Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Maria Agnieszka Derkaczew | -- | 2255 | 2023-12-02 12:56:07 | | | |
| 2 | Lindsay Dong | Meta information modification | 2255 | 2023-12-04 02:36:22 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Derkaczew, M.; Martyniuk, P.; Hofman, R.; Rutkowski, K.; Osowski, A.; Wojtkiewicz, J. Abnormalities in Metabolic Pathways of Phosphoinositides. Encyclopedia. Available online: https://encyclopedia.pub/entry/52278 (accessed on 07 February 2026).
Derkaczew M, Martyniuk P, Hofman R, Rutkowski K, Osowski A, Wojtkiewicz J. Abnormalities in Metabolic Pathways of Phosphoinositides. Encyclopedia. Available at: https://encyclopedia.pub/entry/52278. Accessed February 07, 2026.
Derkaczew, Maria, Piotr Martyniuk, Robert Hofman, Krzysztof Rutkowski, Adam Osowski, Joanna Wojtkiewicz. "Abnormalities in Metabolic Pathways of Phosphoinositides" Encyclopedia, https://encyclopedia.pub/entry/52278 (accessed February 07, 2026).
Derkaczew, M., Martyniuk, P., Hofman, R., Rutkowski, K., Osowski, A., & Wojtkiewicz, J. (2023, December 02). Abnormalities in Metabolic Pathways of Phosphoinositides. In Encyclopedia. https://encyclopedia.pub/entry/52278
Derkaczew, Maria, et al. "Abnormalities in Metabolic Pathways of Phosphoinositides." Encyclopedia. Web. 02 December, 2023.
Copy Citation
Myo-inositol belongs to one of the sugar alcohol groups known as cyclitols. Phosphatidylinositols are one of the derivatives of Myo-inositol, and constitute important mediators in many intracellular processes such as cell growth, cell differentiation, receptor recycling, cytoskeletal organization, and membrane fusion. They also have even more functions that are essential for cell survival. Mutations in genes encoding phosphatidylinositols and their derivatives can lead to many disorders.
myo-inositol
phosphoinositides
phosphatidylinositol
phosphatidylinositol phosphate
1. Introduction
Myo-inositol (MI) is the most common stereoisomer of inositol in eukaryotic cells [1]. MI was discovered by Scherer in 1850, and to this day its properties are still being investigated [2]. The physiological pool of myo-inositol is derived from diet, catabolism of phosphatidylinositols (PIs), phosphatidylinositol phosphates (PIPs)—inositol phosphates (IPs), and form various glucose-included enzymatic reactions [3][4][5]. The main physiological role of myoinositol stands as the precursor of the inositol phospholipids, which after modification by the hormone-stimulated inositol-phospholipid-specific phospholipase C (PLC), generate inositol 1,4,5-trisphosphate (Ins(1,4,5)P3), diacylglycerol (DAG), PI, PIP, IP, glycosylphosphatidylinositols (GPIs), Inositol trisphosphate (IP3), and inositol-phosphoglycans (IPGs) [1][3]. These molecules are used as the ubiquitous second messengers, conveying signals derived by various hormones, e.g., thyroid stimulating hormone (TSH), luteinizing hormone (LH), follicle-stimulating hormone (FSH), and insulin [1][4][6][7]. The interconversions between this group of molecules are conducted by crucial enzymes, whose dysfunction can lead to severe abnormalities, disorders, and illnesses [4][6].
2. The Family of Phosphoinositol and Phosphoinositides
Phosphatidylinositol (PtdIns), the starting point of PIP metabolism, is a ubiquitous phospholipid in eukaryotic cells present in various proportions according to the type of membrane. PIPs are all metabolized directly or sequentially from PIs [8]. The structural formulas of phosphoinositol and phosphoinositides are shown in Table 1.
Table 1. Structural formulas of phosphoinositol and phosphoinositides.
