate preference from IAA to benzoate/SA, while an Arg130-Thr substitution favors JA over IAA.
3. The Modulation of IAA Homeostasis by Small Chemical Molecules via the Inhibition of GH3 Enzyme Activity
3.1. AIEP, the First Chemical Inhibitor of Auxin Conjugation
The GH3-dependent IAA conjugation initiates when the unbound GH3 enzyme interacts with ATP and IAA, leading to the formation of IAA•AMP. Böttcher et al. engineered and synthesized a stable analogue of IAA•AMP, named AIEP. This molecule competes with ATP and IAA for the binding sites on the GH3 enzyme at the onset of catalysis [22]. The competitive inhibitory effect of AIEP on ATP and IAA binding was validated through substrate velocity experiments involving VvGH3.1 and VvGH3.6 from grape.
3.2. KKI, a Specific Inhibitor of IAA-Conjugating GH3 Enzymes
In the quest to identify inhibitors of IAA-conjugating GH3 enzymes, researchers leveraged Arabidopsis AtGH3.6 overexpression plants as a biological assay system. They screened a synthetic chemical library comprising 10,000 compounds for agents capable of reverting the altered root hair growth phenotype of AtGH3.6-overexpressed lines. This led to the initial identification of compound ‘1’, followed by the synthesis of 25 derivatives of ‘1’. Among these, kakeimide (KKI) emerged as a highly potent inhibitor. (Figure 1B). KKI functions by directly interacting with the IAA binding site within the GH3•ATP complex, forming a stable GH3•ATP•KKI ternary complex that impedes the synthesis of IAA-amino acid conjugates [23]. Validation experiments demonstrated KKI’s effectiveness in targeting the IAA binding sites of various GH3 enzymes, notably VvGH3.1, AtGH3.5, and OsGH3.8, while sparing the IBA binding site of AtGH3.15. This specificity, coupled with KKI’s lack of interference in jasmonic acid (JA) homeostasis, underscores its role as a specific inhibitor of IAA-conjugating GH3 enzymes [23].
3.3. Nalacin, a Potent Inhibitor Targeting Group II GH3 Enzymes
Nalacin was identified from a chemical screen by observing the auxin-related root phenotypes in the Arabidopsis wild-type Col-0 (Figure 1B). Subsequent studies have shown that nalacin competitively inhibits substrate acceptance by AtGH3.6 and AtGH3.11 through trifluoromethyl phenyl occupancy of the IAA binding site of AtGH3s, suggesting that nalacin also functions in the first step of the ‘Bi Uni Uni Bi Ping Pong’ reaction of GH3 enzymes. Unlike KKI, which selectively inhibits only class II GH3 enzymes, nalacin also impedes the formation of JA amino acid conjugates mediated by AtGH3.11, albeit through a distinct binding mode [24].
4. The Transcriptional Control of GH3 Enzymes in Plant Growth, Development, and Stress Adaptation
4.1. GH3-Dependent IAA Conjugation Is Involved in Regulating Multiple Developmental Processes
In Arabidopsis, eight Group II GH3 genes are involved in catalyzing IAA conjugation to amino acids. Due to redundant gene functions, mutations in single genes result in only subtle phenotypic changes and modified sensitivity to exogenous IAA [25]. In contrast, mutants with overexpressed GH3 genes, obtained through activation tag insertion, provide a more discernible phenotype for study. For instance, AtGH3.2 and AtGH3.6 were identified through the screening of their overexpressed mutants, ydk1-D and dfl1-D, respectively [25][26]. Interestingly, however, despite all overexpressing genes being closely related to GH3 Group II family members, they still showed inconsistent phenotypes. Under various light conditions, the dfl1-D mutant displayed shortened hypocotyls exclusively in light, while the ydk1-D mutant showed this phenotype under both light and dark conditions. Additionally, the ydk1-D mutant had a shorter primary root but did not exhibit significant difference in susceptibility to auxin-mediated root growth inhibition. In contrast, the dfl1-D mutant was resistant to IAA-mediated root growth inhibition and did not present a short-root phenotype compared with the wild type [25][26].
As early auxin response genes, Group II GH3 genes play a significant role downstream of auxin response factors (ARFs), which are key elements in the auxin signaling pathway [27]. Research has elucidated the involvement of ARF6, AtARF7, AtARF8, and AtARF17 in the transcriptional regulation of several AtGH3 genes. AtARF7 and AtARF8 are known to positively regulate the transcription of AtGH3.2, AtGH3.5, and AtGH3.6, influencing hypocotyl elongation under different light conditions [25][28]. In contrast, AtARF17 has a negative regulatory role on AtGH3.5 and AtGH3.6 transcription, which is essential for proper plant development. The microRNA AtmiR160 directly targets AtARF17 mRNA, which is crucial for normal leaf and root growth [29]. In terms of adventitious root development, AtARF6 and AtARF8 act as positive regulators, while AtARF17 functions as a negative regulator. They co-regulate the transcription of AtGH3.3, AtGH3.5, and AtGH3.6, impacting JA conjugation with amino acids, but not IAA conjugation [30].
