
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yoshimitsu Kiiriyama | -- | 1614 | 2023-11-22 08:54:41 | | | |
| 2 | Jessie Wu | -1 word(s) | 1613 | 2023-11-22 09:19:10 | | | | |
| 3 | Jessie Wu | + 4 word(s) | 1617 | 2023-11-22 09:28:10 | | |
Video Upload Options
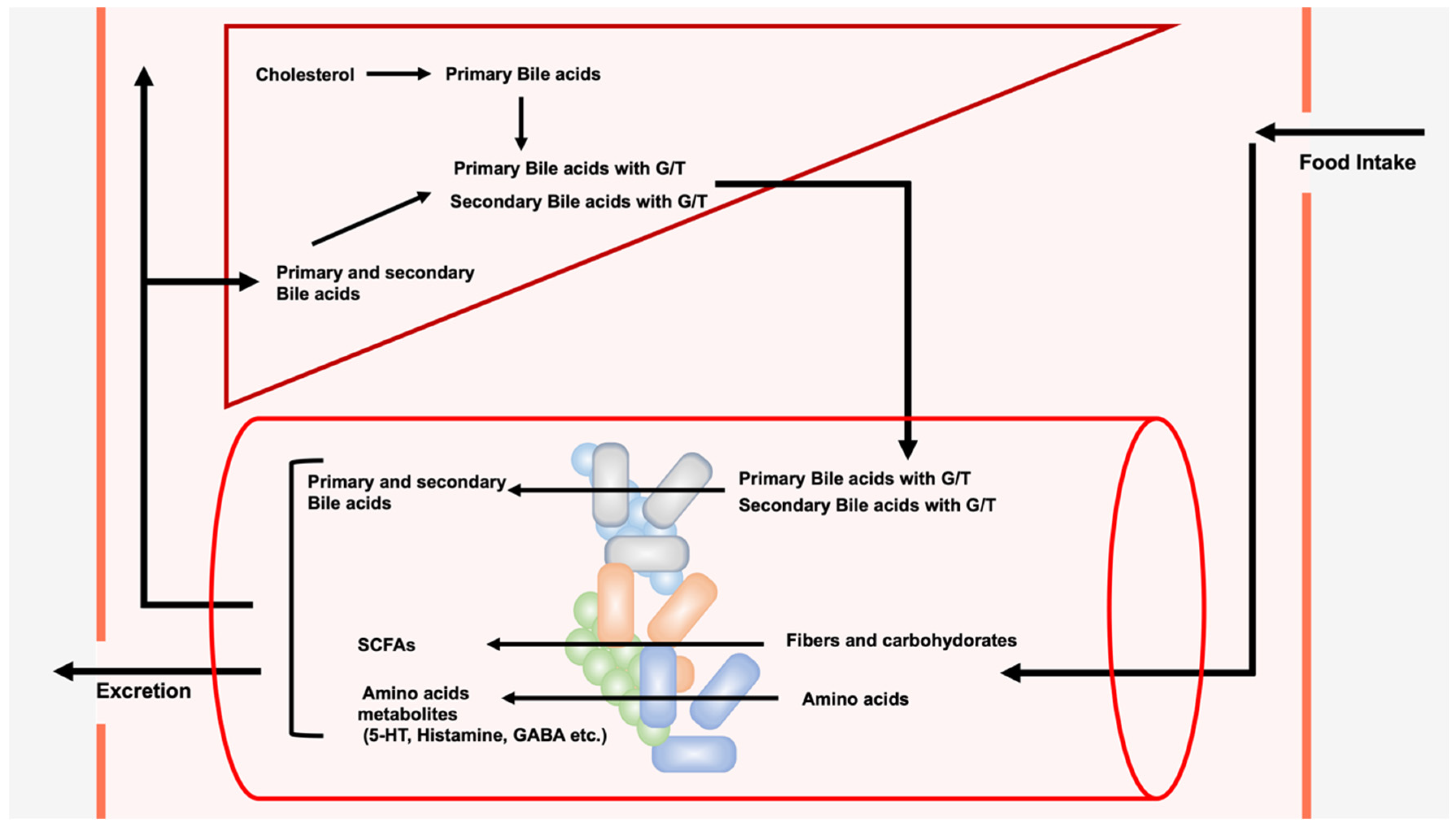
A wide variety and large number of bacterial species live in the gut, forming the gut microbiota. Gut microbiota not only coexist harmoniously with their hosts, but they also induce significant effects on each other. The composition of the gut microbiota can be changed due to environmental factors such as diet and antibiotic intake. In contrast, alterations in the composition of the gut microbiota have been reported in a variety of diseases, including intestinal, allergic, and autoimmune diseases and cancer. The gut microbiota metabolize exogenous dietary components ingested from outside the body to produce short-chain fatty acids (SCFAs) and amino acid metabolites. Unlike SCFAs and amino acid metabolites, the source of bile acids (BAs) produced by the gut microbiota is endogenous BAs from the liver. The gut microbiota metabolize BAs to generate secondary bile acids, such as lithocholic acid (LCA), deoxycholic acid (DCA), and their derivatives, which have recently been shown to play important roles in immune cells.
