
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Giuseppe Murdaca | -- | 3991 | 2023-11-24 09:00:40 | | | |
| 2 | Lindsay Dong | + 1 word(s) | 3992 | 2023-11-27 06:19:59 | | |
Video Upload Options
Multiple sclerosis (MS), a condition characterised by demyelination and axonal damage in the central nervous system, is due to autoreactive immune cells that recognise myelin antigens. Alteration of the immune balance can promote the onset of immune deficiencies, loss of immunosurveillance, and/or development of autoimmune disorders such as MS. Numerous enzymes, transcription factors, signal transducers, and membrane proteins contribute to the control of immune system activity. The “transcriptional machine” of eukaryotic cells is a complex system composed not only of mRNA but also of non-coding elements grouped together in the set of non-coding RNAs. ncRNAs play a crucial role in numerous cellular functions, gene expression, and the pathogenesis of many immune disorders.
1. Multiple Sclerosis
2. Non-Coding RNAs
3. Multiple Sclerosis and circRNAs
3.1. Epigenetic Mechanisms
3.2. circRNAs Biomarkers in MS
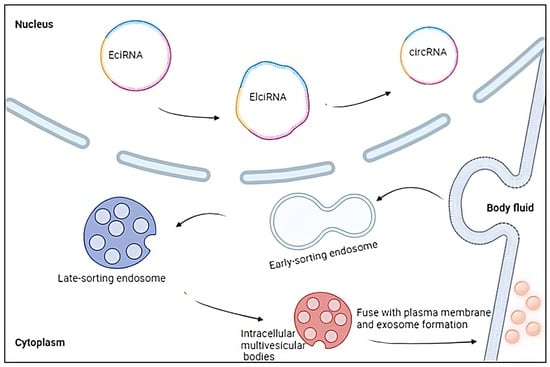
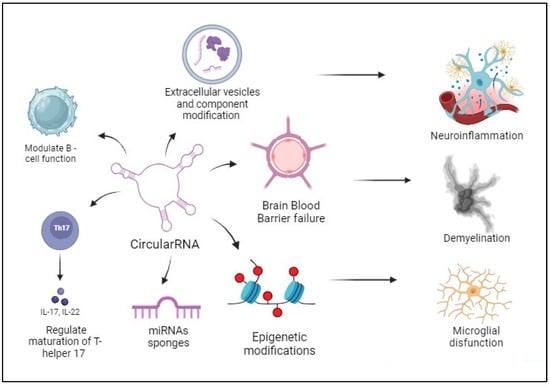
3.3. Extracellular Vesicles and circRNAs
3.4. circRNAs Genetic Variation in MS
3.5. circRNAs and B-Cell Function
3.6. Roles of circRNAs in MS
3.6.1. hsa_circ_0106803 of GSDMB Gene
3.6.2. circ_HECW2 and the Dysfunction of the Blood–Brain Barrier
3.6.3. hsa_circ_0106803 Modulates the Expression of ASIC1a mRNA
3.6.4. circINPP4B Regulates Th17 Cell Differentiation
3.6.5. hsa_circRNA_101145 and hsa_circRNA_001896 in Patients with Relapse-Remitting MS
4. Conclusions
References
- Browne, P.; Chandraratna, D.; Angood, C.; Tremlett, H.; Baker, C.; Taylor, B.V.; Thompson, A.J. Atlas of Multiple Sclerosis 2013: A growing global problem with widespread inequity. Neurology 2014, 83, 1022–1024.
- GBD 2016 Neurology Collaborators. Global, regional, and national burden of neurological disorders, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 459–480.
- Goldenberg, M.M. Multiple sclerosis review. Pharm. Ther. 2012, 37, 175–184.
- Lublin, F.D.; Reingold, S.C.; Cohen, J.A.; Cutter, G.R.; Sørensen, P.S.; Thompson, A.J.; Wolinsky, J.S.; Balcer, L.J.; Banwell, B.; Barkhof, F.; et al. Defining the clinical course of multiple sclerosis: The 2013 revisions. Neurology 2014, 83, 278–286.
- Cree, B.A.C.; Arnold, D.L.; Chataway, J.; Chitnis, T.; Fox, R.J.; Pozo Ramajo, A.; Murphy, N.; Lassmann, H. Secondary Progressive Multiple Sclerosis: New Insights. Neurology 2021, 97, 378–388.
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173.