4.2. The Integration of Hormonal Signals in GH3-Dependent IAA Conjugation’s Responses to Abiotic Stresses
4.2.1. Drought Stress
In Arabidopsis, AtGH3.5 has been observed to respond rapidly to drought conditions, with the wes1-D mutant, which overexpresses AtGH3.5, exhibiting enhanced drought resistance [31]. A subsequent study revealed that AtMYB96 modulates the expression of several AtGH3 genes, including AtGH3.3, AtGH3.5, and AtGH3.6, under drought stress through an abscisic acid (ABA)-dependent pathway [32]. This finding underscores the importance of ABA signaling in modulating GH3 gene expression during drought response. Additionally, the gh3oct mutant, with knockouts of all Group II GH3 genes (GH3.1,2,3,4,5,6,9,17), exhibits increased drought tolerance, further highlighting the role of these genes in drought response mechanisms [33]. In rice, the activation of OsGH3.13 has been linked to a reduction in free IAA levels, leading to a structural adaptation in the leaves, such as thicker blades, which enhance drought tolerance by minimizing water loss [34].
4.2.2. Temperature (Heat/Cold/Freezing) Stress
In Arabidopsis, the transcription of AtGH3.5 is notably responsive to temperature extremes, showing increased levels under both low (4 °C) and high (37 °C) temperature conditions. The wes1-D mutant, characterized by the overexpression of AtGH3.5, shows increased survival after exposure to freezing temperatures (−7 °C). This suggests a broad regulatory role for AtGH3.5 across a spectrum of temperature stresses [35]. In rice, the overexpression of OsGH3.2 leads to a reduction in free IAA levels, thereby activating cold-responsive genes and enhancing the plant’s ability to scavenge reactive oxygen species (ROS).
4.2.3. Salt and Osmotic Stress
All root-expressed Group II GH3 genes in Arabidopsis are upregulated following treatment with NaCl at concentrations of 75 mM and 150 mM. The Atgh3oct mutant, with combined knockouts of all Group II GH3 genes, exhibits greater resilience to NaCl stress compared to the wild type [33]. This enhanced tolerance also extends to sorbitol and mannitol exposure, suggesting that Group II GH3s may confer broad osmotic stress resistance, inclusive of salinity stress. Further investigation reveals that NaCl treatment increases AtACS2 transcription, leading to the accumulation of the ethylene precursor ACC, which in turn downregulates AtGH3.5 and AtGH3.9 transcription, maintaining free IAA levels and primary root growth [36].
4.2.4. Ammonium (NH4+) Stress
NH4+ serves as a vital nitrogen source for plants, but when available in excess, it can be detrimental to growth [37][38]. Prior research indicates that high NH4+ levels lead to a reduction in free IAA [39][40][41]. In Arabidopsis, elevated NH4+ conditions trigger the induction of nearly all Group II GH3 genes, which in turn accelerates the conjugation of free IAA to amino acids [39].
4.2.5. Pathogen Stress
In response to pathogen attacks, plants activate a comprehensive defense strategy: (1) they initiate a hypersensitive response leading to rapid programmed cell death at the infection site alongside other defense responses; (2) they activate systemic-acquired resistance (SAR) in distal tissues; and (3) they activate basal immunity to limit pathogen growth [42]. GH3-dependent IAA conjugation is intricately involved in these plant defense mechanisms. In Arabidopsis, the pathogens B. cinerea and P. syringae pv tomato (Pst) DC3000 significantly upregulate AtGH3.2 and AtGH3.3 transcription. Loss-of-function mutations in AtGH3.2 or AtGH3.4 enhance resistance to both pathogens, suggesting that AtGH3.2, AtGH3.3, and AtGH3.4 may negatively influence the plant’s response to B. cinerea and Pst DC3000 [43]. The overexpression of mutant gh3.5-1D shows a compromised hypersensitive response but retains normal SAR and basal immunity, whereas the Atgh3.5 mutant exhibits a defective SAR response yet maintains a typical hypersensitive response and basal immunity. In contrast, the dfl1-D mutant displays altered hypersensitive and basal immune responses [42].