1. Introduction
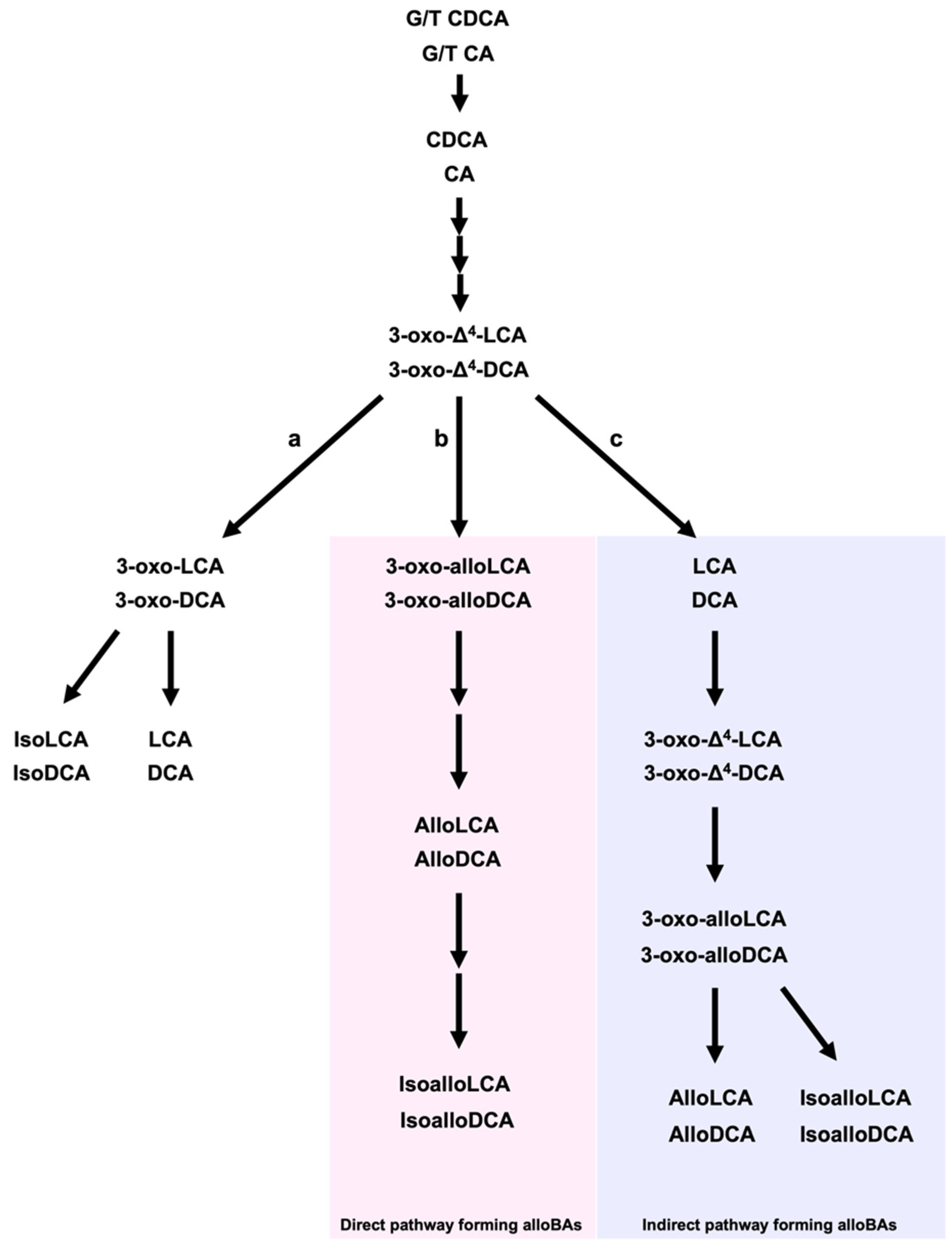
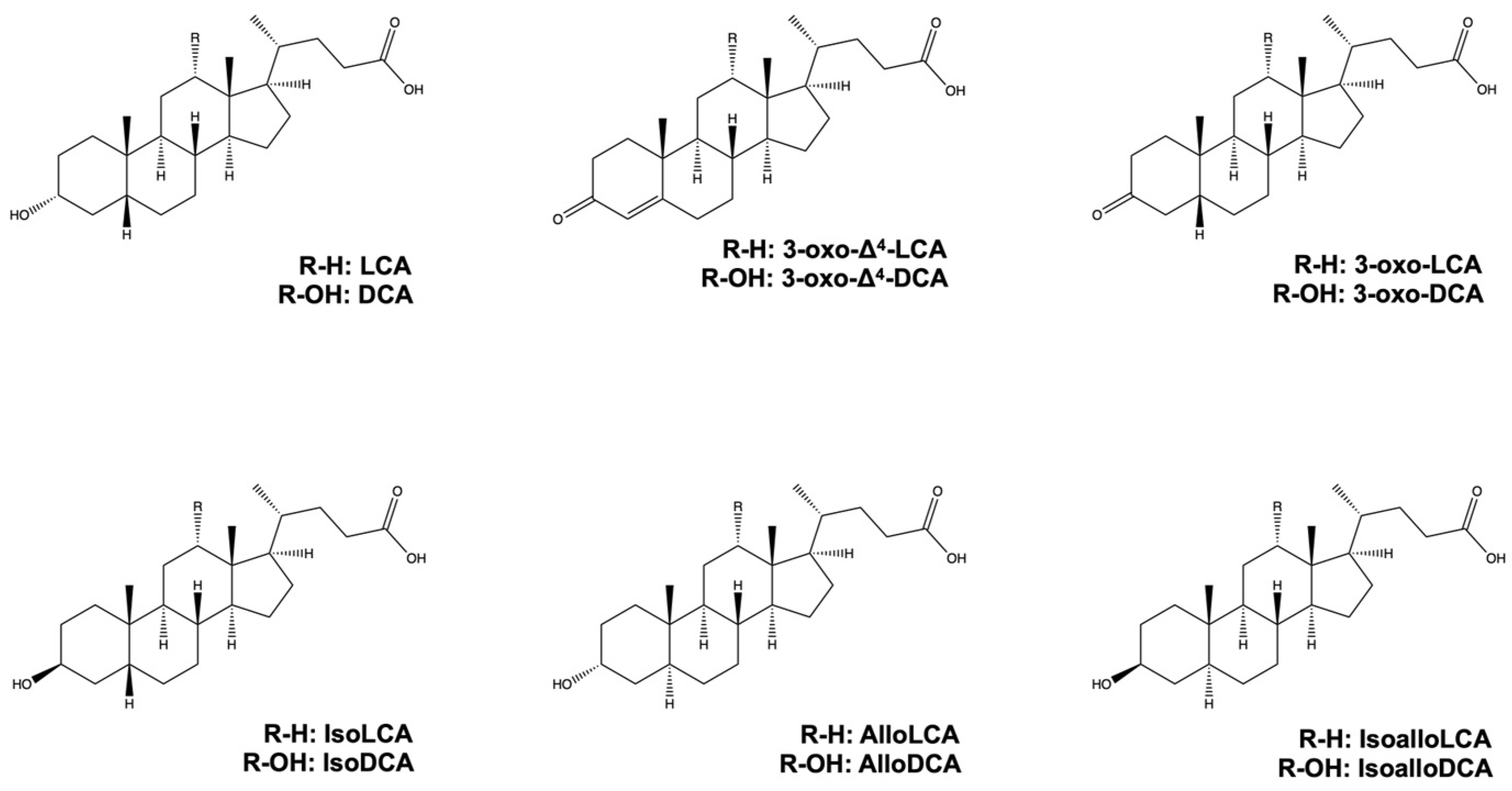
2. Lithocholic Acid, Deoxycholic Acid, and Their Derivatives from Gut Microbiota
References
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533.
- Dekaboruah, E.; Suryavanshi, M.V.; Chettri, D.; Verma, A.K. Human microbiome: An academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 2020, 202, 2147–2167.
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32.
- Mishra, K.; Bukavina, L.; Ghannoum, M. Symbiosis and Dysbiosis of the Human Mycobiome. Front. Microbiol. 2021, 12, 636131.
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target Ther. 2022, 7, 135.
- Zhao, M.; Chu, J.; Feng, S.; Guo, C.; Xue, B.; He, K.; Li, L. Immunological mechanisms of inflammatory diseases caused by gut microbiota dysbiosis: A review. Biomed. Pharmacother. 2023, 164, 114985.