| PI Phosphatidylinositol |
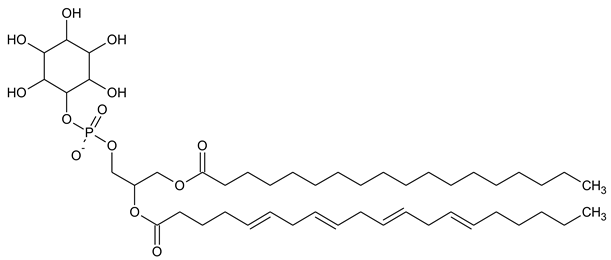 |
| PI3P Phosphatidylinositol 3-phosphate |
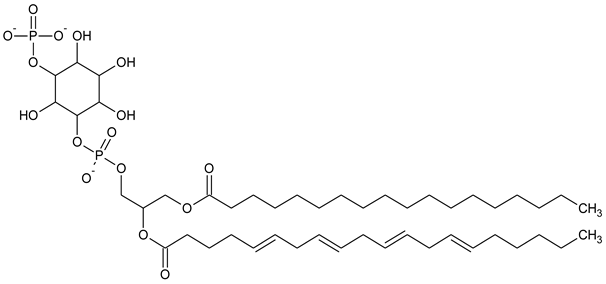 |
| PI(3,5)P2 Phosphatidylinositol 3,5-bisphosphate |
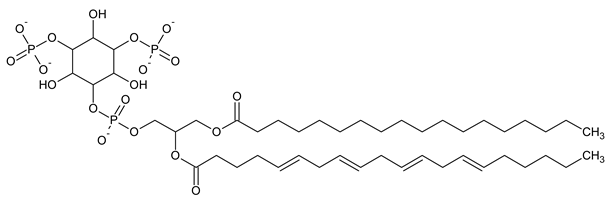 |
| PI4P Phosphatidylinositol 5-phosphate |
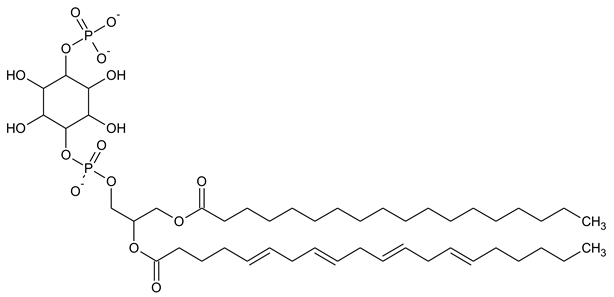 |
| PI(3,4)P2 Phosphatidylinositol 3,4-bisphosphate |
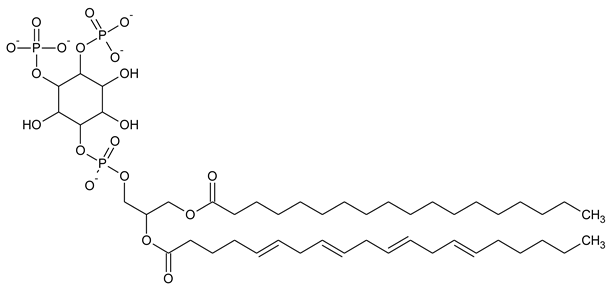 |
| PI(3,4,5)P3 Phosphatidylinositol 3,4,5-trisphosphate |
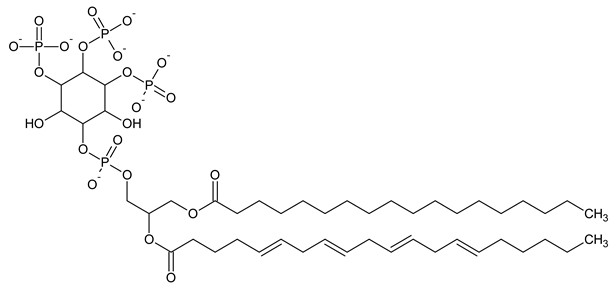 |
| PI5P Phosphatidylinositol 5-phosphate |
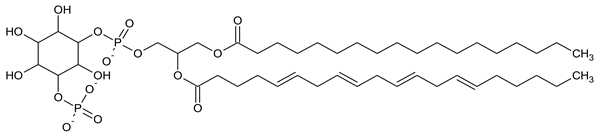 |
| PI(4,5)P2 Phosphatidylinositol 4,5-bisphosphate |
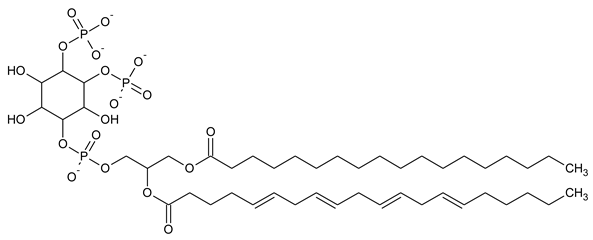 |
3. Routes and Interconversions of PIs
As previously mentioned, PI is a key compound and precursor of PIPs, which are all metabolized directly or sequentially from PI [8]. The scheme below presents detailed metabolic routes and interconversions of the PIPs family. The detailed analysis of genes and encoded enzymes is described in the table below Figure 1.
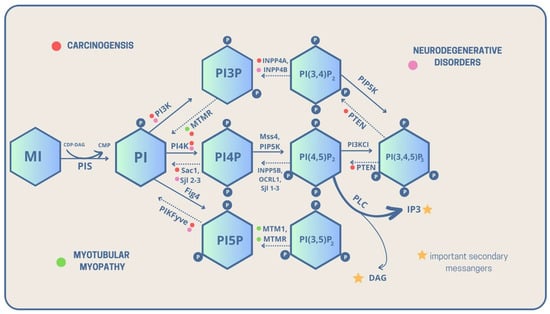
Figure 1. Metabolic interconversions of PIPs with their linkage to carcinogenesis, neurodegenerative diseases, and myotubular myopathies, with highlighted important intracellular secondary messengers [solid lines—phosphorylation, dashed lines—dephosphorylation, CDP-DAG—Cytidine diphosphate diacyloglycerol, CMP—Cytidine monophosphate, DAG—Diacylglycerol, P—phosphate group, PI—Phosphatidylinositol, PI3P—Phosphatidylinositol 3-phosphate, PI4P—Phosphatidylinositol 4-phosphate, PI5P—Phosphatidylinositol 5-phosphate, PI(3,5)P2—Phosphatidylinositol 3,5-bisphosphate, PI(3,4)P2—Phosphatidylinositol 3,4-bisphosphate, PI(4,5)P2—Phosphatidylinositol 4,5-bisphosphate, PI(3,4,5)P3—Phosphatidylinositol 3,4,5-trisphosphate, PIS—PI synthase, PLC—Phospholipase C].
