Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | muhammad sulaiman yousafzai | -- | 2950 | 2023-11-02 21:38:44 | | | |
| 2 | Camila Xu | Meta information modification | 2950 | 2023-11-03 01:31:33 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Yousafzai, M.S.; Hammer, J.A. Optical Sensors in Mechanobiology. Encyclopedia. Available online: https://encyclopedia.pub/entry/51115 (accessed on 08 February 2026).
Yousafzai MS, Hammer JA. Optical Sensors in Mechanobiology. Encyclopedia. Available at: https://encyclopedia.pub/entry/51115. Accessed February 08, 2026.
Yousafzai, Muhammad Sulaiman, John A. Hammer. "Optical Sensors in Mechanobiology" Encyclopedia, https://encyclopedia.pub/entry/51115 (accessed February 08, 2026).
Yousafzai, M.S., & Hammer, J.A. (2023, November 02). Optical Sensors in Mechanobiology. In Encyclopedia. https://encyclopedia.pub/entry/51115
Yousafzai, Muhammad Sulaiman and John A. Hammer. "Optical Sensors in Mechanobiology." Encyclopedia. Web. 02 November, 2023.
Copy Citation
Optical sensors play a central role in the study of mechanobiology by enabling the accurate detection and measurement of mechanical forces and their effects on biological systems. Mechanobiology explores how mechanical forces influence cellular processes, tissue development, and overall physiological functions.
biosensors
organoids
spheroids
extracellular matrix (ECM)
mechanobiology
1. Introduction
The culture of cells in 2D has powered tremendous advances in the biological sciences and in the biotech industry [1]. Two-dimensional cultures are easy to make and manipulate, relatively inexpensive, and for immortal cells last indefinitely. That said, major differences exist between cells grown in 2D and in 3D at both the single cell and collective cell levels. For example, invasive cells use different modes of migration in 3D culture than in 2D culture [2]. Similarly, epithelial cells undergo geometrical and structural transformations in 3D culture that do not occur in 2D cell monolayers [3]. As a case in point, the modeling of brain development is possible using 3D organoids but is not possible using 2D cultures [4]. One major disadvantage of 2D cultures is that they lack many of the cell–cell and cell–matrix interactions seen in tissues. Such interactions likely influence responses to pharmacological perturbations. The fact that cells in 2D are usually homogenous, i.e., not in contact with other cells types as in tissues and many types of organoids, may also influence responses to pharmacological perturbations [5]. On the other hand, 3D cultures provide in vivo-like cell–cell and cell–matrix interactions, and the associated signaling pathways responsible for affecting cell phenotypes. However, 3D cultures are more costly to establish and maintain than 2D cultures. They also rely largely on imaging and manipulation modalities designed for 2D, and so, there are some methodological challenges to overcome [6]. These prominent differences can lead to many differences in biological functionalities, and hence, the fundamental understanding of in vivo processes (Figure 1).
Different terminologies are used by the research community for 3D cellular structures. These include organoids, enteroids, spheroids, cell-aggregates, and organotypic cultures. Importantly, some of these structures differ as regards their structure and function, as well as their phenotypic and genotypic complexity [7]. That said, we can broadly categorize 3D structures as organoids and spheroids, as both are 3D cell culture systems that mimic to varying degrees the structure and function of an in vivo tissue. An organoid is a 3D cell culture system derived from stem cells or tissue progenitor cells. They have the ability to self-organize, regenerate, and recapitulate to varying degrees a tissue-specific phenotype [8]. Organoids are broadly used to study organogenesis of homeostatic and diseased organ tissues [9]. A spheroid is a 3D culture derived from a cell line that under the right conditions will aggregate to form a 3D spherical structure [10]. While spheroids can be grown with or without a matrix, they are most commonly created using nonadherent conditions to achieve high yield and rapid, synchronous formation. Spheroids are commonly used as simple tumor models [11][12][13] (Figure 2).
Cells in vivo are constantly probing their mechanical environment through integrin-based adhesions to the extracellular matrix (ECM), cadherin-based adhesions to surrounding cells, and mechanosensitive ion channels. They then integrate all of these signals to trigger an appropriate response [14]. As a result, tissues exhibit characteristic mechanical features that ensure proper tissue functionality. For example, brain tissues are soft with elastic moduli of ~100 Pa, while bones are very stiff with a modulus of over 100 kPa [15]. While the mechanical features of tissues are mostly invariant under homeostatic conditions, that can change in pathological conditions. For example, tumors are typically stiffer than the surrounding healthy tissues [16]. A good example of this is breast cancer, where normal breast tissue is in the range ~500 Pa and increases to more than 1000 Pa in breast tumors. Importantly, 3D spheroids and organoid models can be exploited to understand the connection between the mechanical signature of a tissue and its functionality in vivo. Generally, Matrigel and Collagen type-I are the most common matrices used for generating 3D cultures [17]. Matrigel is mostly used for growing organoids while collagen I is mostly used in tumor invasion studies as it better mimics the tumor microenvironment [7][18]. Of note, there are batch-to-batch variabilities in the composition of these matrices and, hence, variabilities in their mechanical properties. Moreover, because they are not amenable to chemical and mechanical alterations, it is hard to fine-tune matrix mechanics to promote a specific cell behavior. Given this, there is a clear need to design mechanically controllable matrices with precise chemical compositions and mechanical properties [19][20].
