Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Behnam Kiani Kalejahi | -- | 1278 | 2023-11-09 11:04:29 | | | |
| 2 | Jessie Wu | + 3 word(s) | 1281 | 2023-11-10 04:24:38 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kiani Kalejahi, B.; Meshgini, S.; Danishvar, S. Brain Tumor Segmentation, Deep Learning and GAN Network. Encyclopedia. Available online: https://encyclopedia.pub/entry/51351 (accessed on 07 February 2026).
Kiani Kalejahi B, Meshgini S, Danishvar S. Brain Tumor Segmentation, Deep Learning and GAN Network. Encyclopedia. Available at: https://encyclopedia.pub/entry/51351. Accessed February 07, 2026.
Kiani Kalejahi, Behnam, Saeed Meshgini, Sebelan Danishvar. "Brain Tumor Segmentation, Deep Learning and GAN Network" Encyclopedia, https://encyclopedia.pub/entry/51351 (accessed February 07, 2026).
Kiani Kalejahi, B., Meshgini, S., & Danishvar, S. (2023, November 09). Brain Tumor Segmentation, Deep Learning and GAN Network. In Encyclopedia. https://encyclopedia.pub/entry/51351
Kiani Kalejahi, Behnam, et al. "Brain Tumor Segmentation, Deep Learning and GAN Network." Encyclopedia. Web. 09 November, 2023.
Copy Citation
Images of brain tumors may only show up in a small subset of scans, so important details may be missed. Further, because labeling is typically a labor-intensive and time-consuming task, there are typically only a small number of medical imaging datasets available for analysis.
generative adversarial networks
brain tumor
medical image segmentation
computer aided diagnosis
1. Introduction
Various imaging methods in medicine are designed for specific purposes, among which the most widely used are angiography (using X-rays), computer tomography (CT) scans (using X-rays), sonography (using ultrasound waves), magnetic resonance imaging (MRI) (using radio waves and magnetic amplification), and radiology (using X-rays). Each of the mentioned imaging techniques is specific to certain tissues in the body. The MRI technique provides clinicians with crucial information on the type, size, shape, and location of brain tumors without exposing the patient to dangerous radiation. Meningioma and glioma are fatal brain tumors that can be discovered via magnetic resonance imaging studies [1]. If tumors are not diagnosed in their early stages, they can be seriously dangerous in the future. T1, T1c, T2, and FLAIR MRI sequences provide detailed information about brain tumors:
- (1)
-
T1-weighted scans that distinguish healthy tissues from those with tumors.
- (2)
-
T2-weighted scans to delineate the tumor area, which creates a bright image area.
- (3)
-
T1-c scans use a contrast agent that builds up at the edge of the tumor and gives a bright signal.
- (4)
-
The water molecule suppression signal is used in FLAIR scans.
Glioma is the most common kind of brain cancer in humans. The World Health Organization (WHO) divides tumors into four types [1]. Low-level tumors such as meningioma are classified as grade one and two cancers, whereas gliomas are classified as grade three and four cancers. Meningiomas account for around 20% of all brain tumors. This type of tumor has a spherical shape and grows at a slower rate. Even though meningioma is a low-risk tumor with a modest growth rate, it can cause considerable harm to the patient if not treated early. Since lesions are typically tiny and have varying color, shape, and texture alterations, interpreting MRI images to detect brain tumors is a time-consuming, difficult, and delicate process. Neurologists and surgeons sometimes struggle to make the right call. Noisy images and exhausted doctors can also cause misinterpretations of images. Analysis with the help of computer algorithms is one of the most promising methods for facing such problems in MRI images. In the meantime, deep learning (DL) architectures are prominent as a method and work well in this field.
