Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alan Herbert | -- | 2704 | 2023-11-21 19:17:10 | | | |
| 2 | Rita Xu | -13 word(s) | 2691 | 2023-11-22 03:07:35 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Herbert, A. Directed Cycles Evolve with Junk DNA. Encyclopedia. Available online: https://encyclopedia.pub/entry/51875 (accessed on 08 February 2026).
Herbert A. Directed Cycles Evolve with Junk DNA. Encyclopedia. Available at: https://encyclopedia.pub/entry/51875. Accessed February 08, 2026.
Herbert, Alan. "Directed Cycles Evolve with Junk DNA" Encyclopedia, https://encyclopedia.pub/entry/51875 (accessed February 08, 2026).
Herbert, A. (2023, November 21). Directed Cycles Evolve with Junk DNA. In Encyclopedia. https://encyclopedia.pub/entry/51875
Herbert, Alan. "Directed Cycles Evolve with Junk DNA." Encyclopedia. Web. 21 November, 2023.
Copy Citation
Cell responses are usually viewed as transitive events with fixed inputs and outputs that are regulated by feedback loops. In contrast, directed cycles (DCs) have all nodes connected, and the flow is in a single direction. Consequently, DCs can regenerate themselves and implement intransitive logic. DCs are able to couple unrelated chemical reactions to each edge. The output depends upon which node is used as input.
evolution
flipons
kolmogorov complexity
dissipative structures
logic
junk DNA
RNA processing
reverse transcription
1. Introduction
Contrary to widely held perceptions, what has been called junk DNA provides an evolutionary advantage by expanding the Kolmogorov complexity of genomes. Those repeat elements that adopt alternative DNA conformations under physiological conditions, called flipons, potentially enable new adaptations by altering the flow of genetic information into RNA. A subset of genomic repeats enables the assembly of novel cellular machines by encoding peptide patches that vary over time in length, composition, and chromosomal location. They promote the protein interactions essential for cells to regenerate, recalibrate, reset, repair, rewrite, and reproduce themselves into the next generation.
Normally, the analysis of how cells evolve focuses on the linear pathways (LPs) that connect substrates with products and the regulatory mechanisms involved. The evolution of systems can instead be viewed from a different perspective. This approach is based on directed cycles (DCs), in which all nodes are connected and where the path between adjacent nodes is directional. In a DC, the path taken between nodes depends on which node act as the input (Figure 1). There are a minimum of two directed paths between each pair of nodes that enable DCs to regenerate themselves. Each path can couple with different cellular processes to produce unique outputs. As I will discuss, the logic is intransitive, in contrast to the LP approach, where the logic is transitive. The design greatly increases the computations that a system can perform, a concept captured by the Kolmogorov complexity of a system, which is a fundamental measure of the information encoded by that system. To understand DCs in biological systems, it is also necessary to analyze them from a thermodynamic perspective and the way natural selection serves to optimize their energy efficiency. DCs are primary units of evolution that enhance the adaptability of cells through the computations they perform. These self-referential circuits act to contextually optimize responses. From a practical perspective, interventions targeting DCs will help in the design and engineering of new therapeutics and the development of bioprocesses that deploy specific chemistries.
