
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tsvetelina Velikova | -- | 1849 | 2023-11-21 06:49:47 | | | |
| 2 | Catherine Yang | Meta information modification | 1849 | 2023-11-21 07:20:07 | | |
Video Upload Options
Over the past century, our genetic background has not changed, but chronic diseases are on the rise globally. In addition to the genetic component, the critical factors for many diseases are lifestyle, eating changes, exposure to drugs, xenobiotics, alcohol, smoking, polluted air, etc. These techniques include genome-wide association studies (GWAS) that allow the identification of unknown genetic risk factors, positional cloning of unknown genes associated with different diseases, the gene tests for single nucleotide variants (SNVs), and next-generation sequencing (NGS) of selected genes or/and the entire genome. Gut microbiome composition and its metabolites are not only regulating factors in carcinogenesis (including de novo after liver transplantation) but also in xenobiotics and anticancer treatment failure, observations that may be related to the genetic background of the individuals.
1. GWAS Loci for Predisposition and Susceptibility of NAFLD
2. NAFLD GWAS Loci Overlap with GWAS Loci for Liver Enzymes, ALD and HCC
3. GWAS Loci for Predisposition and Susceptibility of ALD
4. GWAS Loci for Predisposition and Susceptibility of ALC
5. GWAS Loci with Significant Association with HCC
6. Other Genetic Loci Related to Different Forms of Liver Diseases
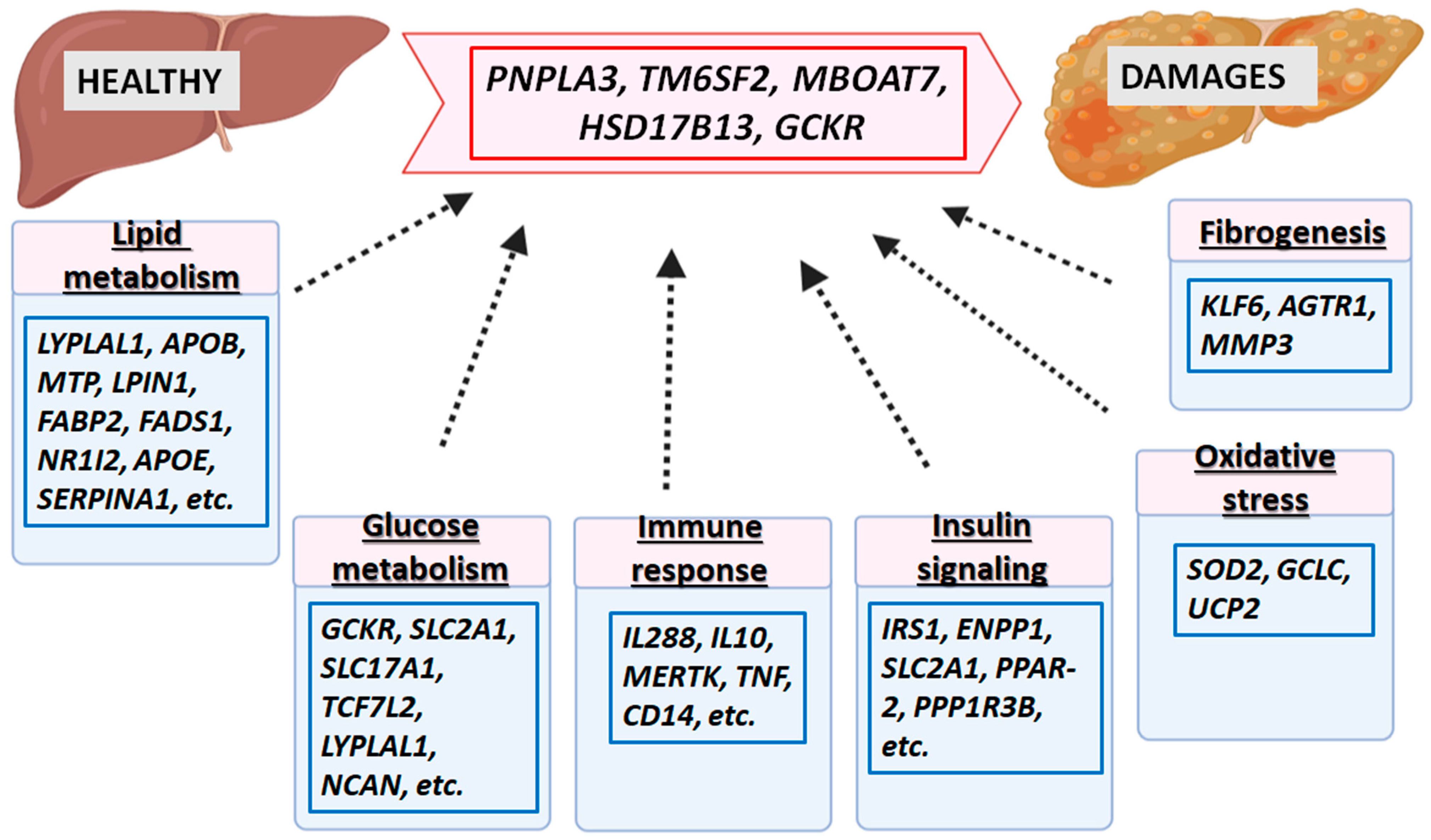
References
- Loomba, R.; Sanyal, A.J. The global NAFLD epidemic. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 686–690.
- Eslam, M.; Sanyal, A.J.; George, J.; Sanyal, A.; Neuschwander-Tetri, B.; Tiribelli, C.; Kleiner, D.E.; Brunt, E.; Bugianesi, E.; Yki-Järvinen, H.; et al. International Consensus Panel MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020, 158, 1999–2014.e1.