To understand cell behavior in 3D environments, and to develop mechanically tunable matrices for such environments, one must possess tools capable of accurately measuring forces in 3D.
Organoid growth involves complex shape transformations driven by mechanical forces generated in large part by actomyosin-dependent contractions. These forces are constantly adjusted by cell–cell and cell–matrix crosstalk to establish the correct morphological structure for the given environmental condition. In organoid cultures, the structure emerges stochastically as there is no control over stem cell differentiation patterns or mechanical forces. To remedy this, organ-on-a-chip (OoC) platforms can be used to precisely control structure by defining the position of stem cells and by allowing the application forces with increased spatiotemporal resolution.
Due to their potential for both basic and translational research, many research groups have begun using 3D culture platforms. A survey of PubMed (www.ncbi.nlm.nih.gov, accessed on 27 June 2023) (Figure 1A) over the last two decades shows a corresponding surge in research publications. Systems complexities increase as get closer to physiological conditions in vitro and throughput decreases (Figure 1B).
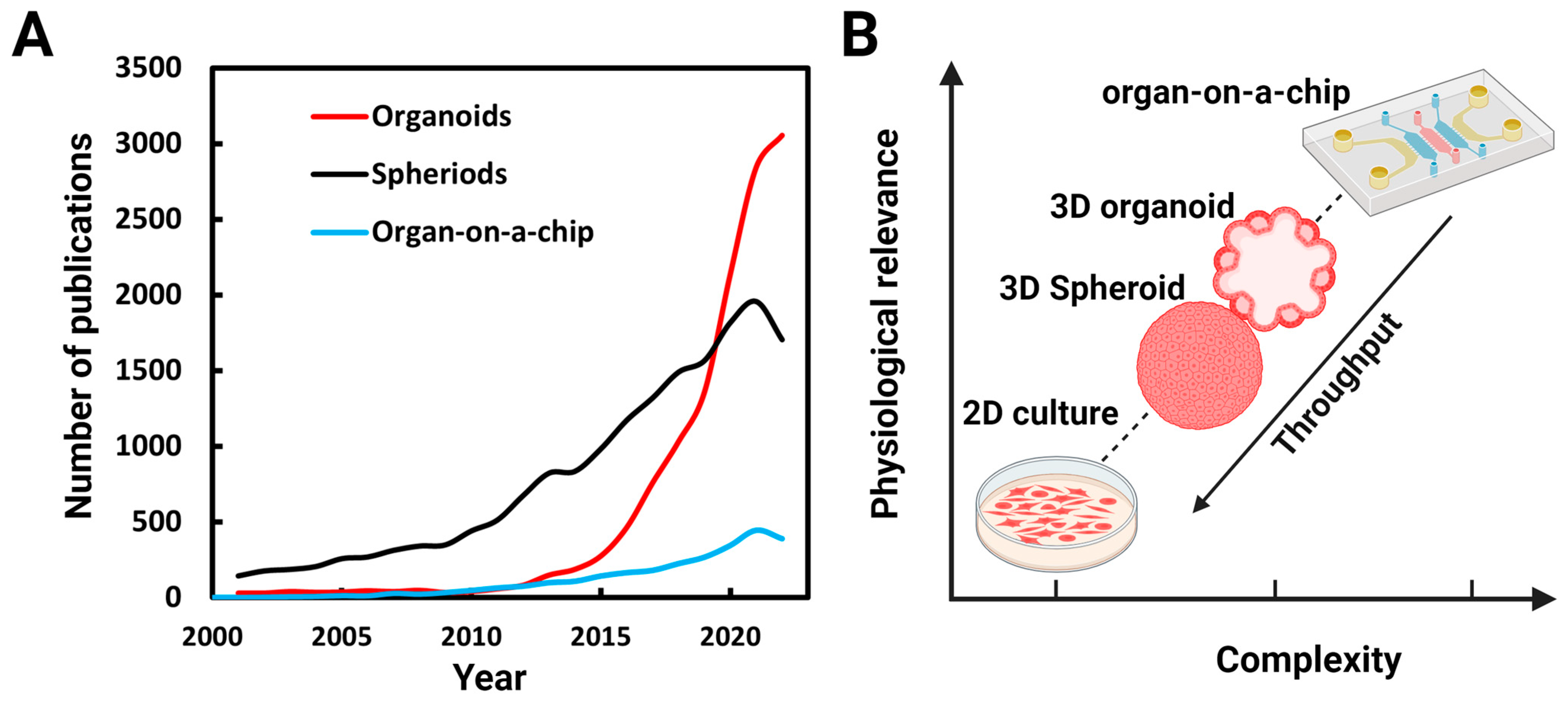
Figure 1. Numbers of publications on organoids, spheroids, and organs-on-a-chip: (A) Number of publications per year found identified by a PubMed search using the terms organoids, spheroids (spheroids and cell aggregates), and organ-on-a-chip between 2001 and 2022. (B) As get closer to in vivo conditions, the complexities of the systems increases and throughput decreases.