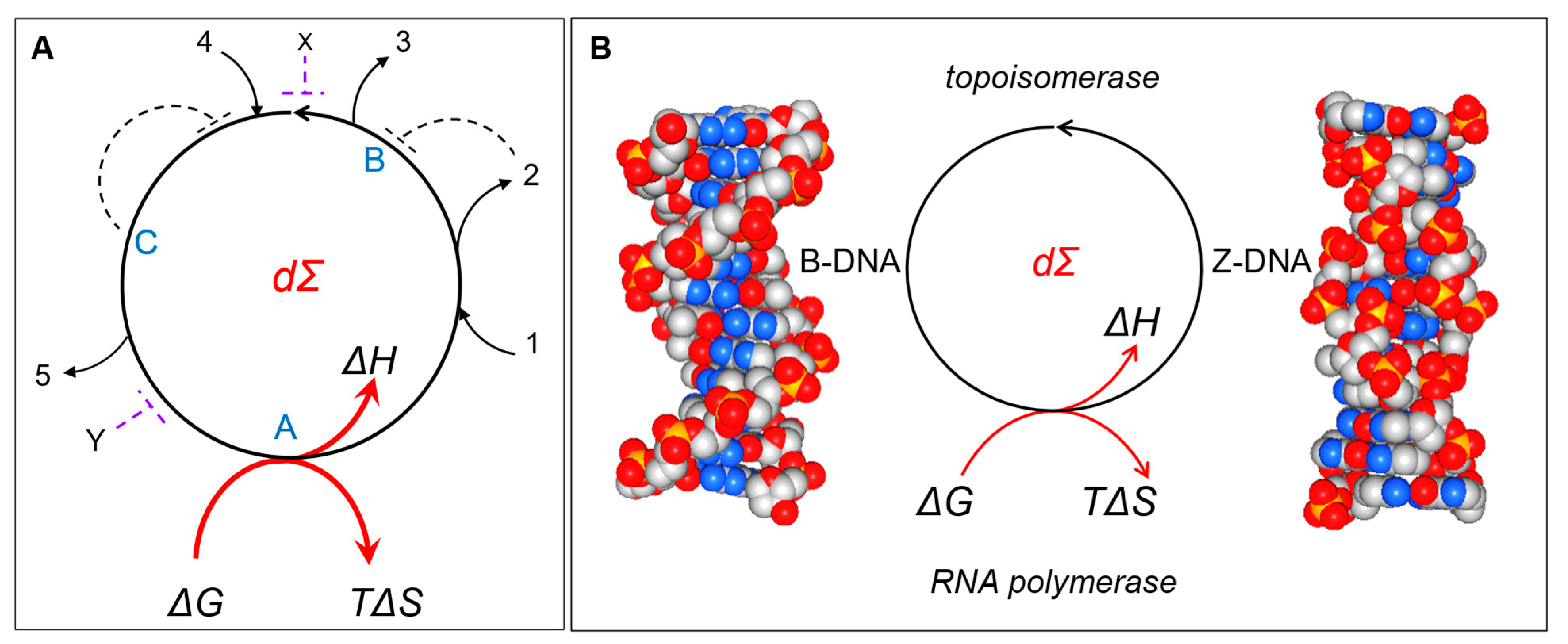
Figure 1. Directed cycle that implements intransitive logic. (A) It is possible to enter and leave the DC at multiple points. They each capture the relationship A > B > C > A. There is no beginning or end to the cycle. The letters A, B, and C could represent the rock, scissors, paper response and the numbers 1 to 5 refer to various environmental inputs and outputs. The cycle depends on the available energy (ΔG). The DC maximizes work (ΔH) by minimizing entropy loss (TΔS). The dotted lines represent a subset of possible paths that allow the negative regulation of the cycle through elements B and C or through points X and Y. In nature, these cycles are quite stable and can be described as a class of dissipative structures (dΣ). (B) Z-flipons are dissipative structures. They represent a DC for the transition between right-handed B-DNA and Z-DNA conformations. RNA polymerases can provide the energy to initiate the flip from B-DNA to Z-DNA. The energy cost depends on the DNA sequence and modifications to bases. The dissipation of energy by topoisomerases relaxes the Z-DNA to the B-DNA conformation.
2. Evolvability of Directed Cycles through Junk Sequences
The analysis suggests that junk DNA extends the Kolmogorov complexity of programs that can be generated by the human genome through its effects on the flow of information from DNA to RNA. In humans, much of the junk arises from endogenous retroelements (EREs) that have contributed sequences to over 50% of the genome. The rewriting of genomic information by EREs occurs through the reverse transcription of the RNAs they transcribe into DNA. While initially dismissed as genomic fluff, it is now appreciated that EREs are essential regulatory components of genes. Sequences derived from Alu retroelements, of which there are over a million copies in the human genome, can change both the splicing and polyadenylation of nascent RNA (for recent reviews, see [1][2]). The EREs involved alter the transcript produced, depending on the position of their insertion, by implementing simple programming rules to change how RNA is processed. Rather than all being hard-wired into the genome, the outcomes are soft-wired and conditional on context. The splicing cascade that programs the sex of flies indicates the potential complexity of these events [3].