- Liu, Q.; Su, Q.; Zhang, F.; Tun, H.M.; Mak, J.W.Y.; Lui, G.C.; Ng, S.S.S.; Ching, J.Y.L.; Li, A.; Lu, W.; et al. Multi-kingdom gut microbiota analyses define COVID-19 severity and post-acute COVID-19 syndrome. Nat. Commun. 2022, 13, 6806.
- Zhang, F.; Lau, R.I.; Liu, Q.; Su, Q.; Chan, F.K.L.; Ng, S.C. Gut microbiota in COVID-19: Key microbial changes, potential mechanisms and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 323–337.
- Wang, B.; Zhang, L.; Wang, Y.; Dai, T.; Qin, Z.; Zhou, F.; Zhang, L. Alterations in microbiota of patients with COVID-19: Potential mechanisms and therapeutic interventions. Signal Transduct. Target Ther. 2022, 7, 143.
- Van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 2021, 29, 700–712.
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105.
- Jameson, K.G.; Olson, C.A.; Kazmi, S.A.; Hsiao, E.Y. Toward Understanding Microbiome-Neuronal Signaling. Mol. Cell 2020, 78, 577–583.
- Sun, P.; Su, L.; Zhu, H.; Li, X.; Guo, Y.; Du, X.; Zhang, L.; Qin, C. Gut Microbiota Regulation and Their Implication in the Development of Neurodegenerative Disease. Microorganisms 2021, 9, 2281.
- Kiriyama, Y.; Nochi, H. The Biosynthesis, Signaling, and Neurological Functions of Bile Acids. Biomolecules 2019, 9, 232.
- Li, J.; Dawson, P.A. Animal models to study bile acid metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 895–911.
- Straniero, S.; Laskar, A.; Savva, C.; Hardfeldt, J.; Angelin, B.; Rudling, M. Of mice and men: Murine bile acids explain species differences in the regulation of bile acid and cholesterol metabolism. J. Lipid Res. 2020, 61, 480–491.
- Sun, J.; Li, M.; Zhou, H.; Chong, J.; Zhang, J.; Yu, B.; Chen, D.; Ge, L. Importance of gut microbiota for bile acid composition and concentration in pigs. Front. Anim. Sci. 2022, 3, 951840.
- Kiriyama, Y.; Nochi, H. Physiological Role of Bile Acids Modified by the Gut Microbiome. Microorganisms 2021, 10, 68.
- Lee, J.W.; Cowley, E.S.; Wolf, P.G.; Doden, H.L.; Murai, T.; Caicedo, K.Y.O.; Ly, L.K.; Sun, F.; Takei, H.; Nittono, H.; et al. Formation of secondary allo-bile acids by novel enzymes from gut Firmicutes. Gut. Microbes 2022, 14, 2132903.
- Doden, H.L.; Ridlon, J.M. Microbial Hydroxysteroid Dehydrogenases: From Alpha to Omega. Microorganisms 2021, 9, 469.
- Kiriyama, Y.; Nochi, H. Role of Microbiota-Modified Bile Acids in the Regulation of Intracellular Organelles and Neurodegenerative Diseases. Genes 2023, 14, 825.
- Che, Y.; Xu, W.; Ding, C.; He, T.; Xu, X.; Shuai, Y.; Huang, H.; Wu, J.; Wang, Y.; Wang, C.; et al. Bile acids target mitofusin 2 to differentially regulate innate immunity in physiological versus cholestatic conditions. Cell Rep. 2023, 42, 112011.
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128.
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.B.; Guo, C.J.; Violante, S.; et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020, 581, 475–479.
- Guo, C.; Xie, S.; Chi, Z.; Zhang, J.; Liu, Y.; Zhang, L.; Zheng, M.; Zhang, X.; Xia, D.; Ke, Y.; et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 2016, 45, 802–816.
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 2019, 576, 143–148.
- Hu, J.; Wang, C.; Huang, X.; Yi, S.; Pan, S.; Zhang, Y.; Yuan, G.; Cao, Q.; Ye, X.; Li, H. Gut microbiota-mediated secondary bile acids regulate dendritic cells to attenuate autoimmune uveitis through TGR5 signaling. Cell Rep. 2021, 36, 109726.
- Hu, J.; Zhang, Y.; Yi, S.; Wang, C.; Huang, X.; Pan, S.; Yang, J.; Yuan, G.; Tan, S.; Li, H. Lithocholic acid inhibits dendritic cell activation by reducing intracellular glutathione via TGR5 signaling. Int. J. Biol. Sci. 2022, 18, 4545–4559.