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20.
- Sookoian, S.; Pirola, C.J. Genetic predisposition in non-alcoholic fatty liver disease. Clin. Mol. Hepatol. 2017, 23, 1–12.
- Juanola, O.; Martínez-López, S.; Francés, R.; Gómez-Hurtado, I. Non-Alcoholic Fatty Liver Disease: Metabolic, Genetic, Epigenetic and Environmental Risk Factors. Int. J. Environ. Res. Public. Health 2021, 18, 5227.
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to non-alcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465.
- Speliotes, E.K.; LYerges-Armstrong, M.; Wu, J.; Hernaez, R.; Kim, L.J.; Palmer, C.D.; Gudnason, V.; Eiriksdottir, G.; Garcia, M.E.; Launer, L.J.; et al. NASH CRN, GIANT Consortium, MAGIC Investigators, GOLD Consortium, Genome-wide association analysis identifies variants associated with non-alcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011, 7, e1001324.
- Eslam, M.; Valenti, L.; Romeo, S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J. Hepatol. 2018, 68, 268–279.
- Chalasani, N.; Guo, X.; Loomba, R.; Goodarzi, M.O.; Haritunians, T.; Kwon, S.; Cui, J.; Taylor, K.D.; Wilson, L.; Cummings, O.W.; et al. Non-alcoholic Steatohepatitis Clinical Research Network, Genome-wide association study identifies variants associated with histologic features of non-alcoholic Fatty liver disease. Gastroenterology 2010, 139, 1567–1576.
- Liu, Y.L.; Reeves, H.L.; Burt, A.D.; Tiniakos, D.; McPherson, S.; Leathart, J.B.S.; Allison, M.E.D.; Alexander, G.J.; Piguet, A.C.; Anty, R.; et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 4309.
- Kitamoto, T.; Kitamoto, A.; Yoneda, M.; Hyogo, H.; Ochi, H.; Nakamura, T.; Teranishi, H.; Mizusawa, S.; Ueno, T.; Chayama, K.; et al. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of non-alcoholic fatty liver disease in Japan. Hum. Genet. 2013, 132, 783–792.