PI itself is a product of the synthesis of cytidine diphosphate diacylglycerol (CPD-DAG) and MI. The reaction is conducted by PI synthase, also called phosphatidylinositol synthase 1 (PIS1) [9][10]. Then, PI phosphorylates are converted into phosphatidylinositol 3-phosphate (PI3P/PtdIns3P) [11]. The conversion is catalyzed by phosphatidylinositol 3 kinase (PI3K) and class III PI 3-kinase—vacuolar protein sorting 34 (Vps34) [12].
In the opposite direction, dephosphorylation occurs, which is conducted by PI3 phosphatases: Yeast myotubularin-related 1 (Ymr1) and Synaptojanin-like proteins 2-3 (Sjl2-3) [13][14]. Next, PI3P is phosphorylated into phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) by PI3P 5-kinase encoded by the Saccharomyces cerevisiae FAB1/PIKfyve genes [15][16][17].
On the other hand, dephosphorylation is conducted by Phosphoinositide 5-phosphatase—FIG4 [18]. Then, PI(3,5)P2 can transform into PtdIns5P by dephosphorylation conducted by PI3-phosphatase, MTM1/MTMR1-4, 6-8 [19]. Subsequently, phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2) can turn into PI3P by dephosphorylation catalyzed by PI4-phosphatase: phosphatidylinositol 4,5-bisphosphate 5-phosphatase A and B (INPP4A, INPP4B) [20].
Afterward, PI can also be metabolized into PtdIns4P during phosphorylation conducted by PI4-kinases: Pik1/Stt4 and PI4Kalfa/PI4Kbeta [21][22]. In dephosphorylation, enzymes such as PI4-phosphatases take part: Sjl2-3/Sac1 and SAC1 are similar to the domain of synaptojanin 1 [21][23].
Finally, PtdIns4P can turn into phosphatidylinositol 4,5-bisphosphate PI(4,5)P2 during phosphorylation conducted by PI4P 5-kinase: PIP5K α, β, and γ [24]. As for dephosphorylation, it is conducted by PI5-phosphatases: Sjl1-3, INPP5B, and OCRL1 [25][26][27][28].
4. Myotubular Myopathy
Centronuclear myopathy (CNM) is one of the disorders affecting the nervous and muscular systems characterized by two main criteria: symptoms of clinically congenital myopathy and multiple centrally located nuclei in muscle cells confirmed by biopsy [29].
Myotubularin 1 (MTM1) is an enzyme involved in the regulation of phosphoinositides, which are important molecules in cell signaling and membrane trafficking. Mutations in the myotubularin gene MTM1 which cause human myotubular myopathy dramatically reduce the phosphatase’s ability to dephosphorylate PI3P, affecting the levels of inositol lipid PI3P in myogenesis. In addition, it inhibits the transport of EGFRs to lysosomes, causing the formation of large endosomal vacuoles through the effects of myotubular phosphatase and its interaction with PI(3,5)P2. In most patients, mutations in the MTM1 gene are associated with the recessive X chromosome form (Xq28) [30][31][32][33], while autosomal dominant and recessive forms primarily involve mutations in the dynamin 2 (DNM2) gene on chromosome 19p13.2 and the amphisin 2 gene (BIN1) on chromosome 2q14 [29].
XLMTM is a specific subtype of myotubular myopathy, which is a rare genetic neuromuscular disorder characterized by severe muscle weakness and hypotonia (low muscle tone) caused by mutations in the MTM1 gene. As a result, the condition is X-linked, and therefore mainly affects males (2/100,000 male births) [29][33][34]. The symptoms of XLMTM are generally similar to those of other myotubular myopathy subtypes, but they tend to be more severe in males. Newborn males with XLMTM often have profound muscle weakness, leading to severe respiratory difficulties and respiratory failure. The weakened muscles can affect an individual’s ability to breathe, swallow, and move, which makes it essential for affected infants to receive immediate medical attention and respiratory support.
BIN1 modulation as well as DMN2 reduction may also be an effective treatment strategy for XLCNM, as it restores adequate myofibrillar integrity [35][36].
Normal cell function is regulated by myotubularins belonging to the phosphatidylinositol 3-phosphate phosphatase family PI3P, identified by positional cloning of the MTM1 gene in patients suffering from X-chromosome-associated myotubular myopathy (with reported specificity toward PI3P) and the myotubularin-related protein 2 (MTMR2) gene (hydrolyzes both PI3P and PI(3,5)P2 together with myotubularin-related protein 3 (MTMR3) in patients suffering from CMT4B) [37]. Myotubularin-related protein 1 (MTMR1) was shown to use PI3P and/or PI(3,5)P2 as substrates. The overall structure was very similar to the previously described structure of MTMR2 [38][39]. It turns out that MTMR2, whose abnormal function is revealed in the neurodegenerative Charcot-Marie-Tooth disease type 4B2, is also highly specific for PI3P as a substrate. In addition, the myotubularin-related phosphatases MTMR1, MTMR3, and myotubularin-related protein 6 (MTMR6) also dephosphorylate PI3P [39].
