
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yuji Sasaki | -- | 1460 | 2023-11-16 10:19:00 | | | |
| 2 | Sirius Huang | Meta information modification | 1460 | 2023-11-17 09:15:03 | | |
Video Upload Options
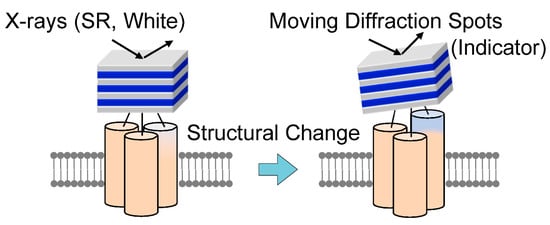
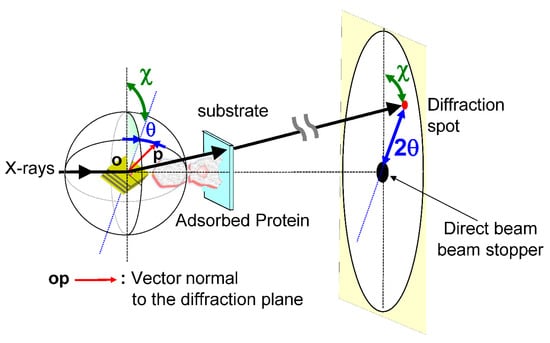
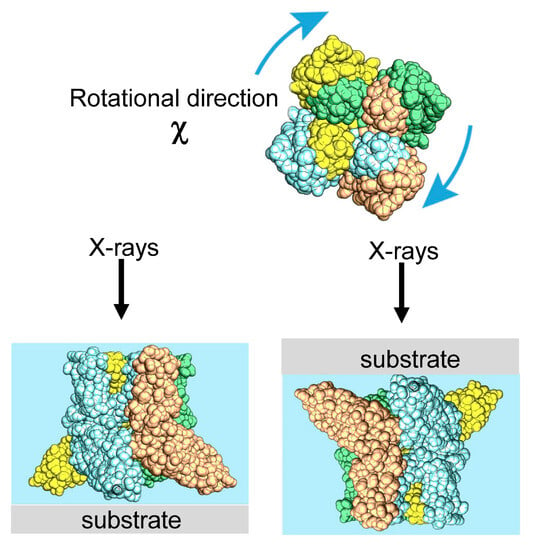
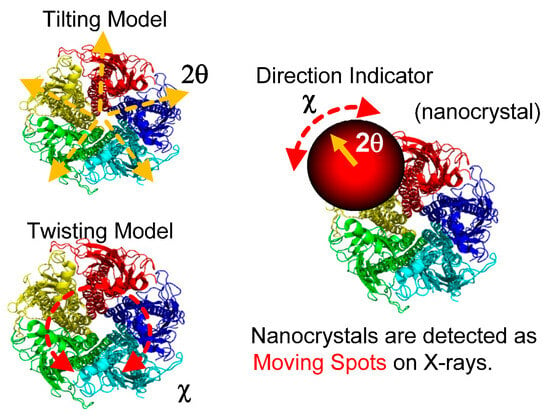
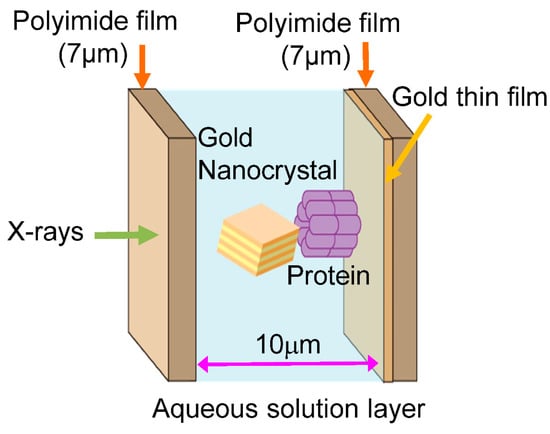
In 1998, the diffracted X-ray tracking (DXT) method pioneered the attainment of molecular dynamics measurements within individual molecules. This breakthrough revolutionized the field by enabling unprecedented insights into the complex workings of molecular systems. Similar to the single-molecule fluorescence labeling technique used in the visible range, DXT uses a labeling method and a pink beam to closely track the diffraction pattern emitted from the labeled gold nanocrystals. Moreover, by utilizing X-rays with extremely short wavelengths, DXT has achieved unparalleled accuracy and sensitivity, exceeding initial expectations. As a result, this remarkable advance has facilitated the search for internal dynamics within many protein molecules.