- Li, W.; Hang, S.; Fang, Y.; Bae, S.; Zhang, Y.; Zhang, M.; Wang, G.; McCurry, M.D.; Bae, M.; Paik, D.; et al. A bacterial bile acid metabolite modulates T(reg) activity through the nuclear hormone receptor NR4A1. Cell Host Microbe 2021, 29, 1366–1377.e9.
- Roggeri, A.; Schepers, M.; Tiane, A.; Rombaut, B.; van Veggel, L.; Hellings, N.; Prickaerts, J.; Pittaluga, A.; Vanmierlo, T. Sphingosine-1-Phosphate Receptor Modulators and Oligodendroglial Cells: Beyond Immunomodulation. Int. J. Mol. Sci. 2020, 21, 7537.
- Pandak, W.M.; Kakiyama, G. The acidic pathway of bile acid synthesis: Not just an alternative pathway. Liver Res. 2019, 3, 88–98.
- Russell, D.W. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 2009, 50, S120–S125.
- Chiang, J.Y.L.; Ferrell, J.M. Up to date on cholesterol 7 alpha-hydroxylase (CYP7A1) in bile acid synthesis. Liver Res. 2020, 4, 47–63.
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu. Rev. Nutr. 2019, 39, 175–200.
- Xue, R.; Su, L.; Lai, S.; Wang, Y.; Zhao, D.; Fan, J.; Chen, W.; Hylemon, P.B.; Zhou, H. Bile Acid Receptors and the Gut-Liver Axis in Nonalcoholic Fatty Liver Disease. Cells 2021, 10, 2806.
- Kenna, J.G.; Taskar, K.S.; Battista, C.; Bourdet, D.L.; Brouwer, K.L.R.; Brouwer, K.R.; Dai, D.; Funk, C.; Hafey, M.J.; Lai, Y.; et al. Can Bile Salt Export Pump Inhibition Testing in Drug Discovery and Development Reduce Liver Injury Risk? An International Transporter Consortium Perspective. Clin. Pharmacol. Ther. 2018, 104, 916–932.
- Daly, J.W.; Keely, S.J.; Gahan, C.G.M. Functional and Phylogenetic Diversity of BSH and PVA Enzymes. Microorganisms 2021, 9, 732.
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585.
- O’Flaherty, S.; Briner Crawley, A.; Theriot, C.M.; Barrangou, R. The Lactobacillus Bile Salt Hydrolase Repertoire Reveals Niche-Specific Adaptation. mSphere 2018, 3, 10–1128.
- Clarke, G.; Sandhu, K.V.; Griffin, B.T.; Dinan, T.G.; Cryan, J.F.; Hyland, N.P. Gut Reactions: Breaking Down Xenobiotic-Microbiome Interactions. Pharmacol. Rev. 2019, 71, 198–224.
- Riviere, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979.
- Guo, X.; Okpara, E.S.; Hu, W.; Yan, C.; Wang, Y.; Liang, Q.; Chiang, J.Y.L.; Han, S. Interactive Relationships between Intestinal Flora and Bile Acids. Int. J. Mol. Sci. 2022, 23, 8343.
- Begley, M.; Hill, C.; Gahan, C.G. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738.
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut. Microbes 2016, 7, 22–39.
- Ridlon, J.M.; Daniel, S.L.; Gaskins, H.R. The Hylemon-Bjorkhem pathway of bile acid 7-dehydroxylation: History, biochemistry, and microbiology. J. Lipid Res. 2023, 64, 100392.
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259.
- Paik, D.; Yao, L.; Zhang, Y.; Bae, S.; D’Agostino, G.D.; Zhang, M.; Kim, E.; Franzosa, E.A.; Avila-Pacheco, J.; Bisanz, J.E.; et al. Human gut bacteria produce Tau(Eta)17-modulating bile acid metabolites. Nature 2022, 603, 907–912.
- Sato, Y.; Atarashi, K.; Plichta, D.R.; Arai, Y.; Sasajima, S.; Kearney, S.M.; Suda, W.; Takeshita, K.; Sasaki, T.; Okamoto, S.; et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature 2021, 599, 458–464.
- Camilleri, M. Bile acid detergency: Permeability, inflammation, and effects of sulfation. Am. J. Physiol. Gastrointest. Liver Physiol. 2022, 322, G480–G488.
- Sheng, W.; Ji, G.; Zhang, L. The Effect of Lithocholic Acid on the Gut-Liver Axis. Front. Pharmacol. 2022, 13, 910493.