5. Atypical Roles of Group II GH3 and IAA-Amino Acids
5.1. The Roles of Group II GH3 beyond the Catalyzation of IAA-Amino Acid Conjugate Formation
The capacity of Group II GH3 enzymes extends beyond the synthesis of IAA-amino acid conjugates. Research using recombinant GH3 IAA-amino acid synthetase from pea has revealed the enzyme’s ability to conjugate IAA not only to aspartate but also to proteins in immature seeds. The proposition that IAA conjugation to proteins may serve a regulatory function acting as a prosthetic group and influencing protein activity via posttranslational modifications is a compelling avenue for further exploration [44][45].
5.2. The Specialized Functions of IAA-Amino Acid Conjugates beyond Their Role as Auxin Stock
Previous studies found that the exogenous addition of IAA-aa and IAA both rapidly increased content-free IAA levels and exhibited similar high growth factor phenotypes, suggesting that IAA-aa is a storage form of IAA [46]. Subsequent studies found that these IAA-aa, IAA-Leu, and IAA-Ala could be reversibly converted to free IAA by the action of the hydrolases, IAA-LEUCINE RESIS TANT1 (ILR1), ILR1-LIKE proteins (ILLs), and IAA-ALANINE RESISTANT3 (IAR3) [5][47][48]. IAA-Glu and IAA-Asp, once considered only as degradation intermediates, are now recognized as reversible storage forms [49]. Beyond storage, IAA-aa have been identified as possessing unique biological functions. IAA-Trp, for instance, acts as a ‘super inactivator’ by not only consuming free IAA for its synthesis but also antagonizing the activity of residual IAA, with IAA-Trp significantly mitigating root inhibition effects caused by IAA [50]. IAA-Asp has been reported to have more diverse roles: (1) correcting the temperature sensitivity of henbane (Hyoscyamus muticus) XIlB2 (temperature-sensitive variant) cells [51]; (2) IAA-Asp directly and specifically enhance the pea (Pisum sativum) responses to abiotic stress by increasing the antioxidant enzyme activity and then reducing the H2O2 concentration [52]; (3) IAA-Asp as a ripening signal in grapes (Vitis vinifera) can be perceived at a certain stage of fruit development; however, the mechanism of sensing remains unknown [53]; and (4) IAA-Asp promotes pathogen development in plants by regulating the transcription of virulence genes [43]. These insights suggest that IAA-amino acids are not just byproducts of GH3 activity but are biologically active molecules with specific roles.
References
- Di, D.W.; Zhang, C.; Luo, P.; An, C.W.; Guo, G.Q. The biosynthesis of auxin: How many paths truly lead to IAA? Plant Growth Regul. 2016, 78, 275–285.
- Fiedler, L.; Friml, J. Rapid auxin signaling: Unknowns old and new. Curr. Opin. Plant Biol. 2023, 75, 102443.
- Jing, H.; Wilkinson, E.G.; Sageman-Furnas, K.; Strader, L.C. Auxin and abiotic stress responses. J. Exp. Bot. 2023, 74, 7000–7014.
- Luo, P.; Di, D.W. Precise regulation of the TAA1/TAR-YUCCA auxin biosynthesis pathway in plants. Int. J. Mol. Sci. 2023, 24, 8514.
- LeClere, S.; Tellez, R.; Rampey, R.A.; Matsuda, S.P.T.; Bartel, B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002, 277, 20446–20452.
- Mellor, N.; Band, L.R.; Pencík, A.; Novák, O.; Rashed, A.; Holman, T.; Wilson, M.H.; Voss, U.; Bishopp, A.; King, J.R.; et al. Dynamic regulation of auxin oxidase and conjugating enzymes and modulates auxin homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 11022–11027.
- Porco, S.; Pencík, A.; Rashed, A.; Voss, U.; Casanova-Sáez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R.; Swarup, R.; Swarup, K.; et al. Dioxygenase-encoding gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021.
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627.
- Yang, Y.; Xu, R.; Ma, C.J.; Vlot, A.C.; Klessig, D.F.; Pichersky, E. Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AMES esterase family of Arabidopsis. Plant Physiol. 2008, 147, 1034–1045.
- Zhang, J.; Lin, J.E.; Harris, C.; Pereira, F.C.M.; Wu, F.; Blakeslee, J.J.; Peer, W.A. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in. Proc. Natl. Acad. Sci. USA 2016, 113, 11010–11015.
- Casanova-Saez, R.; Mateo-Bonmati, E.; Ljung, K. Auxin Metabolism in Plants. CSH Perspect. Biol. 2021, 13, a039867.