MTMR3 and myotubularin-related protein 4 (MTMR4) are protein tyrosine phosphatases that dephosphorylate position 3 in PI and generate PI5P from PI(3,5)P2; as well as PI from PI3P. These regulate the production of PI3P, which plays a key role in inhibiting the DNA immune response by regulating the transport of STING, which is an activator of the protein tank-binding kinase (TBK1) that catalyzes the phosphorylation of interferon regulatory factor 3 (IRF3) [40]. Myotubularin-related protein 4 (MTMR4) present in endosomes and regulating their recirculation process has been identified as a novel factor interacting with the ubiquitin ligase neural precursor cell expression protein 4 (Nedd4), which is downregulated during development. MTMR4 expression decreased in atrophied muscles, while Nedd4 expression increased, and MTMR4 was ubiquitinated by Nedd4, indicating that this new relationship between MTMR4 and Nedd4 may underlie the biological process of muscle degradation [41][42][43]. MTMR8/R9 complex controls a cellular pool of PI3P that has been proposed to be essential in autophagy, a conserved intracellular process for the degradation of cytoplasmic proteins or organelles. Overexpression of both MTMR8 and MTMR9 resulted in a significant increase in the level of p62, a protein that is degraded in autophagosomes and is used to monitor autophagy. Mutations in both active and inactive myotubularins (which may play a regulatory role) are associated with diseases such as myotubular myopathy, Charcot-Marie-Tooth (CMT), and others [44][45].
CMT4B is a large group of heterogeneous diseases that are inherited in an autosomal recessive manner and have a progressive sensorimotor neuropathy. The etiopathological basis of MTMR2 and MTMR13, which affect vesicular transport in Schwann cells, is one where the loss of these proteins can lead to uncontrolled myelin folding and, ultimately, to the development of CMT4B disease. Among them, CMT4B is distinguished as having three forms associated with myotubularin family genes: CMT4B1 (MTMR2 located on chromosome 11q22), CMT4B2 (MTMR13/SBF2 located on chromosome 11p15) and CMT4B3 (MTMR5/SBF1) [46][47].
5. Neurodegenerative Diseases
Neurodegenerative diseases are still a significant problem in our aging society [48]. Their occurrence is associated with many factors, such as genetic defects and disturbances in immunological processes. Many neurodegenerative diseases have their cause in the accumulation of extra- and intracellular deposits in the nervous system [49].
Abnormalities in PIP metabolism and their linkage to neurodegenerative disorders has, to date, been mentioned by many authors. Researchers have described the influence of PIPs on the incidence of central nervous system (CNS) diseases through changes in the levels of selected PIPs as well as the enzymes catalyzing their interconversions. The vacuole 14 protein homolog (Vac14) is a frequently mentioned protein, also known as the Associated Regulator of PIKfyve (ArPIKfyve) [50][51].
Zhang et al. postulate that PI(3,5)P2 is critical to neuronal health. Vac14 protein is a regulator of the signaling lipid PI(3,5)P2 synthesis. The loss of Vac14 results in neurodegeneration processes in the midbrain and peripheral sensory neurons of mice models [52]. Other proteins, such as Fab1/PIKfyve and Fig4/Sac3, also have a regulatory role in the PI(3,5)P2 biosynthesis and its relationship with PI5P. Mutations in the genes encoding these proteins lead to the occurrence of neurological diseases, including amyotrophic lateral sclerosis (ALS) and CMT syndrome [53]. For example, Fab1 binds the PI3P and allows it to be converted to PI(3,5)P2, and consequently, the appropriate levels of PI(3,5)P2 affect the proper functioning of cells in the nervous system. The complex responsible for the Fig1 activity consists of Vac14, Vac7, Fig4, and Atg18 proteins.
Other authors have suggested that changes in the regulation of the phosphatidylinositol-3 kinase (PI3-K) are also linked to many neurodegenerative diseases. One of them is a Nieman-Pick type C disease (NPC), caused by mutations in the NPC1 and NPC2 genes. This disorder results in the deposition of neurofibrillary tangles in the CNS, the occurrence of which has been suggested to be associated with increased levels of specific kinases such as phosphatidylinositol 3-kinase (PI3K), glycogen synthase kinase (GSK-3β), and protein kinase B (Akt/PKB). The PI3K cascade leads to the activation of Akt and the inactivation of GSK-3β. Research on the NPC1-deficient mice model has shown major disruptions in the PI3K cascade. The inactivated GSK-3 and phosphorylated Akt were elevated in the neuronal cells, which indicated an unusual level of activity of PI3K in the NPC1-deficient mice brains [54]. Cathepsin D (CD) is one of the key lysosomal proteases. A lack of CD results in a neurodegenerative pediatric disease known as neuronal ceroid lipofuscinosis (NCL/Batten disease). In the research on CD-deficient brains in mice by Walls et al., a decrease in PI3K was observed [55].
6. Carcinogenesis
Carcinogenesis is a highly complex process involving environmental factors and gene mutations [56]. Critical mutations involve proto-oncogenes, tumor-suppressor genes, and DNA-repair genes. Key principles of cancer are uncontrolled proliferation, metastasis, apoptotic loss, and angiogenesis [57].
Mutation of myotubularin-related protein 7 gene (MTMR7) has been described as contributing to colorectal cancer (CRC) development. MTMR7 itself participates in decreasing insulin-mediated activation of Akt and ERK1/2 signaling, resulting in proliferation reduction of human CRC cells. In human colorectal cancers, MTMR7 has been down-regulated, which has been related to a poor prognosis [58].