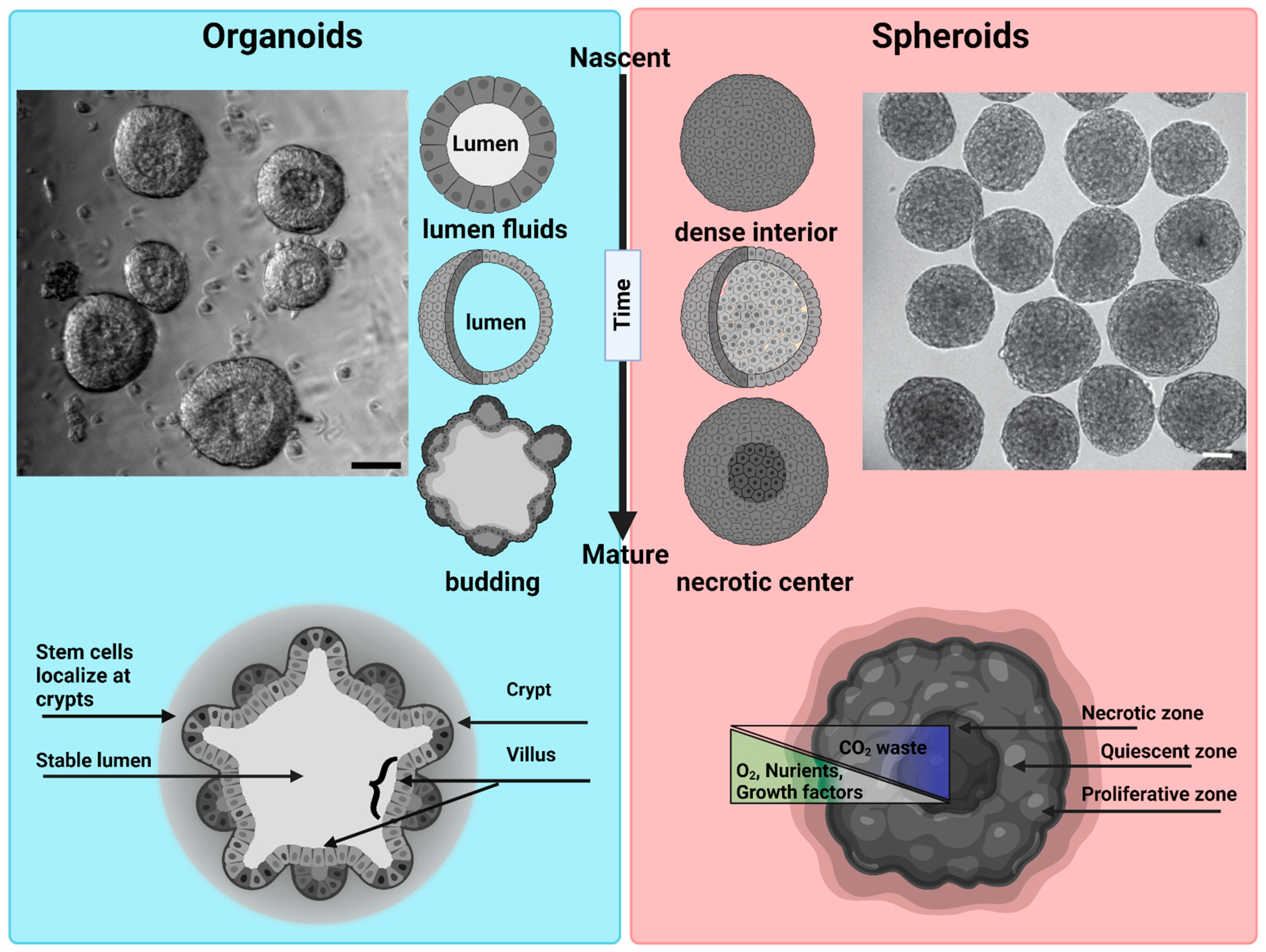
Figure 2. Morphological differences between organoids and spheroids: Organoids: a representative bright field image of intestinal organoids grown in Matrigel from an intestinal crypt. Nascent organoids have a spherical morphology, are composed of a monolayer of a columnar stem cells and transient amplifying cells, and contain a lumen filled with fluid and extruded cells. As they mature, organoids grow in size and develop a crypt and villus morphology just like intestinal tissue in vivo. The stem cells are localized within the crypts, while the nutrient-absorbing enterocytes form the villus. Intestinal organoids can be used to study mechanisms of barrier function, lumenogenesis, cell division, and cell extrusion. Spheroids: a representative DIC image of spheroids formed by Sarcoma S-180 cells formed via a spinning method within a non-adherent platform. Adopted with permission from [21]. Nascent spheroids are initially roughly spherical, and then merge together to form big spheroids. They are dense structures where cells are connected initially through cell–cell-adhesion. As they mature, they produce necrotic centers. Spheroids are very useful for measuring tumor growth dynamics, mechanical and immuno-resistant barriers, drug responses, and solid stresses. Scale bar is 50 µm.