- Alcina, A.; Abad-Grau Mdel, M.; Fedetz, M.; Izquierdo, G.; Lucas, M.; Fernández, O.; Ndagire, D.; Catalá-Rabasa, A.; Ruiz, A.; Gayán, J.; et al. Multiple sclerosis risk variant HLA-DRB1*1501 associates with high expression of DRB1 gene in different human populations. PLoS ONE 2012, 7, e29819.
- Sintzel, M.B.; Rametta, M.; Reder, A.T. Vitamin D and Multiple Sclerosis: A Comprehensive Review. Neurol. Ther. 2018, 7, 59–85.
- Guan, Y.; Jakimovski, D.; Ramanathan, M.; Weinstock-Guttman, B.; Zivadinov, R. The role of Epstein-Barr virus in multiple sclerosis: From molecular pathophysiology to in vivo imaging. Neural Regen. Res. 2019, 14, 373–386.
- Olsson, T.; Barcellos, L.F.; Alfredsson, L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat. Rev. Neurol. 2017, 13, 25–36.
- Liu, R.; Du, S.; Zhao, L.; Jain, S.; Sahay, K.; Rizvanov, A.; Lezhnyova, V.; Khaibullin, T.; Martynova, E.; Khaiboullina, S.; et al. Autoreactive lymphocytes in multiple sclerosis: Pathogenesis and treatment target. Front. Immunol. 2022, 13, 996469.
- Mahad, D.H.; Trapp, B.D.; Lassmann, H. Pathological mechanisms in progressive multiple sclerosis. Lancet Neurol. 2015, 14, 183–193.
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108.
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563.
- Rodriguez, P.D.; Paculova, H.; Kogut, S.; Heath, J.; Schjerven, H.; Frietze, S. Non-Coding RNA Signatures of B-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2021, 22, 2683.
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491.
- Frankish, A.; Diekhans, M.; Ferreira, A.M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773.
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745.
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314.
- Seal, R.L.; Chen, L.L.; Griffiths-Jones, S.; Lowe, T.M.; Mathews, M.B.; O’Reilly, D.; Pierce, A.J.; Stadler, P.F.; Ulitsky, I.; Wolin, S.L.; et al. A guide to naming human non-coding RNA genes. EMBO J. 2020, 39, e103777.
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139.
- Dozmorov, M.G.; Giles, C.B.; Koelsch, K.A.; Wren, J.D. Systematic classification of non-coding RNAs by epigenomic similarity. BMC Bioinform. 2013, 14 (Suppl. S14), S2.
- Avenoso, A.; Campo, S.; Scuruchi, M.; Mania, M.; Innao, V.; D’Ascola, A.; Mandraffino, G.; Allegra, A.G.; Musolino, C.; Allegra, A. Quantitative polymerase Chain reaction profiling of microRNAs in peripheral lymph-monocytes from MGUS subjects. Pathol. Res. Pract. 2021, 218, 153317.
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639.
- Chen, Q.; Meng, X.; Liao, Q.; Chen, M. Versatile interactions and bioinformatics analysis of noncoding RNAs. Brief. Bioinform. 2019, 20, 1781–1794.
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027.
- Adachi, H.; Yu, Y.T. Insight into the mechanisms and functions of spliceosomal snRNA pseudouridylation. World J. Biol. Chem. 2014, 5, 398–408.
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992.
- Kumar, P.; Kuscu, C.; Dutta, A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 2016, 41, 679–689.
- Peschansky, V.J.; Wahlestedt, C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2014, 9, 3–12.
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565.
- Heffler, E.; Allegra, A.; Pioggia, G.; Picardi, G.; Musolino, C.; Gangemi, S. MicroRNA Profiling in Asthma: Potential Biomarkers and Therapeutic Targets. Am. J. Respir. Cell Mol. Biol. 2017, 57, 642–650.
- Caserta, S.; Gangemi, S.; Murdaca, G.; Allegra, A. Gender Differences and miRNAs Expression in Cancer: Implications on Prognosis and Susceptibility. Int. J. Mol. Sci. 2023, 24, 11544.
- Han, B.W.; Zamore, P.D. piRNAs. Curr. Biol. 2014, 24, R730–R733.
- Musolino, C.; Oteri, G.; Allegra, A.; Mania, M.; D’Ascola, A.; Avenoso, A.; Innao, V.; Allegra, A.G.; Campo, S. Altered microRNA expression profile in the peripheral lymphoid compartment of multiple myeloma patients with bisphosphonate-induced osteonecrosis of the jaw. Ann. Hematol. 2018, 97, 1259–1269.