- Ostin, A.; Kowalyczk, M.; Bhalerao, R.P.; Sandberg, G. Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 1998, 118, 285–296.
- Rampey, R.A.; LeClere, S.; Kowalczyk, M.; Ljung, K.; Sandberg, G.; Bartel, B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004, 135, 978–988.
- Feng, S.; Yue, R.; Tao, S.; Yang, Y.; Zhang, L.; Xu, M.; Wang, H.; Shen, C. Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J. Integr. Plant Biol. 2015, 57, 783–795.
- Hagen, G.; Kleinschmidt, A.; Guilfoyle, T. Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta 1984, 162, 147–153.
- Jain, M.; Kaur, N.; Tyagi, A.K.; Khurana, J.P. The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 36–46.
- Jiang, W.; Yin, J.; Zhang, H.; He, Y.; Shuai, S.; Chen, S.; Cao, S.; Li, W.; Ma, D.; Chen, H. Genome-wide identification, characterization analysis and expression profiling of auxin-responsive GH3 family genes in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2020, 47, 3885–3907.
- Ludwig-Muller, J.; Julke, S.; Bierfreund, N.M.; Decker, E.L.; Reski, R. Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol. 2009, 181, 323–338.
- Sherp, A.M.; Westfall, C.S.; Alvarez, S.; Jez, J.M. Arabidopsis thaliana GH3.15 acyl acid amido synthetase has a highly specific substrate preference for the auxin precursor indole-3-butyric acid. J. Biol. Chem. 2018, 293, 4277–4288.
- Chen, Q.; Westfall, C.S.; Hicks, L.M.; Wang, S.; Jez, J.M. Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J. Biol. Chem. 2010, 285, 29780–29786.
- Xu, G.L.; Zhang, Y.K.; Li, M.Y.; Jiao, X.; Zhou, L.; Ming, Z.H. Crystal structure of the acyl acid amido synthetase GH3-8 from Oryza sativa. Biochem. Biophys. Res. Commun. 2021, 534, 266–271.
- Bottcher, C.; Dennis, E.G.; Booker, G.W.; Polyak, S.W.; Boss, P.K.; Davies, C. A novel tool for studying auxin-metabolism: The inhibition of grapevine indole-3-acetic acid-amido synthetases by a reaction intermediate analogue. PLoS ONE 2012, 7, e37632.
- Fukui, K.; Arai, K.; Tanaka, Y.; Aoi, Y.; Kukshal, V.; Jez, J.M.; Kubes, M.F.; Napier, R.; Zhao, Y.; Kasahara, H.; et al. Chemical inhibition of the auxin inactivation pathway uncovers the roles of metabolic turnover in auxin homeostasis. Proc. Natl. Acad. Sci. USA 2022, 119, e2206869119.
- Xie, Y.; Zhu, Y.; Wang, N.; Luo, M.; Ota, T.; Guo, R.; Takahashi, I.; Yu, Z.; Aizezi, Y.; Zhang, L.; et al. Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation. Proc. Natl. Acad. Sci. USA 2022, 119, e2209256119.
- Takase, T.; Nakazawa, M.; Ishikawa, A.; Kawashima, M.; Ichikawa, T.; Takahashi, N.; Shimada, H.; Manabe, K.; Matsui, M. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004, 37, 471–483.
- Nakazawa, M.; Yabe, N.; Ichikawa, T.; Yamamoto, Y.Y.; Yoshizumi, T.; Hasunuma, K.; Matsui, M. DFL2, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001, 25, 213–221.
- Di, D.W.; Zhang, C.G.; Guo, G.Q. Involvement of secondary messengers and small organic molecules in auxin perception and signaling. Plant Cell Rep. 2015, 34, 895–904.
- Tian, C.; Muto, H.; Higuchi, K.; Matamura, T.; Tatematsu, K.; Koshiba, T.; Yamamoto, K.T. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004, 40, 333–343.
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375.
- Gutierrez, L.; Mongelard, G.; Flokova, K.; Pacurar, D.I.; Novak, O.; Staswick, P.; Kowalczyk, M.; Pacurar, M.; Demailly, H.; Geiss, G.; et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527.
- Park, J.E.; Park, J.Y.; Kim, Y.S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.Y.; Kim, J.; Lee, Y.H.; Park, C.M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046.
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289.
- Casanova-Saez, R.; Mateo-Bonmati, E.; Simura, J.; Pencik, A.; Novak, O.; Staswick, P.; Ljung, K. Inactivation of the entire Arabidopsis group II GH3s confers tolerance to salinity and water deficit. New Phytol. 2022, 235, 263–275.