- Chung, G.E.; Lee, Y.; Yim, J.Y.; Choe, E.K.; Kwak, M.S.; Yang, J.I.; Park, B.; Lee, J.E.; Kim, J.A.; Kim, J.S. Genetic polymorphisms of PNPLA3 and SAMM50 are associated with non-alcoholic fatty liver disease in a Korean population. Gut Liver 2018, 12, 316–323.
- Kitamoto, A.; Kitamoto, T.; Nakamura, T.; Ogawa, Y.; Yoneda, M.; Hyogo, H.; Ochi, H.; Mizusawa, S.; Ueno, T.; Nakao, K.; et al. Association of polymorphisms in GCKR and TRIB1 with non-alcoholic fatty liver disease and metabolic syndrome traits. Endocr. J. 2014, 61, 683–689.
- Emdin, C.A.; Haas, M.E.; Khera, A.V.; Aragam, K.; Chaffin, M.; Klarin, D.; Hindy, G.; Jiang, L.; Wei, W.Q.; Feng, Q.; et al. Million Veteran Program, A missense variant in Mitochondrial Amidoxime Reducing Component 1 gene and protection against liver disease. PLoS Genet. 2020, 16, e1008629.
- Anstee, Q.M.; Darlay, R.; Cockell, S.; Meroni, M.; Govaere, O.; Tiniakos, D.; Burt, A.D.; Bedossa, P.; Palmer, J.; Liu, Y.L.; et al. EPoS Consortium Investigators, Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterized cohort. J. Hepatol. 2020, 73, 505–515.
- Miao, Z.; Garske, K.M.; Pan, D.Z.; Koka, A.; Kaminska, D.; Männistö, V.; Sinsheimer, J.S.; Pihlajamäki, J.; Pajukanta, P. Identification of 90 NAFLD GWAS loci and establishment of NAFLD PRS and causal role of NAFLD in coronary artery disease. HGG Adv. 2021, 3, 100056.
- Fairfield, C.J.; Drake, T.M.; Pius, R.; Bretherick, A.D.; Campbell, A.; Clark, D.W.; Fallowfield, J.A.; Hayward, C.; Henderson, N.C.; Joshi, P.K.; et al. Genome-wide Association Study of NAFLD Using Electronic Health Records. Hepatol. Commun. 2022, 6, 297–308.
- Namjou, B.; Lingren, T.; Huang, Y.; Parameswaran, S.; Cobb, B.L.; Stanaway, I.B.; Connolly, J.J.; Mentch, F.D.; Benoit, B.; Niu, X.; et al. GWAS and enrichment analyses of non-alcoholic fatty liver disease identify new trait-associated genes and pathways across eMERGE Network. BMC Med. 2019, 17, 135.
- Anstee, Q.M.; Day, C.P. The Genetics of Non-alcoholic Fatty Liver Disease: Spotlight on PNPLA3 and TM6SF2. Semin. Liver Dis. 2015, 35, 270–290.
- Mancina, R.M.; Dongiovanni, P.; Petta, S.; Pingitore, P.; Meroni, M.; Rametta, R.; Boren, J.; Montalcini, T.; Pujia, A.; Wiklund, O.; et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Non-alcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 2016, 150, 1219–1230.e1216.
- Donati, B.; Dongiovanni, P.; Romeo, S.; Meroni, M.; McCain, M.; Miele, L.; Petta, S.; Maier, S.; Rosso, C.; De Luca, L.; et al. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci. Rep. 2017, 7, 4492.
- Buch, S.; Stickel, F.; Trépo, E.; Way, M.; Herrmann, A.; Nischalke, H.D.; Brosch, M.; Rosendahl, J.; Berg, T.; Ridinger, M.; et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 2015, 47, 1443–1448.