- Allegra, A.; Cicero, N.; Tonacci, A.; Musolino, C.; Gangemi, S. Circular RNA as a Novel Biomarker for Diagnosis and Prognosis and Potential Therapeutic Targets in Multiple Myeloma. Cancers 2022, 14, 1700.
- Chen, C.Y.; Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995, 268, 415–417.
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856.
- Kos, A.; Dijkema, R.; Arnberg, A.C.; van der Meide, P.H.; Schellekens, H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986, 323, 558–560.
- Arnberg, A.C.; Van Ommen, G.J.; Grivell, L.A.; Van Bruggen, E.F.; Borst, P. Some yeast mitochondrial RNAs are circular. Cell 1980, 19, 313–319.
- Cocquerelle, C.; Daubersies, P.; Majérus, M.A.; Kerckaert, J.P.; Bailleul, B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992, 11, 1095–1098.
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160.
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030.
- Bos, S.D.; Page, C.M.; Andreassen, B.K.; Elboudwarej, E.; Gustavsen, M.W.; Briggs, F.; Quach, H.; Leikfoss, I.S.; Bjølgerud, A.; Berge, T.; et al. Genome-wide DNA methylation profiles indicate CD8+ T cell hypermethylation in multiple sclerosis. PLoS ONE 2015, 10, e0117403.
- Ferreira, H.J.; Davalos, V.; de Moura, M.C.; Soler, M.; Perez-Salvia, M.; Bueno-Costa, A.; Setien, F.; Moran, S.; Villanueva, A.; Esteller, M. Circular RNA CpG island hypermethylation-associated silencing in human cancer. Oncotarget 2018, 9, 29208–29219.
- Shayevitch, R.; Askayo, D.; Keydar, I.; Ast, G. The importance of DNA methylation of exons on alternative splicing. RNA 2018, 24, 1351–1362.
- Xu, T.; Wang, L.; Jia, P.; Song, X.; Zhao, Z. An Integrative Transcriptomic and Methylation Approach for Identifying Differentially Expressed Circular RNAs Associated with DNA Methylation Change. Biomedicines 2021, 9, 657.
- Cardamone, G.; Paraboschi, E.M.; Soldà, G.; Liberatore, G.; Rimoldi, V.; Cibella, J.; Airi, F.; Tisato, V.; Cantoni, C.; Gallia, F.; et al. The circular RNA landscape in multiple sclerosis: Disease-specific associated variants and exon methylation shape circular RNA expression profile. Mult. Scler. Relat. Disord. 2023, 69, 104426.
- Kular, L.; Liu, Y.; Ruhrmann, S.; Zheleznyakova, G.; Marabita, F.; Gomez-Cabrero, D.; James, T.; Ewing, E.; Lindén, M.; Górnikiewicz, B.; et al. DNA methylation as a mediator of HLA-DRB1*15:01 and a protective variant in multiple sclerosis. Nat. Commun. 2018, 9, 2397.
- Wang, Y.; Murakami, Y.; Yasui, T.; Wakana, S.; Kikutani, H.; Kinoshita, T.; Maeda, Y. Significance of glycosylphosphatidylinositol-anchored protein enrichment in lipid rafts for the control of autoimmunity. J. Biol. Chem. 2013, 288, 25490–25499.
- Gerke, V.; Moss, S.E. Annexins: From structure to function. Physiol. Rev. 2002, 82, 331–371.
- El-Abd, N.; Fawzy, A.; Elbaz, T.; Hamdy, S. Evaluation of annexin A2 and as potential biomarkers for hepatocellular carcinoma. Tumour Biol. 2016, 37, 211–216.
- Cañas, F.; Simonin, L.; Couturaud, F.; Renaudineau, Y. Annexin A2 autoantibodies in thrombosis and autoimmune diseases. Thromb. Res. 2015, 135, 226–230.
- Murakami, H.; Wang, Y.; Hasuwa, H.; Maeda, Y.; Kinoshita, T.; Murakami, Y. Enhanced response of T lymphocytes from Pgap3 knockout mouse: Insight into roles of fatty acid remodeling of GPI anchored proteins. Biochem. Biophys. Res. Commun. 2012, 417, 1235–1241.
- Iparraguirre, L.; Muñoz-Culla, M.; Prada-Luengo, I.; Castillo-Triviño, T.; Olascoaga, J.; Otaegui, D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum. Mol. Genet. 2017, 26, 3564–3572.
- Fang, W.; Fa, Z.Z.; Xie, Q.; Wang, G.Z.; Yi, J.; Zhang, C.; Meng, G.X.; Gu, J.L.; Liao, W.Q. Complex Roles of Annexin A2 in Host Blood-Brain Barrier Invasion by Cryptococcus neoformans. CNS Neurosci. Ther. 2017, 23, 291–300.