- Zhang, S.W.; Li, C.H.; Cao, J.; Zhang, Y.C.; Zhang, S.Q.; Xia, Y.F.; Sun, D.Y.; Sun, Y. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009, 151, 1889–1901.
- Park, J.E.; Seo, P.J.; Lee, A.K.; Jung, J.H.; Kim, Y.S.; Park, C.M. An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol. 2007, 48, 1236–1241.
- Han, S.; Jia, M.Z.; Yang, J.F.; Jiang, J. The integration of ACS2-generated ACC with GH3-mediated IAA homeostasis in NaCl-stressed primary root elongation of Arabidopsis seedlings. Plant Growth Regul. 2019, 88, 151–158.
- Xiao, C.; Fang, Y.; Wang, S.; He, K. The alleviation of ammonium toxicity in plants. J. Integr. Plant Biol. 2023, 65, 1362–1368.
- Di, D.W. New molecular mechanisms of plant response to ammonium nutrition. Appl. Sci. 2023, 13, 11570.
- Di, D.W.; Li, G.J.; Sun, L.; Wu, J.J.; Wang, M.; Kronzucker, H.J.; Fang, S.; Chu, J.F.; Shi, W.M. High ammonium inhibits root growth in Arabidopsis thaliana by promoting auxin conjugation rather than inhibiting auxin biosynthesis. J. Plant Physiol. 2021, 261, 153415.
- Di, D.W.; Sun, L.; Zhang, X.N.; Li, G.J.; Kronzucker, H.J.; Shi, W.M. Involvement of auxin in the regulation of ammonium tolerance in rice (Oryza sativa L.). Plant Soil. 2018, 432, 373–387.
- Dziewit, K.; Pencik, A.; Dobrzynska, K.; Novak, O.; Szal, B.; Podgorska, A. Spatiotemporal auxin distribution in Arabidopsis tissues is regulated by anabolic and catabolic reactions under long-term ammonium stress. BMC Plant Biol. 2021, 21, 602.
- Zhang, Z.Q.; Li, Q.; Li, Z.M.; Staswick, P.E.; Wang, M.Y.; Zhu, Y.; He, Z.H. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007, 145, 450–464.
- González-Lamothe, R.; El Oirdi, M.; Brisson, N.; Bouarab, K. The conjugated auxin indole-3-acetic acid-aspartic acid promotes plant disease development. Plant Cell 2012, 24, 762–777.
- Ostrowski, M.; Ciarkowska, A. Pea GH3 acyl acid amidosynthetase conjugates IAA to proteins in immature seeds of Pisum sativum L.—A new perspective on formation of high-molecular weight conjugates of auxin. J. Plant Physiol. 2021, 256, 153312.
- Seidel, C.; Walz, A.; Park, S.; Cohen, J.D.; Ludwig-Muller, J. Indole-3-acetic acid protein conjugates: Novel players in auxin homeostasis. Plant Biol. 2006, 8, 340–345.
- Wang, Q.; De Gernier, H.; Duan, X.L.; Xie, Y.M.; Geelen, D.; Hayashi, K.; Xuan, W.; Geisler, M.; ten Tusscher, K.; Beeckman, T.; et al. GH3-mediated auxin inactivation attenuates multiple stages of lateral root development. New Phytol. 2023, 240, 1900–1912.
- Bartel, B.; Fink, G.R. Ilr1, an amidohydrolase that releases active indole-3-acetic-acid from conjugates. Science 1995, 268, 1745–1748.
- Davies, R.T.; Goetz, D.H.; Lasswell, J.; Anderson, M.N.; Bartel, B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 1999, 11, 365–376.
- Hayashi, K.I.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.; Hu, Y.; Ge, C.; Zhao, Y.; Kasahara, H.; et al. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 2021, 12, 6752.
- Staswick, P.E. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 2009, 150, 1310–1321.
- Oetiker, J.H.; Aeschbacher, G. Temperature-sensitive plant cells with shunted indole-3-acetic acid conjugation. Plant Physiol. 1997, 114, 1385–1395.
- Ostrowski, M.; Ciarkowska, A.; Jakubowska, A. The auxin conjugate indole-3-acetyl-aspartate affects responses to cadmium and salt stress in Pisum sativum L. J. Plant Physiol. 2016, 191, 63–72.
- Böttcher, C.; Burbidge, C.A.; Boss, P.K.; Davies, C. Interactions between ethylene and auxin are crucial to the control of grape (Vitis vinifera L.) berry ripening. BMC Plant Biol. 2013, 13, 222.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
872
Revisions:
2 times
(View History)
Update Date:
18 Dec 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