2. Optical Sensors in Mechanobiology
Optical sensors play a central role in the study of mechanobiology by enabling the accurate detection and measurement of mechanical forces and their effects on biological systems. The history of biosensors dates back several decades to when the term “biosensor” was first introduced by Clark and Lyons in 1962 to describe a device that used an enzyme electrode to measure glucose levels [22]. An enormous advance in the field of biosensors was the discovery of Green Fluorescent Protein (GFP) [23]. By allowing one to observe the dynamic activities of proteins within living cells in real time, GFP profoundly reshaped the landscapes of imaging, cell biology, and optical biosensing. Mechanobiology explores how mechanical forces influence cellular processes, tissue development, and overall physiological functions [24]. Unlike chemical sensors, which can be studied from the analyte concentrations, force cannot be directly measured; it can only be inferred from material deformations [25]. Harris, Wild, and Stopak published a seminal paper in 1980 in which they measured cell-generated forces by observing wrinkles in a silicone substratum, thereby laying the groundwork for traction force microscopy (TFM) [26]. In 1995, Guilford and colleagues employed micro rheology to measure internal cellular forces by utilizing magnetic beads that were internalized by macrophages [27]. The integration of Förster Resonance Energy Transfer (FRET) in cells [28], the development of force spectroscopy techniques (optical tweezers, magnetic tweezers, atomic force microscopy), and super-resolution microscopy (SIM, STED, STORM, PALM) have all changed the course of mechanobiology.
While the field of mechanobiology has experienced enormous growth over the last few decades, there is still ongoing debate regarding the time and length scales over which cell mechanics actively contribute to biological processes [29]. Cells have the ability to sense (outside-in), generate (inside-out), and integrate mechanical forces over a range of time and length scales [30]. The cell’s plasma membrane is the primary site of force transmission. Moreover, it is decorated with mechanosensitive channels (i.e., Piezo1, Piezo2) and transmembrane receptors (e.g., integrins) that link the cell’s interior to the outside word. When subjected to mechanical stimuli, mechanosensitive channels respond within milliseconds. Conversely, integrin-mediated adhesion interactions take tens of seconds, and the cytoskeletal alterations that occur downstream of force transmission through integrins occurs over the span of many minutes. Likewise, cells also receive mechanical forces from neighboring cells in the tissue [31]. Cell integrates these mechanical forces to organize and control global responses like cell migration, proliferation, cell extrusion, and structural/morphological changes [32]. How cells integrate these forces to create global responses is only partially understood. Efforts to quantify mechanotransduction at different time and length scales, and to correlate these measurements with physiological responses, will provide major insights.
Prominent mechanobiological techniques that utilize optical sensors include TFM, optical tweezers, and FRET-based tension sensors. While it is not possible to fully understand mechanotransduction using just one technique, results obtained using multiple techniques can be combined to obtain deeper understanding of any particular event.
TFM has played a major role in the growth of the field of mechanobiology. This technique is used to quantify the forces that cells exert on their surrounding environment [33]. It is most commonly used to study cell–substrate interactions, where cells adhered to a deformable substrate like a hydrogel can generate mechanical forces. These forces are quantified by imaging fluorescent microbeads embedded within the substrate. As the cell applies force to the substrate, the beads in the substrate are displaced. By tracking bead displacements using particle image velocimetry (PIV), the distribution and magnitude of the forces exerted by the cells on the substrate can be calculated. While TFM is typically used to measure forces produced by single cells [34], and even forces generated by individual focal adhesions [35], it can also be used to measure forces produced by collections of cells (e.g., 2D organoids) [36]. Studies employing TFM have provided major insights into the mechanobiology of cell migration [37], cell division [38], wound healing [39], and jamming- unjamming transitions [40]. That said, the implementation of TFM in 3D systems will be challenging as the technique relies on displacements fields of ECMs with known mechanical properties, which is not the case for 3D models like organoids and spheroids. TFM in 3D is further discussed in Section 6.
Optical tweezers use focused laser beams to trap and manipulate microscopic objects such as cells and beads [41]. By harnessing radiation pressure and momentum, optical tweezers create a localized region of higher intensity where particles experience a trapping force. This force can be precisely controlled to manipulate and measure the displacements of individual cells, particles, or even single molecules in the range of 1 nm to 1 mm [42]. In the context of mechanobiology, optical tweezers are used to apply and measure forces at the pico-newton (pN) level. This powerful technique has provided major insights into the biomechanical properties of single cells and the role of mechanical forces in various biological processes. The high sensitivity of advanced optical tweezers has also provided a powerful tool for investigating the biophysical properties of single molecules like motor proteins [42][43]. The application of optical tweezers to 3D cell culture models is so far limited, although it has been used successfully to measure the viscoelasticity of cells within organoids [44].