- Lopez-Ramirez, M.A.; Wu, D.; Pryce, G.; Simpson, J.E.; Reijerkerk, A.; King-Robson, J.; Kay, O.; de Vries, H.E.; Hirst, M.C.; Sharrack, B.; et al. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J. 2014, 28, 2551–2565.
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066.
- Selmaj, I.; Cichalewska, M.; Namiecinska, M.; Galazka, G.; Horzelski, W.; Selmaj, K.W.; Mycko, M.P. Global exosome transcriptome profiling reveals biomarkers for multiple sclerosis. Ann. Neurol. 2017, 81, 703–717.
- Fanale, D.; Taverna, S.; Russo, A.; Bazan, V. Circular RNA in Exosomes. Adv. Exp. Med. Biol. 2018, 1087, 109–117.
- Lukiw, W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 2013, 4, 307.
- Iparraguirre, L.; Alberro, A.; Hansen, T.B.; Castillo-Triviño, T.; Muñoz-Culla, M.; Otaegui, D. Profiling of Plasma Extracellular Vesicle Transcriptome Reveals That circRNAs Are Prevalent and Differ between Multiple Sclerosis Patients and Healthy Controls. Biomedicines 2021, 9, 1850.
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606.
- Iranifar, E.; Seresht, B.M.; Momeni, F.; Fadaei, E.; Mehr, M.H.; Ebrahimi, Z.; Rahmati, M.; Kharazinejad, E.; Mirzaei, H. Exosomes and microRNAs: New potential therapeutic candidates in Alzheimer disease therapy. J. Cell Physiol. 2019, 234, 2296–2305.
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984.
- Mattingly, J.; Li, Y.; Bihl, J.C.; Wang, J. The promise of exosome applications in treating central nervous system diseases. CNS Neurosci. Ther. 2021, 27, 1437–1445.
- Paraboschi, E.M.; Cardamone, G.; Soldà, G.; Duga, S.; Asselta, R. Interpreting Non-coding Genetic Variation in Multiple Sclerosis Genome-Wide Associated Regions. Front. Genet. 2018, 9, 647.
- Brucklacher-Waldert, V.; Stuerner, K.; Kolster, M.; Wolthausen, J.; Tolosa, E. Phenotypical and functional characterization of T helper 17 cells in multiple sclerosis. Brain 2009, 132 Pt 12, 3329–3341.
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312.
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383.
- Krishna Swaroop, A.; Krishnan Namboori, P.K.; Esakkimuthukumar, M.; Praveen, T.K.; Nagarjuna, P.; Patnaik, S.K.; Selvaraj, J. Leveraging decagonal in-silico strategies for uncovering IL-6 inhibitors with precision. Comput. Biol. Med. 2023, 163, 107231.
- Najafi, S.; Aghaei Zarch, S.M.; Majidpoor, J.; Pordel, S.; Aghamiri, S.; Fatih Rasul, M.; Asemani, Y.; Vakili, O.; Mohammadi, V.; Movahedpour, A.; et al. Recent insights into the roles of circular RNAs in human brain development and neurologic diseases. Int. J. Biol. Macromol. 2023, 225, 1038–1048.
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429.
- Berger, T.; Rubner, P.; Schautzer, F.; Egg, R.; Ulmer, H.; Mayringer, I.; Dilitz, E.; Deisenhammer, F.; Reindl, M. Antimyelin antibodies as a predictor of clinically definite multiple sclerosis after a first demyelinating event. N. Engl. J. Med. 2003, 349, 139–145.
- Allegra, A.; Caserta, S.; Genovese, S.; Pioggia, G.; Gangemi, S. Gender Differences in Oxidative Stress in Relation to Cancer Susceptibility and Survival. Antioxidants 2023, 12, 1255.
- Magliozzi, R.; Howell, O.; Vora, A.; Serafini, B.; Nicholas, R.; Puopolo, M.; Reynolds, R.; Aloisi, F. Meningeal B-cell follicles in secondary progressive multiple sclerosis associate with early onset of disease and severe cortical pathology. Brain 2007, 130 Pt 4, 1089–1104.
- Hauser, S.L.; Bar-Or, A.; Cohen, J.A.; Comi, G.; Correale, J.; Coyle, P.K.; Cross, A.H.; de Seze, J.; Leppert, D.; Montalban, X.; et al. Ofatumumab versus Teriflunomide in Multiple Sclerosis. N. Engl. J. Med. 2020, 383, 546–557.