1. History and Concept of Single Molecule Dynamics Using X-rays
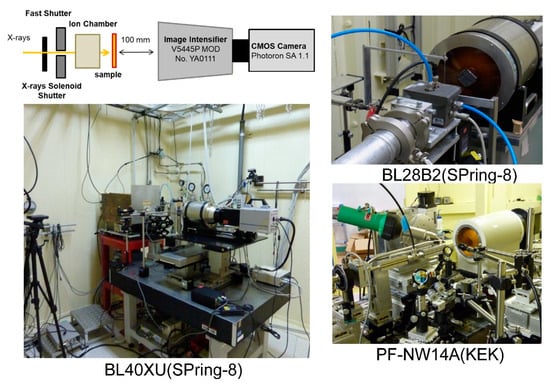
2. Measuring Single Molecule Dynamics Using Diffracted X-ray Tracking
References
- Michaelis, J.; Hettich, C.; Mlynek, J.; Sandoghdar, V. Optical microscopy using a single-molecule light source. Nature 2000, 405, 325–328.
- Celebrano, M.; Kukura, P.; Renn, A.; Sandoghdar, V. Single-molecule imaging by optical absorption. Nat. Photonics 2011, 5, 95–98.
- Weiss, S. Fluorescence Spectroscopy of Single Biomolecules. Science 1999, 283, 1676–1683.
- Fratalocchi, A.; Ruocco, G. Single-Molecule Imaging with X-Ray Free-Electron Lasers: Dream or Reality? Phys. Rev. Lett. 2011, 106, 105504.
- Sasaki, Y.C.; Suzuki, Y.; Ishibashi, T.; Satoh, I. Interference Effect of Electron-Capture X-Rays from an 125I-Labeled Protein Monolayer in an Aqueous Solution. Anal. Sci. 1995, 11, 545–548.
- Sasaki, Y.C.; Suzuki, Y.; Yasuda, K.; Tomioka, Y.; Ishibashi, T.; Satoh, I. Site determination of radioactive atoms from the interference effect of electron-capture rays: Structural change of 111In-labelled azobenzene derivative. Thin Solid Films 1996, 248–285, 456–458.
- Sasaki, Y.C.; Suzuki, Y.; Tomioka, Y.; Ishibashi, T.; Takahashi, M.; Satoh, I. Observation of Nanometer-Level Structural Changes by the Trans−Cis Transition of an Azobenzene Derivative Monolayer with a Radioactive Tracer. Langmuir 1996, 12, 4173–4175.
- Sasaki, Y.C.; Suzuki, Y.; Yamanashi, H.; Arai, A.; Yanagihara, M. Time-resolved fluorescent X-ray interference. J. Synchrotron Radiat. 1998, 5, 1075–1078.
- Sasaki, Y.C.; Yasuda, K.; Takahashi, M.; Satoh, I.; Ishiwata, S. Structural information from the interference effect of electron-capture X-rays. J. Radioanal. Nucl. Chem. 1999, 239, 341–344.
- Adrian, M.; Dubochet, J.; Lepault, J.; McDowall, A.W. Cryo-electron microscopy of viruses. Nature 1984, 308, 32–36.
- Frank, J. Single-Particle Imaging of Macromolecules by Cryo-Electron Microscopy. Annual Review of Biophysics and Biomolecular Structure. Annu. Rev. 2002, 31, 303–319.
- Assaiya, A.; Burada, A.P.; Dhingra, S.; Kumar, J. An overview of the recent advances in cryo-electron microscopy for life sciences. Bose, K., editor. Emerg. Top. Life Sci. 2021, 5, 151–168.
- Sasaki, Y.C.; Suzuki, Y.; Yagi, N.; Adachi, S.; Ishibashi, M.; Suda, H.; Toyota, K.; Yanagihara, M. Tracking of individual nanocrystals using diffracted x rays. Phys. Rev. E 2000, 62, 3843–3847.
- Sasaki, Y.C.; Okumura, Y.; Adachi, S.; Suzuki, Y.; Yagi, N. Diffracted X-ray tracking: New system for single molecular detection with X-rays. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2001, 467–468, 1049–1052.