FRET has evolved from a theoretical concept [45] to a widely used experimental technique with diverse applications in cellular and molecular research [46]. Indeed, FRET sensors have revolutionized the ability to interrogate mechanical forces and dynamics within living cells [47] by providing insights into how cells respond to mechanical cues, how forces are transmitted across cellular structures, and how mechanical processes contribute to cellular behaviors. Moreover, the constant improvements being made to FRET-based technologies should ensure even deeper insights into the mechanics of tissues, cells and molecules. FRET is a fundamental molecular phenomenon that involves the transfer of energy from an excited donor fluorophore to an acceptor fluorophore when they are in close proximity [48]. Because energy transfer is extremely sensitive to the distance between the donor and acceptor molecules, FRET provides a very powerful tool to study interactions between different molecules, as well as conformational changes within individual molecules [49]. These genetically encoded tension sensors can be used to measure mechanical forces within living cells by revealing mechanically responsive domains within individual proteins tagged appropriately with both the donor and acceptor probes as changes in fluorescence intensity, spectral shift, or other measurable signals [48]. These sensors have been used extensively to visualize interactions and quantify mechanical forces exerted on specific cellular components such as cadherin-based cell–cell adhesions, integrin-based cell–substrate adhesions [50], membrane–membrane interactions [51][52], and cytoskeletal structure interactions [53]. Although FRET has proven very valuable for investigating molecular interactions, its utilization in the context of 3D mechanobiology presents challenges as regards spatial resolution, penetration depth for optical imaging, quantification accuracy, background noise, microenvironment variations, cellular diversity, and other technical complexities.
While the advanced methods discussed above have provided many insights into molecular aspects of mechanotransduction, interpreting these results in the context of tissues (e.g., tissue function, differentiation, and remodeling) poses significant challenges. Indeed, the development of techniques that allow biosensors to be used in 3D cellular structures like organoids and spheroids will be particularly important. For example, measuring cellular-scale forces within organoids, spheroids, and tissues remains very challenging because 3D ECMs are quite complex. The optical biosensors currently employed to study the mechanobiology of organoids and spheroids are summarized in Figure 3.
YAP/TAZ: YAP-TAZ (Yes-associated protein (YAP) and WW domain-containing transcription regulator protein 1 WWTR1, also known as TAZ) are transcriptional coactivators that play a pivotal role in several fundamental cellular processes, including cell proliferation and tissue development. When cells are subjected to mechanical stimuli or experience changes in their mechanical environment, YAP translocate into the cell’s nucleus where it acts together with various transcription factors to regulate gene expression. Because YAP/TAZ responds to mechanical cues, fluorescent versions can be used as a mechanosensors to define the cell’s mechanical state [54]. YAP has been shown to regulate intestinal stem cells [55] (YAP activation is higher in the stem-cell rich crypt than in the villus [56]), and transient YAP activation is necessary for symmetry breaking in organoids [57].
Nuclear deformation: Forces impinging on the nucleus can alter nuclear shape, chromatin structure, and, ultimately, the cell’s transcriptional profile. The dynamic visualization of cell nuclei using GFP or RFP tagged with a nuclear localization sequence (NLS), GFP-tagged Histone H2B, or even the DNA dye DAPI have the potential to serve as mechanical sensors in 3D cell models. Because nuclear volume scales with cell volume [58], the orientation, size, and shape of the nucleus can be employed as an indicator of the cell’s mechanical surroundings [21][59].
Pressure sensors: Cells present within 3D structures are exposed to a wide variety of forces that include hydrostatic pressure, compression, tension, and shear stress. These forces are created by cell compaction, by fluid filled lumens, and by changes in the microenvironment. The understanding of how these stresses and pressures alter cellular behavior and influence 3D morphology is held back by a lack of proper tools to accurately measure such forces within tissues. Of note, cell-sized mechanical sensors have used been to detect internal stresses within spheroids [60]. These sensors are made of fluorescent polyacrylamide [61] or oil droplets [62], both with known mechanical properties.
In addition to the optical sensors discussed above, Brillouin microscopy and Elastic Resonator Interference Stress Microscopy (ERISM) are increasingly being used to measure the mechanical properties of tissues.
Brillouin microscopy is an advanced imaging technique that utilizes the phenomenon of Brillouin scattering to characterize the mechanical properties of cells and tissues [63]. By measuring the frequency shift of scattered light due to acoustic waves propagating through the sample, this method can be used to map variations in material stiffness and elasticity at the microscopic level [64]. Brillouin microscopy offers a non-invasive and non-destructive means to evaluate intracellular stiffness at submicron resolution [65].
ERISM is an innovative microscopy technique that employs a micro-resonator (typically made of an elastic material) as a probe to measure the mechanical properties of cells [66]. The interaction between the resonator and the cell creates interference patterns that reveal variations in stress and mechanical properties.
Importantly, comprehensive knowledge regarding the composition and mechanical properties of the matrices that support 3D structures is essential for the proper analysis and interpretation of biosensor data. In this regard, the development of 3D matrices with tunable mechanical properties would greatly accelerate efforts to characterize the mechanobiology of organoids and spheroids. Such an effort will require accurate measurements of the mechanical properties of 3D cell cultures and determining how these properties can be harnessed to advance biosensor development.