- Montalban, X.; Hauser, S.L.; Kappos, L.; Arnold, D.L.; Bar-Or, A.; Comi, G.; de Seze, J.; Giovannoni, G.; Hartung, H.P.; Hemmer, B.; et al. Ocrelizumab versus Placebo in Primary Progressive Multiple Sclerosis. N. Engl. J. Med. 2017, 376, 209–220.
- Zurawska, A.E.; Mycko, M.P.; Selmaj, I.; Raine, C.S.; Selmaj, K.W. Multiple Sclerosis: circRNA Profile Defined Reveals Links to B-Cell Function. Neurol. Neuroimmunol. Neuroinflamm. 2021, 8, e1041.
- Chou, J.; Alazami, A.M.; Jaber, F.; Hoyos-Bachiloglu, R.; Jones, J.; Weeks, S.; Alosaimi, M.F.; Bainter, W.; Cangemi, B.; Badran, Y.R.; et al. Hypomorphic variants in AK2 reveal the contribution of mitochondrial function to B-cell activation. J. Allergy Clin. Immunol. 2020, 146, 192–202.
- Heizmann, B.; Kastner, P.; Chan, S. The Ikaros family in lymphocyte development. Curr. Opin. Immunol. 2018, 51, 14–23.
- Keshari, P.K.; Harbo, H.F.; Myhr, K.M.; Aarseth, J.H.; Bos, S.D.; Berge, T. Allelic imbalance of multiple sclerosis susceptibility genes IKZF3 and IQGAP1 in human peripheral blood. BMC Genet. 2016, 17, 59.
- Cortés, M.; Georgopoulos, K. Aiolos is required for the generation of high affinity bone marrow plasma cells responsible for long-term immunity. J. Exp. Med. 2004, 199, 209–219.
- Marinho, S.; Custovic, A.; Marsden, P.; Smith, J.A.; Simpson, A. 17q12-21 variants are associated with asthma and interact with active smoking in an adult population from the United Kingdom. Ann. Allergy Asthma. Immunol. 2012, 108, 402–411.e9.
- Barrett, J.C.; Clayton, D.G.; Concannon, P.; Akolkar, B.; Cooper, J.D.; Erlich, H.A.; Julier, C.; Morahan, G.; Nerup, J.; Nierras, C.; et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009, 41, 703–707.
- Tamura, M.; Tanaka, S.; Fujii, T.; Aoki, A.; Komiyama, H.; Ezawa, K.; Sumiyama, K.; Sagai, T.; Shiroishi, T. Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics 2007, 89, 618–629.
- Evsyukova, I.; Somarelli, J.A.; Gregory, S.G.; Garcia-Blanco, M.A. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biol. 2010, 7, 462–473.
- Cardamone, G.; Paraboschi, E.M.; Rimoldi, V.; Duga, S.; Soldà, G.; Asselta, R. The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis. Int. J. Mol. Sci. 2017, 18, 576.
- Qu, X.; Han, J.; Zhang, Y.; Wang, X.; Fan, H.; Hua, F.; Yao, R. TLR4-RelA-miR-30a signal pathway regulates Th17 differentiation during experimental autoimmune encephalomyelitis development. J. Neuroinflamm. 2019, 16, 183.
- Quinn, J.L.; Kumar, G.; Agasing, A.; Ko, R.M.; Axtell, R.C. Role of TFH Cells in Promoting T Helper 17-Induced Neuroinflammation. Front. Immunol. 2018, 9, 382.
- Qu, X.; Zhou, J.; Wang, T.; Han, J.; Ma, L.; Yu, H.; Geng, D.; Fan, H.; Zhang, Q.; Hua, F.; et al. MiR-30a inhibits Th17 differentiation and demyelination of EAE mice by targeting the IL-21R. Brain Behav. Immun. 2016, 57, 193–199.
- Baecher-Allan, C.; Kaskow, B.J.; Weiner, H.L. Multiple Sclerosis: Mechanisms and Immunotherapy. Neuron 2018, 97, 742–768.
- Hemmer, B.; Kerschensteiner, M.; Korn, T. Role of the innate and adaptive immune responses in the course of multiple sclerosis. Lancet Neurol. 2015, 14, 406–419.
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370.
- Cencioni, M.T.; Mattoscio, M.; Magliozzi, R.; Bar-Or, A.; Muraro, P.A. B cells in multiple sclerosis—From targeted depletion to immune reconstitution therapies. Nat. Rev. Neurol. 2021, 17, 399–414.