Flipons also offer the opportunity to modulate their conformations to alter downstream events. The single-stranded regions they form in the DNA duplex as they flip from one conformation to another expose binding sites that allow the sequence-specific docking of small RNAs, especially those derived from the same family of repeats as the flipon. The binding of these small RNAs to flipons allows the targeting of the cellular machinery to these genomic locations to edit and modify the transcripts produced. The proteins involved can be generic and bind in a structure-specific manner. They need not specifically recognize any particular nucleic acid sequence. The assembly of these complexes is directed only by the sequence-specificity of the RNA. This design has a number of evolutionary advantages. Importantly, the RNA sequence space available to target flipons in a sequence-specific manner is much larger than for developing sequence-specific proteins, where problems with folding and loss of function constrain the span of possible protein variations.
Changes in the RNA space are also not all or nothing, so there is no loss of any adaptations that proved successful in the past. The altered processing just increases the number of isoforms produced. In contrast, protein variants abandon the previous versions. Similarly, the spread of flipons through the genome creates many possible ways to alter the local DNA conformation to generate variant transcripts. The digital nature of flipons enables many different combinations. It is unlikely that the genomes of any two cells are set identically. As a consequence, the selection of cells at the tissue level can enable the responses that are most adaptable to local stressors (recently reviewed [4]). Small RNAs that are transmitted through germ cells also have the potential to bootstrap embryonic development by modulating the flipon conformation during early embryonic development [5]. These effects are likely modulated through the extraembryonic endoderm, which induces highly conserved programs within the rapidly dividing embryo and could possibly involve the reverse transcription of these small RNAs into the extraembryonic genome.
2.1. Evolvability of Directed Cycles through Peptide Patches
There are other ways to evolve a directed cycle through junk DNA. The peptide patches I discussed earlier as part of the cell’s wetware can act as Velcro to pull proteins together to create new assemblies (Figure 2A). The output from one of the sequestered proteins then potentially acts as an input to another. Eventually, a self-sustaining cycle arises through a set of protein interactions that positively reinforce each other’s output. This strategy assumes that proteins are more multifunctional than is currently presented in textbooks. In reality, the patched-together proteins often contain multiple different domains. Though many domains have well-studied functions, others remain uncharacterized. With the patchwork design just described, peptides with no enzymatic function can create new opportunities to unmask proteins with multiple personalities and are able to perform unexpectedly. Frequently, experimentalists find the newly discovered properties of a well-characterized protein surprising. They then write papers entitled “Hidden protein functions and what they may teach us” [6] and “Protein moonlighting: what is it, and why is it important?” [7].