- Vujkovic, M.; Ramdas, S.; Lorenz, K.M.; Guo, X.; Darlay, R.; Cordell, H.J.; He, J.; Gindin, Y.; Chung, C.; Myers, R.P.; et al. A multiancestry genome-wide association study of unexplained chronic ALT elevation as a proxy for non-alcoholic fatty liver disease with histological and radiological validation. Nat. Genet. 2022, 54, 761–771.
- Stojsavljević, S.; Gomerčić Palčić, M.; Virović Jukić, L.; Smirčić Duvnjak, L.; Duvnjak, M. Adipokines and proinflammatory cytokines, the key mediators in the pathogenesis of non-alcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 18070–18091.
- Dogru, T.; Ercin, C.N.; Erdem, G.; Sonmez, A.; Tapan, S.; Tasci, I. Increased Hepatic and Circulating Interleukin-6 Levels in Human Non-alcoholic Steatohepatitis. Am. J. Gastroenterol. 2008, 103, 3217–3218.
- Park, J.; Zhao, Y.; Zhang, F.; Zhang, S.; Kwong, A.C.; Zhang, Y.; Hoffmann, H.H.; Bushweller, L.; Wu, X.; Ashbrook, A.W.; et al. IL-6/STAT3 axis dictates the PNPLA3-mediated susceptibility to non-alcoholic fatty liver disease. J. Hepatol. 2023, 78, 45–56.
- van Beek, J.H.; de Moor, M.H.; de Geus, E.J.; Lubke, G.H.; Vink, J.M.; Willemsen, G.; Boomsma, D.I. The genetic architecture of liver enzyme levels: GGT, ALT and AST. Behav. Genet. 2013, 43, 329–339.
- Rahmioglu, N.; Andrew, T.; Cherkas, L.; Surdulescu, G.; Swaminathan, R.; Spector, T.; Ahmadi, K.R. Epidemiology and genetic epidemiology of the liver function test proteins. PLoS ONE 2009, 4, e4435.
- Kwo, P.Y.; Cohen, S.M.; Lim, J.K. ACG Clinical Guideline: Evaluation of abnormal liver chemistries. Am. J. Gastroenterol. 2017, 112, 18–35.
- Abul-Husn, N.S.; Cheng, X.; Li, A.H.; Xin, Y.; Schurmann, C.; Stevis, P.; Liu, Y.; Kozlitina, J.; Stender, S.; Wood, G.C.; et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N. Engl. J. Med. 2018, 378, 1096–1106.
- Ward, L.D.; Tu, H.C.; Quenneville, C.B.; Tsour, S.; Flynn-Carroll, A.O.; Parker, M.M.; Deaton, A.M.; Haslett, P.A.J.; Lotta, L.A.; Verweij, N.; et al. GWAS of serum ALT and AST reveals an association of SLC30A10 Thr95Ile with hypermanganesemia symptoms. Nat. Commun. 2021, 12, 4571.
- Chen, V.L.; Du, X.; Chen, Y.; Kuppa, A.; Handelman, S.K.; Vohnoutka, R.B.; Peyser, P.A.; Palmer, N.D.; Bielak, L.F.; Halligan, B.; et al. Genome-wide association study of serum liver enzymes implicates diverse metabolic and liver pathology. Nat. Commun. 2021, 12, 816.
- Tian, C.; Stokowski, R.P.; Kershenobich, D.; Ballinger, D.G.; Hinds, D.A. Variant in PNPLA3 is associated with alcoholic liver disease. Nat. Genet. 2010, 42, 21–23.
- Stickel, F.; Buch, S.; Lau, K.; Zu Schwabedissen, H.M.; Berg, T.; Ridinger, M.; Rietschel, M.; Schafmayer, C.; Braun, F.; Hinrichsen, H.; et al. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology 2011, 58, 86–95.