2. Brain Tumor Segmentation
The present categorization of methods for brain tumor segmentation can be classified into many groups according to unique conceptual frameworks. The categorization of brain tumor segmentation methods is commonly based on the level of human involvement, resulting in three distinct categories: manual, semi-automated, and fully automatic [2]. Proficient manual brain tumor segmentation requires professionals to possess a comprehensive understanding of picture interpretation, brain tumor characteristics, and relevant disciplines such as anatomy. The manual segmentation of brain tumors is the process of manually delineating the boundaries of the tumor and assigning distinct labels to the various regions of the anatomical components. To date, manual segmentation has been extensively utilized in clinical trials. Due to the association of numerous images with the progression of brain tumors, professionals may encounter challenges when manually segmenting various regions of brain tumors, as this process is prone to errors, time-consuming, and yields unsatisfactory results. Consequently, the utilization of more sophisticated segmentation techniques, such as semi-automatic and fully automatic segmentation methods, can effectively address this issue. The process of semi-automated brain tumor segmentation requires the utilization of specialized knowledge, human involvement, and computational tools. In the context of brain tumor diagnosis, semi-automated procedures involve the input of parameters by an expert, the analysis of visual data, and the provision of feedback for software computations. The method category is comprised of three distinct components: initial processing, feedback, and assessment. While semi-automated algorithms for brain tumor segmentation have demonstrated superior performance compared to manual segmentation, it is important to note that discrepancies in findings can arise due to variations among various specialists and over different time points. Consequently, there has been a development of techniques aimed at achieving the fully automated segmentation of brain tumors.
3. Deep Learning
Deep learning is a subfield within the broader discipline of Machine Learning (ML) that is employed to model complex problems and abstract notions. Deep learning (DL) facilitates the training of models that consist of numerous layers of processing, known as deep neural networks, which have the ability to acquire abstract representations of data. The assessment of conceptual qualities as nonlinear functions of low-level features is facilitated by the multilayer nature of DL networks. Convolutional Neural Networks (CNN), Restricted Boltzmann Machines (RBM), Deep Belief Networks (DBN), Deep Auto-Encoders (DAE), Recurrent Neural Networks (RNN), and their derivatives, including Long Short-term Memory (LSTM), are considered to be very valuable Deep Neural Networks (DNNs). These networks are frequently employed to execute extensive operations for a multitude of objectives. The important difference between conventional machine learning (ML) and deep learning (DL) algorithms is due to the aspect of feature engineering. Traditional machine learning (ML) algorithms perform classification tasks by utilizing a predefined set of characteristics. In contrast, deep learning (DL) techniques possess the ability to automatically extract features, resulting in higher accuracy compared to conventional ML models. The effectiveness of these models in addressing large-scale data issues surpasses that of shallow machine learning techniques due to the increased depth of the processing layers [2]. The study applied various machine learning (ML) methodologies, such as the random forest algorithm and Support Vector Machines (SVM), to automate the process of identifying and segmenting lesions using MRI data. Deep learning methods such as Restricted Boltzmann Machines (RBM), Denoising Autoencoders (DAE), Convolutional Neural Networks (CNN), and Long Short-Term Memory (LSTM) have recently been increasingly prominent in the field of medical picture analysis. The Generative Adversarial Network (GAN) is a widely recognized and very efficient deep learning model.
4. Generative Adversarial Network
Generative Adversarial (GAN) networks are generative models that generate high-quality data from a small collection of training datasets. The GAN network is made up of two components: the generator and the discriminator. The generator attempts to learn the model built from the data and, as a result, generates graphics from random noise inputs. The discriminator is a CNN network that attempts to discriminate between actual data (training data) and data generated by the generator, and it assesses the likelihood of a mistake using this approach. Two of the most well-known GAN architectures are the semi-supervised GAN and Laplacian pyramid GAN. Since GAN networks are trained using only non-lesion images, it is expected that the error probability assigned by the network to images with lesions will be significantly different from that assigned to non-lesion images. The general structure of a standard GAN network with a generator and discriminator is shown in Figure 1. In general, GANs have two uses in medical imaging. The first use is concerned with generation, which can aid in understanding the underlying structure of training data and learning to produce new pictures. This aspect of GAN networks holds great promise for coping with data shortages while protecting patient privacy. The second application emphasizes the differentiating feature. Due to the necessity for vast volumes of training data, image creation methods have proliferated in the field of deep learning. Recent research has demonstrated that GAN networks may be utilized for a variety of applications, including image-to-image translation [3] and unsupervised representation learning. GANs have also been demonstrated for unsupervised data domain pattern matching in multimodal medical imaging, indicating their potential for usage with limited medical imaging data.