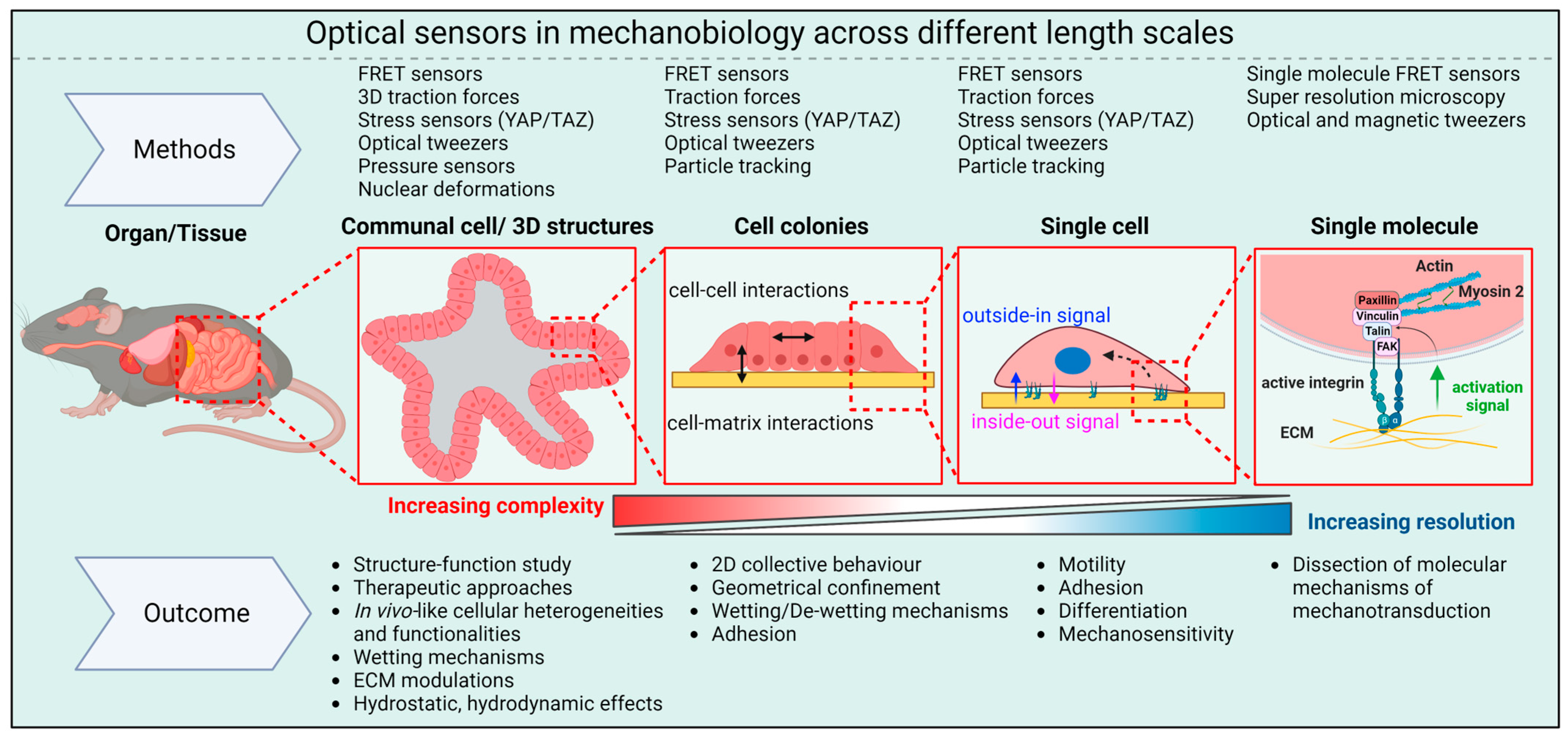
Figure 3. Optical sensors in mechanobiology: Optical sensors can be used across a wide range of length scales. At the molecular level, they provide insights into the pN forces involved in mechanotransduction inside cells. On larger scales, these intracellular forces combine with forces between cells and between cells and the ECM to shape the overall 3D structure of tissues and organs.
References
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures–a comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919.
- Yamada, K.M.; Sixt, M. Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 2019, 20, 738–752.
- Lee, G.Y.; Kenny, P.A.; Lee, E.H.; Bissell, M.J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 2007, 4, 359–365.
- Paşca, S.P. Assembling human brain organoids. Science 2019, 363, 126–127.
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527.
- Jensen, C.; Teng, Y. Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 2020, 7, 33.
- Wood, L.D.; Ewald, A.J. Organoids in cancer research: A review for pathologist-scientists. J. Pathol. 2021, 254, 395–404.
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125.
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420.
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108.
- Gilazieva, Z.; Ponomarev, A.; Rutland, C.; Rizvanov, A.; Solovyeva, V. Promising applications of tumor spheroids and organoids for personalized medicine. Cancers 2020, 12, 2727.
- Shariati, L.; Esmaeili, Y.; Haghjooy Javanmard, S.; Bidram, E.; Amini, A. Organoid technology: Current standing and future perspectives. Stem Cells 2021, 39, 1625–1649.
- Zanoni, M.; Cortesi, M.; Zamagni, A.; Arienti, C.; Pignatta, S.; Tesei, A. Modeling neoplastic disease with spheroids and organoids. J. Hematol. Oncol. 2020, 13, 97.
- Discher, D.E.; Janmey, P.; Wang, Y.-L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143.