- Falleti, E.; Fabris, C.; Cmet, S.; Cussigh, A.; Bitetto, D.; Fontanini, E.; Fornasiere, E.; Bignulin, S.; Fumolo, E.; Bignulin, E.; et al. PNPLA3 rs738409 C/G polymorphism in cirrhosis: Relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int. 2011, 31, 1137–1143.
- Anstee, Q.M.; Daly, A.K.; Day, C.P. Genetics of Alcoholic Liver Disease. Semin. Liver Dis. 2015, 35, 361–374.
- Yang, J.; Trepo, E.; Nahon, P.; Cao, Q.; Moreno, C.; Letouze, E.; Imbeaud, S.; Gustot, T.; Deviere, J.; Debette, S.; et al. PNPLA3 and TM6SF2 variants as risk factors of hepatocellular carcinoma across various etiologies and severity of underlying liver diseases. Int. J. Cancer 2018, 144, 533–544.
- Guyot, E.; Sutton, A.; Rufat, P.; Laguillier, C.; Mansouri, A.; Moreau, R.; Ganne-Carrié, N.; Beaugrand, M.; Charnaux, N.; Trinchet, J.C.; et al. PNPLA3 rs738409, hepatocellular carcinoma occurrence and risk model prediction in patients with cirrhosis. J. Hepatol. 2013, 58, 312–318.
- Falleti, E.; Cussigh, A.; Cmet, S.; Fabris, C.; Toniutto, P. PNPLA3 rs738409 and TM6SF2 rs58542926 variants increase the risk of hepatocellular carcinoma in alcoholic cirrhosis. Dig. Liver Dis. 2016, 48, 69–75.
- He, S.; McPhaul, C.; Li, J.Z.; Garuti, R.; Kinch, L.; Grishin, N.V.; Cohen, J.C.; Hobbs, H.H. A sequence variation (I148M) in PNPLA3 associated with non-alcoholic fatty liver disease disrupts triglyceride hydrolysis. J. Biol. Chem. 2010, 285, 6706–6715.
- Stickel, F.; Buch, S.; Nischalke, H.D.; Weiss, K.H.; Gotthardt, D.; Fischer, J.; Rosendahl, J.; Marot, A.; Elamly, M.; Casper, M.; et al. Genetic variants in PNPLA3 and TM6SF2 predispose to the development of hepatocellular carcinoma in individuals with alcohol-related cirrhosis. Am. J. Gastroenterol. 2018, 113, 1475–1483.
- Surakka, I.; Horikoshi, M.; Mägi, R.; Sarin, A.P.; Mahajan, A.; Lagou, V.; Marullo, L.; Ferreira, T.; Miraglio, B.; Timonen, S.; et al. ENGAGE Consortium. The impact of low-frequency and rare variants on lipid levels. Nat. Genet. 2015, 47, 589–597.
- Anstee, Q.M.; Seth, D.; Day, C.P. Genetic Factors That Affect Risk of Alcoholic and Non-alcoholic Fatty Liver Disease. Gastroenterology 2016, 150, 1728–1744.e7.
- Stewart, S.F.; Leathart, J.B.; Chen, Y.; Daly, A.K.; Rolla, R.; Vay, D.; Mottaran, E.; Vidali, M.; Albano, E.; Day, C.P. Valine-alanine manganese superoxide dismutase polymorphism is not associated with alcohol-induced oxidative stress or liver fibrosis. Hepatology 2002, 36, 1355–1360.
- Lucena, M.I.; Andrade, R.J.; Martínez, C.; Ulzurrun, E.; García-Martín, E.; Borraz, Y.; Fernández, M.C.; Romero-Gomez, M.; Castiella, A.; Planas, R.; et al. Glutathione S-transferase m1 and t1 null genotypes increase susceptibility to idiosyncratic drug-induced liver injury. Hepatology 2008, 48, 588–596.