PI3K mis-activation has been widely reported in cancer diseases [59][60][61]. Its activation can be signaled via various pathways including mTOR, JAK2/STAT5, Akt, or RTK [59], which gives a promising location for the development of target drugs, but can also be responsible for therapy resistance. Phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3), a subunit of the PI3K complex, takes part in the formation, initiation, and maturation of autophagosomes [62]. It has been shown that, in the condition of oncogenic herpesvirus KSHV infection, the expression of PIK3C3 is upregulated, and takes part in tumor progression and metastasis [63]. It is worth mentioning that various drugs have been developed to interfere with the PI3K/AKT/mTOR axis.
The PKR/PI4K2A axis, which takes part in the clearance of misfolded proteins in lysosomes, has been revealed to be a potential drug target—inhibiting tumor growth in the lung and breast [64].
The phosphatase and tensin homolog (PTEN) is a commonly known cancer suppressor that acts mainly via inhibiting PI3K/Akt activation [65]. This molecule controls the cell cycle, driving apoptosis among pathological conditions [66]. Mutations of PTEN are commonly known for the link to the PTEN hamartoma tumor syndrome (PHTS), which is characterized by a greater risk of cancer occurrence, includes Cowden syndrome (CS), Bannayan-Riley-Ruvalcaba syndrome (BRRS), and PTEN-related Proteus syndrome (PS) [67][68]. To date, there has been some effort put into establishing PTEN as a potential drug target.
7. Conclusions
Summing up, MI is a forerunner of many derivatives, including PIs, PIPs, IPs, GPIs, and IPGs, which take part in transmitting various signals in the eukaryotic cells. That is why gene mutations for these molecules can play a pivotal role in the development of many widespread diseases, such as cancers or neurodegenerative disorders. Cancer diseases and neurodegenerative disorders, such as AD, are wide-spread problems among developing countries, and the number of patients has been increasing for many years.
References
- Downes, C.P.; Macphee, C.H. myo-Inositol Metabolites as Cellular Signals. Eur. J. Biochem. 1990, 193, 1–18.
- Derkaczew, M.; Martyniuk, P.; Osowski, A.; Wojtkiewicz, J. Cyclitols: From Basic Understanding to Their Association with Neurodegeneration. Nutrients 2023, 15, 2029.
- Daniel, E.D.; Kew, J.N.; Maycox, P.R. Investigation of the H+– Myo -Inositol Transporter (HMIT) as a Neuronal Regulator of Phosphoinositide Signalling. Biochem. Soc. Trans. 2009, 37, 1139–1143.
- Croze, M.L.; Soulage, C.O. Potential Role and Therapeutic Interests of Myo-Inositol in Metabolic Diseases. Biochimie 2013, 95, 1811–1827.
- Schneider, S. Inositol Transport Proteins. FEBS Lett. 2015, 589, 1049–1058.
- Bizzarri, M.; Fuso, A.; Dinicola, S.; Cucina, A.; Bevilacqua, A. Pharmacodynamics and Pharmacokinetics of Inositol(s) in Health and Disease. Expert Opin. Drug Metab. Toxicol. 2016, 12, 1181–1196.
- Benvenga, S.; Antonelli, A. Inositol(s) in Thyroid Function, Growth and Autoimmunity. Rev. Endocr. Metab. Disord. 2016, 17, 471–484.
- De Craene, J.-O.; Bertazzi, D.; Bär, S.; Friant, S. Phosphoinositides, Major Actors in Membrane Trafficking and Lipid Signaling Pathways. Int. J. Mol. Sci. 2017, 18, 634.
- Elabbadi, N.; Ancelin, M.L.; Vial, H.J. Characterization of Phosphatidylinositol Synthase and Evidence of a Polyphosphoinositide Cycle in Plasmodium-Infected Erythrocytes. Mol. Biochem. Parasitol. 1994, 63, 179–192.
- Cockcroft, S. Phosphatidylinositol Transfer Proteins Couple Lipid Transport to Phosphoinositide Synthesis. Semin. Cell Dev. Biol. 2001, 12, 183–191.
- Devereaux, K.; Dall’Armi, C.; Alcazar-Roman, A.; Ogasawara, Y.; Zhou, X.; Wang, F.; Yamamoto, A.; De Camilli, P.; Di Paolo, G. Regulation of Mammalian Autophagy by Class II and III PI 3-Kinases through PI3P Synthesis. PLoS ONE 2013, 8, e76405.
- Byfield, M.P.; Murray, J.T.; Backer, J.M. HVps34 Is a Nutrient-Regulated Lipid Kinase Required for Activation of P70 S6 Kinase. J. Biol. Chem. 2005, 280, 33076–33082.
- Strahl, T.; Thorner, J. Synthesis and Function of Membrane Phosphoinositides in Budding Yeast, Saccharomyces Cerevisiae. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 353–404.
- Dickson, E.J.; Hille, B. Understanding Phosphoinositides: Rare, Dynamic, and Essential Membrane Phospholipids. Biochem. J. 2019, 476, 1–23.
- Whitley, P.; Hinz, S.; Doughty, J. Arabidopsis FAB1/PIKfyve Proteins Are Essential for Development of Viable Pollen. Plant Physiol. 2009, 151, 1812–1822.