- Irianto, J.; Pfeifer, C.R.; Xia, Y.; Discher, D.E. SnapShot: Mechanosensing matrix. Cell 2016, 165, 1820–1820.e1821.
- Paszek, M.J.; Zahir, N.; Johnson, K.R.; Lakins, J.N.; Rozenberg, G.I.; Gefen, A.; Reinhart-King, C.A.; Margulies, S.S.; Dembo, M.; Boettiger, D. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254.
- Kratochvil, M.J.; Seymour, A.J.; Li, T.L.; Paşca, S.P.; Kuo, C.J.; Heilshorn, S.C. Engineered materials for organoid systems. Nat. Rev. Mater. 2019, 4, 606–622.
- Butcher, D.T.; Alliston, T.; Weaver, V.M. A tense situation: Forcing tumour progression. Nat. Rev. Cancer 2009, 9, 108–122.
- Nguyen-Ngoc, K.-V.; Shamir, E.R.; Huebner, R.J.; Beck, J.N.; Cheung, K.J.; Ewald, A.J. 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. Tissue Morphog. Methods Protoc. 2015, 1189, 135–162.
- Drost, J.; Clevers, H. Translational applications of adult stem cell-derived organoids. Development 2017, 144, 968–975.
- Yousafzai, M.; Yadav, V.; Amiri, S.; Errami, Y.; Murrell, M. Active Regulation of Pressure and Volume Defines an Energetic Constraint on the Size of Cell Aggregates. Phys. Rev. Lett. 2022, 128, 48103.
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45.
- Shimomura, O. Discovery of green fluorescent protein (GFP) (Nobel Lecture). Angew. Chem. Int. Ed. 2009, 48, 5590–5602.
- Ingber, D. Mechanobiology and diseases of mechanotransduction. Ann. Med. 2003, 35, 564–577.
- Trepat, X. When cellular forces became visible. Nat. Rev. Mol. Cell Biol. 2020, 21, 253.
- Harris, A.K.; Wild, P.; Stopak, D. Silicone rubber substrata: A new wrinkle in the study of cell locomotion. Science 1980, 208, 177–179.
- Guilford, W.H.; Lantz, R.C.; Gore, R.W. Locomotive forces produced by single leukocytes in vivo and in vitro. Am. J. Physiol. Cell Physiol. 1995, 268, C1308–C1312.
- Adams, S.R.; Harootunian, A.T.; Buechler, Y.J.; Taylor, S.S.; Tsien, R.Y. Fluorescence ratio imaging of cyclic AMP in single cells. Nature 1991, 349, 694–697.
- Romani, P.; Valcarcel-Jimenez, L.; Frezza, C.; Dupont, S. Crosstalk between mechanotransduction and metabolism. Nat. Rev. Mol. Cell Biol. 2021, 22, 22–38.
- Humphrey, J.D.; Dufresne, E.R.; Schwartz, M.A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 2014, 15, 802–812.
- Yousafzai, M.S.; Coceano, G.; Mariutti, A.; Ndoye, F.; Amin, L.; Niemela, J.; Bonin, S.; Scoles, G.; Cojoc, D. Effect of neighboring cells on cell stiffness measured by optical tweezers indentation. J. Biomed. Opt. 2016, 21, 57004.
- Petridou, N.I.; Spiró, Z.; Heisenberg, C.-P. Multiscale force sensing in development. Nat. Cell Biol. 2017, 19, 581–588.
- Style, R.W.; Boltyanskiy, R.; German, G.K.; Hyland, C.; MacMinn, C.W.; Mertz, A.F.; Wilen, L.A.; Xu, Y.; Dufresne, E.R. Traction force microscopy in physics and biology. Soft Matter 2014, 10, 4047–4055.
- Mertz, A.F.; Banerjee, S.; Che, Y.; German, G.K.; Xu, Y.; Hyland, C.; Marchetti, M.C.; Horsley, V.; Dufresne, E.R. Scaling of traction forces with the size of cohesive cell colonies. Phys. Rev. Lett. 2012, 108, 198101.
- Han, S.J.; Oak, Y.; Groisman, A.; Danuser, G. Traction microscopy to identify force modulation in subresolution adhesions. Nat. Methods 2015, 12, 653–656.
- Pérez-González, C.; Alert, R.; Blanch-Mercader, C.; Gómez-González, M.; Kolodziej, T.; Bazellieres, E.; Casademunt, J.; Trepat, X. Active wetting of epithelial tissues. Nat. Phys. 2019, 15, 79–88.
- Plotnikov, S.V.; Pasapera, A.M.; Sabass, B.; Waterman, C.M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 2012, 151, 1513–1527.
- Gupta, V.K.; Nam, S.; Yim, D.; Camuglia, J.; Martin, J.L.; Sanders, E.N.; O’Brien, L.E.; Martin, A.C.; Kim, T.; Chaudhuri, O. The nature of cell division forces in epithelial monolayers. J. Cell Biol. 2021, 220, e202011106.