- Sasaki, Y.C.; Okumura, Y.; Adachi, S.; Suda, H.; Taniguchi, Y.; Yagi, N. Picometer-Scale Dynamical X-Ray Imaging of Single DNA Molecules. Phys. Rev. Lett. 2001, 87, 248102.
- Sasaki, Y.C. Dynamical Observations of Soft Nanomaterials Using X-rays or High-Energy Probes. 69–107, Chapter 2, Soft Nanomaterial; Nalwa, H.S., Ed.; American Scientific Publishers: Valencia, CA, USA, 2009.
- Sasaki, Y.C. Dynamical Observations of Local Bio-molecular Sites Using Nanocrystals. AIP Conf. Proc. 2004, 705, 1023–1026.
- Sasaki, Y.C. Single protein molecular dynamics determined with ultra-high precision. Biochem. Soc. Trans. 2004, 32, 761–763.
- Ichiyanagi, K.; Sekiguchi, H.; Hoshino, M.; Kajiwara, K.; Hoshisashi, K.; Jae-won, C.; Tokue, M.; Matsushita, Y.; Nishijima, M.; Inoue, Y.; et al. Diffracted X-ray tracking for monitoring intramolecular motion in individual protein molecules using broad band X-ray. Rev. Sci. Instrum. 2013, 84, 103701.
- Sekiguchi, H.; Sasaki, Y.C. Dynamic 3D visualization of active protein’s motion using diffracted X-ray tracking. Jpn. J. Appl. Phys. 2019, 58, 120501.
- Okumura, Y.; Oka, T.; Kataoka, M.; Taniguchi, Y.; Sasaki, Y.C. Picometer-scale dynamical observations of individual membrane proteins: The case of bacteriorhodopsin. Phys. Rev. E 2004, 70, 021917.
- Okumura, Y.; Oka, T.; Taniguchi, Y.; Sasaki, Y.C. Dynamical Observations of Membrane Proteins: The Case of Bacteriorhodopsin. AIP Conf. Proc. 2004, 705, 1174–1177.
- Kawashima, Y.; Sasaki, Y.C.; Sugita, Y.; Yoda, T.; Okamoto, Y. Replica-exchange molecular dynamics simulation of diffracted X-ray tracking. Mol. Simul. 2007, 33, 97–102.
- Sagawa, T.; Azuma, T.; Sasaki, Y.C. Dynamical regulations of protein–ligand bindings at single molecular level. Biochem. Biophys. Res. Commun. 2007, 355, 770–775.
- Shimizu, H.; Iwamoto, M.; Konno, T.; Nihei, A.; Sasaki, Y.C.; Oiki, S. Global Twisting Motion of Single Molecular KcsA Potassium Channel upon Gating. Cell 2008, 132, 67–78.
- Sekiguchi, H.; Suzuki, Y.; Nishino, Y.; Kobayashi, S.; Shimoyama, Y.; Cai, W.; Nagata, K.; Okada, M.; Ichiyanagi, K.; Ohta, N.; et al. Real Time Ligand-Induced Motion Mappings of AChBP and nAChR Using X-ray Single Molecule Tracking. Sci. Rep. 2014, 4, 6384.
- Kozono, H.; Matsushita, Y.; Ogawa, N.; Kozono, Y.; Miyabe, T.; Sekiguchi, H.; Ichiyanagi, K.; Okimoto, N.; Taiji, M.; Kanagawa, O.; et al. Single-Molecule Motions of MHC Class II Rely on Bound Peptides. Biophys. J. 2015, 108, 350–359.
- Matsushita, Y.; Sekiguchi, H.; Ichiyanagi, K.; Ohta, N.; Ikezaki, K.; Goto, Y.; Sasaki, Y.C. Time-resolved X-ray Tracking of Expansion and Compression Dynamics in Supersaturating Ion-Networks. Sci. Rep. 2015, 5, 17647.
- Matsushita, Y.; Sekiguchi, H.; Wong, C.J.; Nishijima, M.; Ikezaki, K.; Hamada, D.; Goto, Y.; Sasaki, Y.C. Nanoscale Dynamics of Protein Assembly Networks in Supersaturated Solutions. Sci. Rep. 2017, 7, 13883.