- Buckley, C.M.; Heath, V.L.; Guého, A.; Bosmani, C.; Knobloch, P.; Sikakana, P.; Personnic, N.; Dove, S.K.; Michell, R.H.; Meier, R.; et al. PIKfyve/Fab1 Is Required for Efficient V-ATPase and Hydrolase Delivery to Phagosomes, Phagosomal Killing, and Restriction of Legionella Infection. PLoS Pathog. 2019, 15, e1007551.
- Sbrissa, D.; Ikonomov, O.C.; Shisheva, A. PIKfyve, a Mammalian Ortholog of Yeast Fab1p Lipid Kinase, Synthesizes 5-Phosphoinositides. J. Biol. Chem. 1999, 274, 21589–21597.
- Rajala, A.; Rajala, R.; Teel, K.; Rajala, R.V.S. Ribosomal Targeting Strategy and Nuclear Labeling to Analyze Photoreceptor Phosphoinositide Signatures. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2022, 1867, 159161.
- Lu, N.; Shen, Q.; Mahoney, T.R.; Neukomm, L.J.; Wang, Y.; Zhou, Z. Two PI 3-Kinases and One PI 3-Phosphatase Together Establish the Cyclic Waves of Phagosomal PtdIns(3)P Critical for the Degradation of Apoptotic Cells. PLoS Biol. 2012, 10, e1001245.
- York, H.M.; Joshi, K.; Wright, C.S.; Kreplin, L.Z.; Rodgers, S.J.; Moorthi, U.K.; Gandhi, H.; Patil, A.; Mitchell, C.A.; Iyer-Biswas, S.; et al. Deterministic Early Endosomal Maturations Emerge from a Stochastic Trigger-and-Convert Mechanism. Nat. Commun. 2023, 14, 4652.
- Godi, A. Regulation and Molecular Composition of the Golgi-Associated Spectrin Skeleton. Ph.D. Thesis, Open University (United Kingdom), Milton Keynes, UK, 2002.
- Foti, M.; Audhya, A.; Emr, S.D. Sac1 Lipid Phosphatase and Stt4 Phosphatidylinositol 4-Kinase Regulate a Pool of Phosphatidylinositol 4-Phosphate That Functions in the Control of the Actin Cytoskeleton and Vacuole Morphology. Mol. Biol. Cell 2001, 12, 2396–2411.
- Böttcher-Sehlmeyer, C. Subcellular Localization and Molecular Interactions of Phosphoinositide 5′-Phosphatases of the Yeast Synaptojanin-like Protein Family. Ph.D. Thesis, University of Stuttgart, Stuttgart, Germany, 2006.
- Ebrahimzadeh, Z. Exploring the Roles of Phosphoinositides in the Biology of the Malaria Parasite Plasmodium Falciparum. Ph.D. Thesis, Université Laval, Quebec City, QC, Canada, 2019.
- Lowe, M. Structure and Function of the Lowe Syndrome Protein OCRL1: Structure and Function of OCRL1. Traffic 2005, 6, 711–719.
- Mehta, Z.B.; Pietka, G.; Lowe, M. The Cellular and Physiological Functions of the Lowe Syndrome Protein OCRL1. Traffic 2014, 15, 471–487.
- Tariq, K.; Luikart, B.W. Striking a Balance: PIP2 and PIP3 Signaling in Neuronal Health and Disease. Explor. Neuroprot. Ther. 2021, 1, 86.
- Antonietta De Matteis, M.; Di Campli, A.; Godi, A. The Role of the Phosphoinositides at the Golgi Complex. Biochim. Biophys. Acta Mol. Cell Res. 2005, 1744, 396–405.
- Jungbluth, H.; Wallgren-Pettersson, C.; Laporte, J. Centronuclear (Myotubular) Myopathy. Orphanet J. Rare Dis. 2008, 3, 26.
- Blondeau, F.; Laporte, J.; Bodin, S.; Superti-Furga, G.; Payrastre, B.; Mandel, J.-L. Myotubularin, a Phosphatase Deficient in Myotubular Myopathy, Acts on Phosphatidylinositol 3-Kinase and Phosphatidylinositol 3-Phosphate Pathway. Hum. Mol. Genet. 2000, 9, 2223–2229.
- Taylor, G.S.; Maehama, T.; Dixon, J.E. Myotubularin, a Protein Tyrosine Phosphatase Mutated in Myotubular Myopathy, Dephosphorylates the Lipid Second Messenger, Phosphatidylinositol 3-Phosphate. Proc. Natl. Acad. Sci. USA 2000, 97, 8910–8915.
- Tsujita, K.; Itoh, T.; Ijuin, T.; Yamamoto, A.; Shisheva, A.; Laporte, J.; Takenawa, T. Myotubularin Regulates the Function of the Late Endosome through the GRAM Domain-Phosphatidylinositol 3,5-Bisphosphate Interaction. J. Biol. Chem. 2004, 279, 13817–13824.
- Lawlor, M.W.; Dowling, J.J. X-Linked Myotubular Myopathy. Neuromuscul. Disord. 2021, 31, 1004–1012.
- Gupta, V.A.; Hnia, K.; Smith, L.L.; Gundry, S.R.; McIntire, J.E.; Shimazu, J.; Bass, J.R.; Talbot, E.A.; Amoasii, L.; Goldman, N.E.; et al. Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-Related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish. PLoS Genet. 2013, 9, e1003583.