- Brind, A.M.; Hurlstone, A.; Edrisinghe, D.; Gilmore, I.; Fisher, N.; Pirmohamed, M.; Fryer, A.A. The role of polymorphisms of glutathione S-transferases GSTM1, M3, P1, T1 and A1 in susceptibility to alcoholic liver disease. Alcohol Alcohol. 2004, 39, 478–483.
- Grove, J.; Daly, A.K.; Bassendine, M.F.; Gilvarry, E.; Day, C.P. Interleukin 10 promoter region polymorphisms and susceptibility to advanced alcoholic liver disease. Gut 2000, 46, 540–545.
- Roy, N.; Mukhopadhyay, I.; Das, K.; Pandit, P.; Majumder, P.P.; Santra, A.; Datta, S.; Banerjee, S.; Chowdhury, A. Genetic variants of TNFα, IL10, IL1β, CTLA4 and TGFβ1 modulate the indices of alcohol-induced liver injury in East Indian population. Gene 2012, 509, 178–188.
- Grove, J.; Daly, A.K.; Pastor, I.J.; Laso, F.J.; Romero, A.; Gonzalez-Sarmiento, R. −238 G>A polymorphism of tumor necrosis factor alpha gene (TNFA) is associated with alcoholic liver cirrhosis in alcoholic Spanish men. Alcohol. Clin. Exp. Res. 2005, 29, 1928–1931.
- Österreicher, C.H.; Halangk, J.; Berg, T.; Patsenker, E.; Homann, N.; Hellerbrand, C.; Seitz, H.K.; Eurich, D.; Stickel, F. Evaluation of the transforming growth factor β1 codon 25 (Arg → Pro) polymorphism in alcoholic liver disease. Cytokine 2008, 42, 18–23.
- Stickel, F.; Österreicher, C.H.; Halangk, J.; Berg, T.; Homann, N.; Hellerbrand, C.; Patsenker, E.; Schneider, V.; Kolb, A.; Friess, H.; et al. No role of matrixmetalloproteinase-3 genetic promoter polymorphism 1171 as a risk factor for cirrhosis in alcoholic liver disease. Alcohol. Clin. Exp. Res. 2008, 32, 959–965.
- Nischalke, H.D.; Lutz, P.; Krämer, B.; Söhne, J.; Müller, T.; Rosendahl, J.; Fischer, J.; Berg, T.; Hittatiya, K.; Fischer, H.P.; et al. A common polymorphism in the NCAN gene is associated with hepatocellular carcinoma in alcoholic liver disease. J. Hepatol. 2014, 61, 1073–1079.
- Kim, K.Y.; Kim, J.O.; Kim, Y.S.; Choi, J.E.; Park, J.M.; Han, K.; Park, D.H.; Park, Y.C.; Kim, B.T.; Hong, K.W. Genome-wide association of individual vulnerability with alcohol-associated liver disease: A Korean genome and epidemiology study. Hepatology 2022, 75, 391–402.
- Stickel, F.; Moreno, C.; Hampe, J.; Morgan, M.Y. The genetics of alcohol dependence and alcohol-related liver disease. J. Hepatol. 2017, 66, 195–211.
- Treutlein, J.; Cichon, S.; Ridinger, M.; Wodarz, N.; Soyka, M.; Zill, P.; Maier, W.; Moessner, R.; Gaebel, W.; Dahmen, N.; et al. Genome-wide association study of alcohol dependence. Pilot Feasibility Stud. 2009, 66, 773–784.
- Gelernter, J.; Kranzler, H.R.; Sherva, R.; Almasy, L.; Koesterer, R.; Smith, A.H.; Anton, R.; Preuss, U.W.; Ridinger, M.; Rujescu, D.; et al. Genome-wide association study of alcohol dependence: Significant findings in African- and European-Americans including novel risk loci. Mol. Psychiatry. 2014, 19, 41–49.