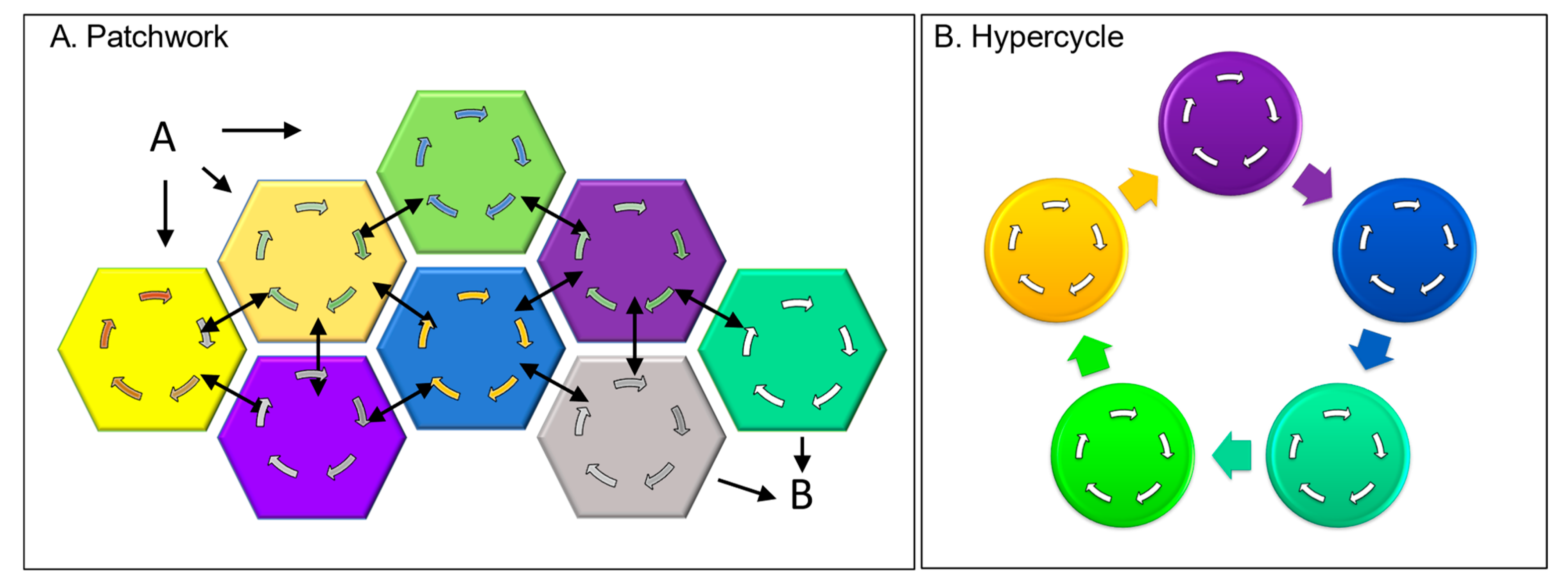
Figure 2. DCs as cellular building blocks. (A) Assembly of DCs into larger complexes through peptide patches, with the multiple connections between the input A and the output B increasing system robustness. (B) Interactions between directed cycles to produce hypercycles that are autocatalytic.
The new cycles established by patching proteins together may initially depend on inputs from the milieux to bridge any missing links. The Krebs cycle that researchers depend upon to extract energy from sugars likely developed in such a way. At an early stage, the reactions depended on environmentally derived metals for catalysis. More efficient reactions arose when binding sites for metals were incorporated into genetically encoded proteins. Many of these strategies based on autocatalytic chemistries, along with the history of this field, have been reviewed [8]. Even today, some DCs still rely on environmentally derived factors to function. The dependency on these essential nutrients is so complete that, without them, certain DCs fail to regenerate. Humans, for example, do not synthesize vitamin C, even though other organisms solved this biochemical challenge long ago.
2.2. Evolvability of Directed Cycles through Hypercycles
The evolution of DCs can proceed through the organization of self-replicating molecules connected in a cyclic, autocatalytic manner, as originally proposed by Manfred Eigen [9] (Figure 2B). Due to the way they interact, the cycles are self-propagating, with each cycle forming a node coupled to a larger cycle (Figure 2). The interactions between different cycles allow them to amplify themselves, each other, and the hypercycle. The hypercycles further favor systems that store the information necessary to continuously regenerate themselves (Figure 2B). In the simplest form, the earliest steps in a pathway did all that was required to produce a particular output. Steps were added that closed the circuit, leading to the self-amplification of that particular cycle. The cycle underwent further elaboration by connecting to other cycles that further assured their mutual perpetuation (Figure 1, dΣc). The creation of genetic systems to transmit this information to subsequent generations was a natural consequence of hypercycle evolution.
2.3. Evolvability of Directed Cycles through Genome Duplication
The rewriting of directed cycles in DNA during evolution can occur in many ways different from those that Eigen imagined. There may be more complex processes involved. On occasion, genes may undergo duplication in ways that Susumu Ohno demonstrated were important during evolution [10]. Fortuitous mutations affecting the level of gene expression, the processing of transcripts, and the non-templated modification of proteins then altered the character of each duplicated gene. At some point, changes to one paralog or the other provided a selective advantage, leading to the creation of new DC variants.