- Cowling, B.S.; Chevremont, T.; Prokic, I.; Kretz, C.; Ferry, A.; Coirault, C.; Koutsopoulos, O.; Laugel, V.; Romero, N.B.; Laporte, J. Reducing Dynamin 2 Expression Rescues X-Linked Centronuclear Myopathy. J. Clin. Investig. 2014, 124, 1350–1363.
- Lionello, V.M.; Nicot, A.-S.; Sartori, M.; Kretz, C.; Kessler, P.; Buono, S.; Djerroud, S.; Messaddeq, N.; Koebel, P.; Prokic, I.; et al. Amphiphysin 2 Modulation Rescues Myotubular Myopathy and Prevents Focal Adhesion Defects in Mice. Sci. Transl. Med. 2019, 11, eaav1866.
- Schaletzky, J.; Dove, S.K.; Short, B.; Lorenzo, O.; Clague, M.J.; Barr, F.A. Phosphatidylinositol-5-Phosphate Activation and Conserved Substrate Specificity of the Myotubularin Phosphatidylinositol 3-Phosphatases. Curr. Biol. 2003, 13, 504–509.
- Kim, S.-A.; Taylor, G.S.; Torgersen, K.M.; Dixon, J.E. Myotubularin and MTMR2, Phosphatidylinositol 3-Phosphatases Mutated in Myotubular Myopathy and Type 4B Charcot-Marie-Tooth Disease. J. Biol. Chem. 2002, 277, 4526–4531.
- Bong, S.M.; Son, K.; Yang, S.-W.; Park, J.-W.; Cho, J.-W.; Kim, K.-T.; Kim, H.; Kim, S.J.; Kim, Y.J.; Lee, B.I. Crystal Structure of Human Myotubularin-Related Protein 1 Provides Insight into the Structural Basis of Substrate Specificity. PLoS ONE 2016, 11, e0152611.
- Dewi Pamungkas Putri, D.; Kawasaki, T.; Murase, M.; Sueyoshi, T.; Deguchi, T.; Ori, D.; Suetsugu, S.; Kawai, T. PtdIns3P Phosphatases MTMR3 and MTMR4 Negatively Regulate Innate Immune Responses to DNA through Modulating STING Trafficking. J. Biol. Chem. 2019, 294, 8412–8423.
- Plant, P.J.; Correa, J.; Goldenberg, N.; Bain, J.; Batt, J. The Inositol Phosphatase MTMR4 Is a Novel Target of the Ubiquitin Ligase Nedd4. Biochem. J. 2009, 419, 57–63.
- Kumar, P.; Munnangi, P.; Chowdary, K.R.; Shah, V.J.; Shinde, S.R.; Kolli, N.R.; Halehalli, R.R.; Nagarajaram, H.A.; Maddika, S. A Human Tyrosine Phosphatase Interactome Mapped by Proteomic Profiling. J. Proteome Res. 2017, 16, 2789–2801.
- Naughtin, M.J.; Sheffield, D.A.; Rahman, P.; Hughes, W.E.; Gurung, R.; Stow, J.L.; Nandurkar, H.H.; Dyson, J.M.; Mitchell, C.A. The Myotubularin Phosphatase MTMR4 Regulates Sorting from Early Endosomes. J. Cell Sci. 2010, 123, 3071–3083.
- Zou, J.; Zhang, C.; Marjanovic, J.; Kisseleva, M.V.; Majerus, P.W.; Wilson, M.P. Myotubularin-Related Protein (MTMR) 9 Determines the Enzymatic Activity, Substrate Specificity, and Role in Autophagy of MTMR8. Proc. Natl. Acad. Sci. USA 2012, 109, 9539–9544.
- Yoo, K.-Y.; Son, J.Y.; Lee, J.U.; Shin, W.; Im, D.-W.; Kim, S.J.; Ryu, S.E.; Heo, Y.-S. Structure of the Catalytic Phosphatase Domain of MTMR8: Implications for Dimerization, Membrane Association and Reversible Oxidation. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 1528–1539.
- Previtali, S.C.; Quattrini, A.; Bolino, A. Charcot–Marie–Tooth Type 4B Demyelinating Neuropathy: Deciphering the Role of MTMR Phosphatases. Expert Rev. Mol. Med. 2007, 9, 1–16.
- Romani, M.; Mehawej, C.; Mazza, T.; Mégarbané, A.; Valente, E.M. “Fork and Bracket” Syndrome Expands the Spectrum of SBF1 -Related Sensory Motor Polyneuropathies. Neurol. Genet. 2016, 2, e61.
- Dugger, B.N.; Dickson, D.W. Pathology of Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2017, 9, a028035.
- Voet, S.; Srinivasan, S.; Lamkanfi, M.; Van Loo, G. Inflammasomes in Neuroinflammatory and Neurodegenerative Diseases. EMBO Mol. Med. 2019, 11, e10248.
- Carmel, L.; Efroni, S.; White, P.D.; Aslakson, E.; Vollmer-Conna, U.; Rajeevan, M.S. Gene Expression Profile of Empirically Delineated Classes of Unexplained Chronic Fatigue. Pharmacogenomics 2006, 7, 375–386.