- Schwantes-An, T.H.; Darlay, R.; Mathurin, P.; Masson, S.; Liangpunsakul, S.; Mueller, S.; Aithal, G.P.; Eyer, F.; Gleeson, D.; Thompson, A.; et al. Genome-wide Association Study and Meta-analysis on Alcohol-Associated Liver Cirrhosis Identifies Genetic Risk Factors. Hepatology 2021, 73, 1920–1931.
- Emdin, C.A.; Haas, M.; Ajmera, V.; Simon, T.G.; Homburger, J.; Neben, C.; Jiang, L.; Wei, W.Q.; Feng, Q.; Zhou, A.; et al. Association of Genetic Variation With Cirrhosis: A Multi-Trait Genome-wide Association and Gene-Environment Interaction Study. Gastroenterology 2021, 160, 1620–1633.e13.
- Kozlitina, J.; Smagris, E.; Stender, S.; Nordestgaard, B.G.; Zhou, H.H.; Tybjærg-Hansen, A.; Vogt, T.F.; Hobbs, H.H.; Cohen, J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to non-alcoholic fatty liver disease. Nat. Genet. 2014, 46, 352–356.
- Innes, H.; Buch, S.; Hutchinson, S.; Guha, I.N.; Morling, J.R.; Barnes, E.; Irving, W.; Forrest, E.; Pedergnana, V.; Goldberg, D.; et al. Genome-wide Association Study for Alcohol-Related Cirrhosis Identifies Risk Loci in MARC1 and HNRNPUL1. Gastroenterology 2020, 159, 1276–1289.
- Wang, Z.; Budhu, A.S.; Shen, Y.; Wong, L.L.; Hernandez, B.Y.; Tiirikainen, M.; Ma, X.; Irwin, M.L.; Lu, L.; Zhao, H.; et al. Genetic susceptibility to hepatocellular carcinoma in chromosome 22q13.31, findings of a genome-wide association study. JGH Open 2021, 5, 1363–1372.
- Trépo, E.; Caruso, S.; Yang, J.; Imbeaud, S.; Couchy, G.; Bayard, Q.; Letouzé, E.; Ganne-Carrié, N.; Moreno, C.; Oussalah, A.; et al. Common genetic variation in alcohol-related hepatocellular carcinoma: A case-control genome-wide association study. Lancet Oncol. 2022, 23, 161–171.
- Trépo, E.; Nahon, P.; Bontempi, G.; Valenti, L.; Falleti, E.; Nischalke, H.D.; Hamza, S.; Corradini, S.G.; Burza, M.A.; Guyot, E.; et al. Association between the PNPLA3 (rs738409 C> G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology 2014, 59, 2170–2177.
- Trépo, E.; Valenti, L. Update on NAFLD genetics: From new variants to the clinic. J. Hepatol. 2020, 72, 1196–1209.
- Hindson, J. GWAS reveals variants for alcohol-related hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 79.
- Yang, T.-H.; Chan, C.; Yang, P.-J.; Huang, Y.-H.; Lee, M.-H. Genetic Susceptibility to Hepatocellular Carcinoma in Patients with Chronic Hepatitis Virus Infection. Viruses 2023, 15, 559.
- Choudhary, N.S.; Duseja, A. Genetic and epigenetic disease modifiers: Non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD). Transl. Gastroenterol. Hepatol. 2021, 6, 2.
- Sakurai, Y.; Kubota, N.; Yamauchi, T.; Kadowaki, T. Role of Insulin Resistance in MAFLD. Int. J. Mol. Sci. 2021, 22, 4156.
- Macaluso, F.S.; Maida, M.; Petta, S. Genetic background in non-alcoholic fatty liver disease: A comprehensive review. World J. Gastroenterol. 2015, 21, 11088–11111.
- Gundogdu, A.; Nalbantoglu, U. Human genome-microbiome interaction: Metagenomics frontiers for the aetiopathology of autoimmune diseases. Microb. Genom. 2017, 3, e000112.