Occasionally, whole genomes undergo duplication. Many plants have a history of expanding their genomes in this manner and are consequently highly polyploid. As a result, they have multiple copies of each gene [11]. The process allows DCs to be reconstituted in different ways or with different combinations to generate new elaborations. The process of genome duplication has also been observed in yeast following a sudden and adverse change in the environment [12]. The high mutation rates that accompany this process drive additional genomic diversity and the elaboration of DCs that enable their regeneration and the survival of progeny in the new environment.
2.4. Evolvability of Directed Cycles through Endosymbiosis
Another way to acquire all of the components necessary to make a new DC is simply by obtaining all of them in one step from another organism. With bacteria, this means gaining an entire operon where all of the genes required for the regulation, expression, and scripting of a cycle are organized into one DNA segment. These outcomes are enabled by bacterial conjugation, the prokaryotic version of sex first observed by Joshua Lederberg and Edward Tatum [13]. To do the same in eukaryotes would require a genomic organization similar to the operons of bacteria and a truly giant virus to transmit the much larger eukaryotic genes that embed all of the required information. It is now possible, using a variety of technologies, to introduce into cells large genomic assemblies with all of the genes required. The most extreme transplant of genes so far performed is the transfer of entire normal mitochondria to replace the defective ones transmitted to an embryo from a parent. Of course, the only reason that eukaryotes have mitochondria in the first place is that at one point in time, the whole set of DCs that another free-living organism had successfully evolved was subsumed to generate energy with available substrates. The most recent proponent of this idea was Lynn Margulis, who also noted that chloroplasts are endosymbiont cyanobacteria [14]. Even today, osteoclasts can source replacement mitochondria from osteomorphs to remain functional [15].
2.5. Evolvability of Directed Cycles through Bioengineering
Experimental approaches aimed at modifying DCs depend on first identifying the minimal set of components required for a DC to regenerate itself. Such studies can be performed in vitro by purifying each element and reconstituting a DC from these parts. These approaches helped elucidate many of the DCs, such as the Krebs cycle, involved in cell metabolism. These studies can also be performed using genetic approaches to identify the different DC components. Over the years, bacteria and yeast have proven particularly powerful in establishing many of the factors that modulate DCs in single cells.
Collectively, these approaches identify the proteins essential for regenerating DCs. The methods also uncover redundancies and scaffolds that enhance the performance and robustness of DCs (Figure 1, dΣb). Further, the results inform on which DC steps can be modulated therapeutically. Drugs to break DCs are part of the pharmacopeia positioned to kill cancer cells. The targeting approach yields valuable insights into the differences between normal and diseased cells. This work identifies multiple pathways between nodes in normal tissue and those that are no longer present in cancer cells. The vulnerability of tumors arises due to mutations that inactivate one or more of the redundant connections between nodes. The tumors are then susceptible to drugs that target the residual pathway. The drugs and mutations synergize to selectively kill the tumor while sparing normal cells.
Drugs that induce synthetic lethality in tumors are important in the clinic. In many cases, tumors are able to mutate and become resistant to most drugs that are used as single agents. The tumors then continue growing [16]. A drug cocktail that targets multiple DCs to induce synthetic lethality through different pathways is often needed to thwart the escape of cancer cells from eradication. The challenges to curing cancers despite the high-precision targeting of molecules underscore the overall resilience of DCs in cells. The intransitive programming based on DCs enhances their adaptability. Winning strategies just require the rewiring of the path between two nodes (Figure 1, dΣb).
The therapeutic potential to alter DC function by programming flipons with small RNAs exists. The interventions can be used to prevent the expression of an essential DC component, to regulate its processing, or to recode the amino acids in key functional domains. There is also the possibility of rewiring connections in DCs to improve their design to engineer new functions. The nature of DCs allows researchers to drive their evolution in cells and to bulk manufacture their outputs by cell culture.