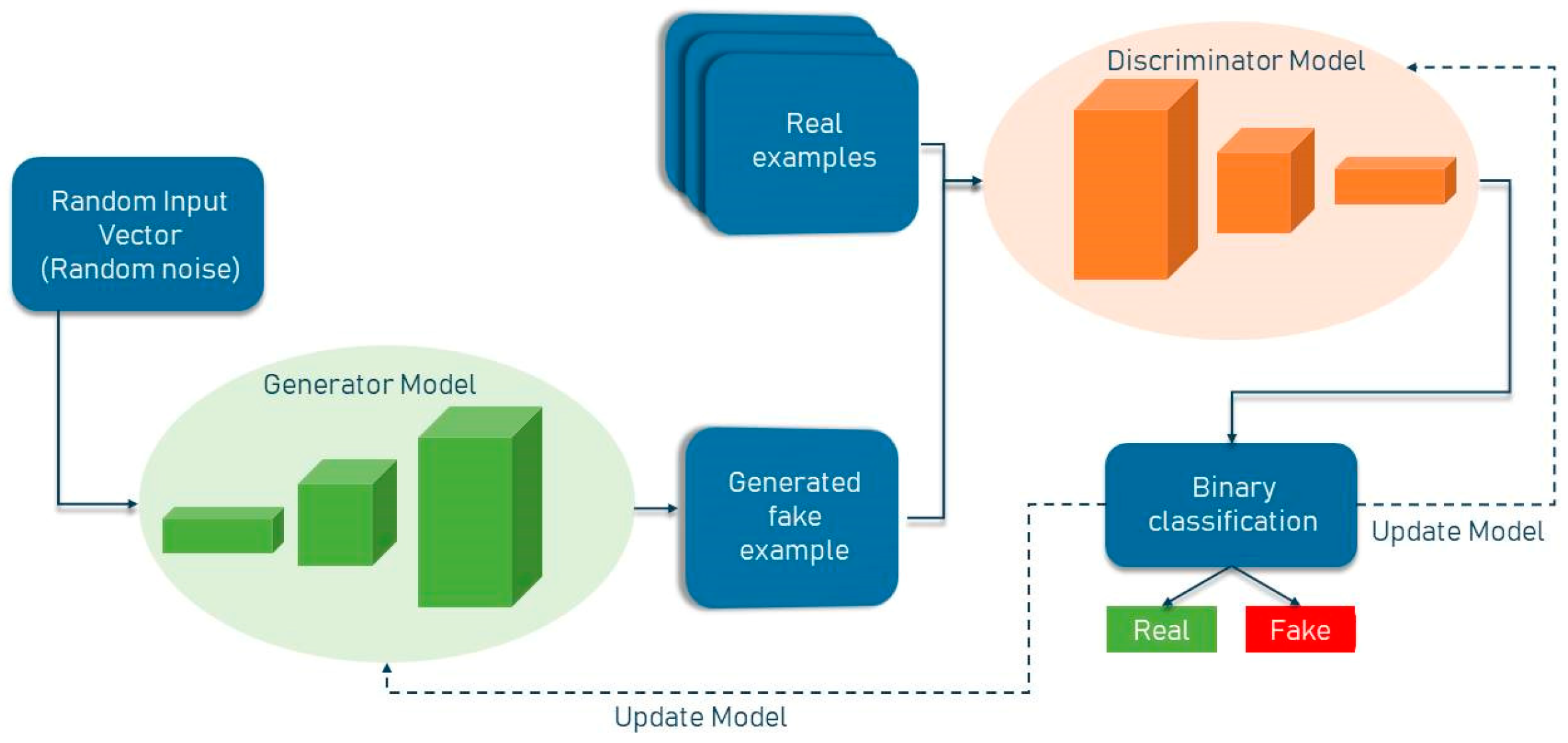
Figure 1. General structure of GAN network.
References
- Gould, S.; Fulton, R.; Koller, D. Decomposing a scene into geometric and semantically consistent regions. In Proceedings of the 2009 IEEE 12th International Conference on Computer Vision, Kyoto, Japan, 29 September–2 October 2009; pp. 1–8.
- Yousefi-Azar, M.; Varadharajan, V.; Hamey, L.; Tupakula, U. Autoencoder-based feature learning for cyber security applications. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 3854–3861.
- Hatcher, W.G.; Yu, W. A Survey of Deep Learning: Platforms, Applications and Emerging Research Trends. IEEE Access 2018, 6, 24411–24432.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
516
Revisions:
2 times
(View History)
Update Date:
10 Nov 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