- Fogarty, K.; Kashem, M.; Bauer, A.; Bernardino, A.; Brennan, D.; Cook, B.; Farrow, N.; Molinaro, T.; Nelson, R. Development of Three Orthogonal Assays Suitable for the Identification and Qualification of PIKfyve Inhibitors. ASSAY Drug Dev. Technol. 2017, 15, 210–219.
- Zhang, Y.; Zolov, S.N.; Chow, C.Y.; Slutsky, S.G.; Richardson, S.C.; Piper, R.C.; Yang, B.; Nau, J.J.; Westrick, R.J.; Morrison, S.J.; et al. Loss of Vac14, a Regulator of the Signaling Lipid Phosphatidylinositol 3,5-Bisphosphate, Results in Neurodegeneration in Mice. Proc. Natl. Acad. Sci. USA 2007, 104, 17518–17523.
- McCartney, A.J.; Zhang, Y.; Weisman, L.S. Phosphatidylinositol 3,5-Bisphosphate: Low Abundance, High Significance: Prospects & Overviews. BioEssays 2014, 36, 52–64.
- Bi, X.; Liu, J.; Yao, Y.; Baudry, M.; Lynch, G. Deregulation of the Phosphatidylinositol-3 Kinase Signaling Cascade Is Associated with Neurodegeneration in Npc1−/− Mouse Brain. Am. J. Pathol. 2005, 167, 1081–1092.
- Walls, K.C.; Klocke, B.J.; Saftig, P.; Shibata, M.; Uchiyama, Y.; Roth, K.A.; Shacka, J.J. Altered Regulation of Phosphatidylinositol 3-Kinase Signaling in Cathepsin D-Deficient Brain. Autophagy 2007, 3, 222–229.
- Peters, J.M.; Gonzalez, F.J. The Evolution of Carcinogenesis. Toxicol. Sci. 2018, 165, 272–276.
- Bertram, J.S. The Molecular Biology of Cancer. Mol. Asp. Med. 2001, 21, 167–223.
- Weidner, P.; Söhn, M.; Schroeder, T.; Helm, L.; Hauber, V.; Gutting, T.; Betge, J.; Röcken, C.; Rohrbacher, F.N.; Pattabiraman, V.R.; et al. Myotubularin-Related Protein 7 Activates Peroxisome Proliferator-Activated Receptor-Gamma. Oncogenesis 2020, 9, 59.
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635.
- Fattahi, S.; Amjadi-Moheb, F.; Tabaripour, R.; Ashrafi, G.H.; Akhavan-Niaki, H. PI3K/AKT/MTOR Signaling in Gastric Cancer: Epigenetics and Beyond. Life Sci. 2020, 262, 118513.
- Ediriweera, M.K.; Tennekoon, K.H.; Samarakoon, S.R. Role of the PI3K/AKT/MTOR Signaling Pathway in Ovarian Cancer: Biological and Therapeutic Significance. Semin. Cancer Biol. 2019, 59, 147–160.
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42.
- Hu, H.; Dong, J.; Liang, D.; Gao, Z.; Bai, L.; Sun, R.; Hu, H.; Zhang, H.; Dong, Y.; Lan, K. Genome-Wide Mapping of the Binding Sites and Structural Analysis of Kaposi’s Sarcoma-Associated Herpesvirus Viral Interferon Regulatory Factor 2 Reveal That It Is a DNA-Binding Transcription Factor. J. Virol. 2016, 90, 1158–1168.
- Pataer, A.; Ozpolat, B.; Shao, R.; Cashman, N.R.; Plotkin, S.S.; Samuel, C.E.; Lin, S.H.; Kabil, N.N.; Wang, J.; Majidi, M.; et al. Therapeutic Targeting of the PI4K2A/PKR Lysosome Network Is Critical for Misfolded Protein Clearance and Survival in Cancer Cells. Oncogene 2020, 39, 801–813.
- Maehama, T.; Dixon, J.E. The Tumor Suppressor, PTEN/MMAC1, Dephosphorylates the Lipid Second Messenger, Phosphatidylinositol 3,4,5-Trisphosphate. J. Biol. Chem. 1998, 273, 13375–13378.
- Chung, J.-H.; Ginn-Pease, M.E.; Eng, C. Phosphatase and Tensin Homologue Deleted on Chromosome 10 (PTEN) Has Nuclear Localization Signal–Like Sequences for Nuclear Import Mediated by Major Vault Protein. Cancer Res. 2005, 65, 4108–4116.
- Li, D.-M.; Sun, H. TEP1, Encoded by a Candidate Tumor Suppressor Locus, Is a Novel Protein Tyrosine Phosphatase Regulated by Transforming Growth Factor. Cancer Res. 1997, 57, 2124–2129.
- Davidson, L.; Maccario, H.; Perera, N.M.; Yang, X.; Spinelli, L.; Tibarewal, P.; Glancy, B.; Gray, A.; Weijer, C.J.; Downes, C.P.; et al. Suppression of Cellular Proliferation and Invasion by the Concerted Lipid and Protein Phosphatase Activities of PTEN. Oncogene 2010, 29, 687–697.
More
Information
Subjects:
Medicine, Research & Experimental
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
625
Revisions:
2 times
(View History)
Update Date:
04 Dec 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