The patchwork approach to generating new DCs also offers opportunities. Researchers do not know how far this strategy can be pushed to engineer new DCs. Experimentally, researchers could ask whether they can tag well-folded functional domains with interacting peptide patches to Velcro together new protein assemblies with defined properties. Can researchers then evolve a DC with a desired output (Figure 2A)? Or can researchers expose existing DCs to alternative chemistries to create completely new reaction schemes that have never before existed in nature? Already, DCs have been adapted to use synthetic chemicals in preference to their natural substrates. For example, Madeleine Bouzon and Philippe Marlière substituted 4-hydroxy-2-oxobutanoic acid for the amino acids serine and glycine as a carbon source for one particular metabolic pathway [17].
An underexplored area is the use of repeat-derived RNAs to build scaffolds. As shown by the assembly of the spliceosome, many proteins exist that bind to simple sequence motifs exposed on single-stranded RNAs. In principle, these motifs could be used in a combinatorial fashion to create novel RNA scaffolds on which to assemble existing proteins in a cell into new assemblies and then select for a phenotype of interest. The targeting of the cellular machinery to triplex-forming flipons by noncoding RNAs through this mechanism has been previously reviewed [1].
References
- Herbert, A. ALU non-B-DNA conformations, flipons, binary codes and evolution. R. Soc. Open Sci. 2020, 7, 200222.
- Gussakovsky, D.; McKenna, S.A. Alu RNA and their roles in human disease states. RNA Biol. 2021, 18 (Suppl. S2), 574–585.
- Herbert, A.; Rich, A. RNA Processing in Evolution: The Logic of Soft-Wired Genomesa. Ann. N. Y. Acad. Sci. 1999, 870, 119–132.
- Herbert, A. Flipons and small RNAs Accentuate the Asymmetries of Pervasive Transcription by the Reset and Sequence-Specific Microcoding of Promoter Conformation. J. Biol. Chem. 2023, 299, 105140.
- Herbert, A.; Pavlov, F.; Konovalov, D.; Poptsova, M. Conserved microRNAs and Flipons Shape Gene Expression during Development by Altering Promoter Conformations. Int. J. Mol. Sci. 2023, 24, 4884.
- Schwille, P.; Frohn, B.P. Hidden protein functions and what they may teach us. Trends Cell Biol. 2022, 32, 102–109.
- Jeffery, C.J. Protein moonlighting: What is it, and why is it important? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160523.
- Bissette, A.J.; Fletcher, S.P. Mechanisms of autocatalysis. Angew. Chem. Int. Ed. Engl. 2013, 52, 12800–12826.
- Eigen, M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971, 58, 465–523.
- Ohno, S. Evolution by Gene Duplication, 1st ed.; Springer: Berlin/Heidelberg, Germany, 1970.
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38.
- Charron, G.; Marsit, S.; Henault, M.; Martin, H.; Landry, C.R. Spontaneous whole-genome duplication restores fertility in interspecific hybrids. Nat. Commun. 2019, 10, 4126.
- Lederberg, J.; Tatum, E.L. Gene recombination in Escherichia coli. Nature 1946, 158, 558.
- Margulis, L. Symbiosis in Cell Evolution: Microbial Communities in the Archean and Proterozoic Eons, 2nd ed.; Freeman: New York, NY, USA, 1993.
- McDonald, M.M.; Khoo, W.H.; Ng, P.Y.; Xiao, Y.; Zamerli, J.; Thatcher, P.; Kyaw, W.; Pathmanandavel, K.; Grootveld, A.K.; Moran, I.; et al. Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption. Cell 2021, 184, 1330–1347.e13.
- Ferrari, E.; Lucca, C.; Foiani, M. A lethal combination for cancer cells: Synthetic lethality screenings for drug discovery. Eur. J. Cancer 2010, 46, 2889–2895.
- Bouzon, M.; Perret, A.; Loreau, O.; Delmas, V.; Perchat, N.; Weissenbach, J.; Taran, F.; Marliere, P. A Synthetic Alternative to Canonical One-Carbon Metabolism. ACS Synth. Biol. 2017, 6, 1520–1533.
More
Information
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
560
Revisions:
2 times
(View History)
Update Date:
22 Nov 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