- Ajeti, V.; Tabatabai, A.P.; Fleszar, A.J.; Staddon, M.F.; Seara, D.S.; Suarez, C.; Yousafzai, M.S.; Bi, D.; Kovar, D.R.; Banerjee, S. Wound healing coordinates actin architectures to regulate mechanical work. Nat. Phys. 2019, 15, 696–705.
- Mitchel, J.A.; Das, A.; O’Sullivan, M.J.; Stancil, I.T.; DeCamp, S.J.; Koehler, S.; Ocaña, O.H.; Butler, J.P.; Fredberg, J.J.; Nieto, M.A. In primary airway epithelial cells, the unjamming transition is distinct from the epithelial-to-mesenchymal transition. Nat. Commun. 2020, 11, 5053.
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505.
- Bustamante, C.J.; Chemla, Y.R.; Liu, S.; Wang, M.D. Optical tweezers in single-molecule biophysics. Nat. Rev. Methods Primers 2021, 1, 25.
- Yousafzai, M.S.; Ndoye, F.; Coceano, G.; Niemela, J.; Bonin, S.; Scoles, G.; Cojoc, D. Substrate-dependent cell elasticity measured by optical tweezers indentation. Opt. Lasers Eng. 2016, 76, 27–33.
- Volpe, G.; Maragò, O.M.; Rubinsztein-Dunlop, H.; Pesce, G.; Stilgoe, A.B.; Volpe, G.; Tkachenko, G.; Truong, V.G.; Chormaic, S.N.; Kalantarifard, F. Roadmap for optical tweezers. J. Phys. Photonics 2023, 5, 022501.
- Forster, T. Energiewanderung und fluoreszenz. Naturwissenschaften 1946, 33, 166–175.
- Pietraszewska-Bogiel, A.; Gadella, T. FRET microscopy: From principle to routine technology in cell biology. J. Microsc. 2011, 241, 111–118.
- Wang, Y.; Wang, N. FRET and mechanobiology. Integr. Biol. 2009, 1, 565–573.
- Fischer, L.S.; Rangarajan, S.; Sadhanasatish, T.; Grashoff, C. Molecular force measurement with tension sensors. Annu. Rev. Biophys. 2021, 50, 595–616.
- Imani, M.; Mohajeri, N.; Rastegar, M.; Zarghami, N. Recent advances in FRET-Based biosensors for biomedical applications. Anal. Biochem. 2021, 630, 114323.
- Grashoff, C.; Hoffman, B.D.; Brenner, M.D.; Zhou, R.; Parsons, M.; Yang, M.T.; McLean, M.A.; Sligar, S.G.; Chen, C.S.; Ha, T. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 2010, 466, 263–266.
- Stabley, D.R.; Jurchenko, C.; Marshall, S.S.; Salaita, K.S. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat. Methods 2012, 9, 64–67.
- Hsu, Y.-Y.; Resto Irizarry, A.M.; Fu, J.; Liu, A.P. Mechanosensitive Channel-Based Optical Membrane Tension Reporter. ACS Sens. 2023, 8, 12–18.
- Meng, F.; Suchyna, T.M.; Lazakovitch, E.; Gronostajski, R.M.; Sachs, F. Real time FRET based detection of mechanical stress in cytoskeletal and extracellular matrix proteins. Cell. Mol. Bioeng. 2011, 4, 148–159.
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183.
- Gregorieff, A.; Liu, Y.; Inanlou, M.R.; Khomchuk, Y.; Wrana, J.L. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature 2015, 526, 715–718.
- Serra, D.; Mayr, U.; Boni, A.; Lukonin, I.; Rempfler, M.; Challet Meylan, L.; Stadler, M.B.; Strnad, P.; Papasaikas, P.; Vischi, D. Self-organization and symmetry breaking in intestinal organoid development. Nature 2019, 569, 66–72.
- Nikolaev, M.; Mitrofanova, O.; Broguiere, N.; Geraldo, S.; Dutta, D.; Tabata, Y.; Elci, B.; Brandenberg, N.; Kolotuev, I.; Gjorevski, N. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 2020, 585, 574–578.
- Guo, M.; Pegoraro, A.F.; Mao, A.; Zhou, E.H.; Arany, P.R.; Han, Y.; Burnette, D.T.; Jensen, M.H.; Kasza, K.E.; Moore, J.R. Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc. Natl. Acad. Sci. USA 2017, 114, E8618–E8627.
- Khavari, A.; Ehrlicher, A.J. Nuclei deformation reveals pressure distributions in 3D cell clusters. PLoS ONE 2019, 14, e0221753.
- Dolega, M.; Delarue, M.; Ingremeau, F.; Prost, J.; Delon, A.; Cappello, G. Cell-like pressure sensors reveal increase of mechanical stress towards the core of multicellular spheroids under compression. Nat. Commun. 2017, 8, 14056.
- Lee, W.; Kalashnikov, N.; Mok, S.; Halaoui, R.; Kuzmin, E.; Putnam, A.J.; Takayama, S.; Park, M.; McCaffrey, L.; Zhao, R. Dispersible hydrogel force sensors reveal patterns of solid mechanical stress in multicellular spheroid cultures. Nat. Commun. 2019, 10, 144.
- Campàs, O.; Mammoto, T.; Hasso, S.; Sperling, R.A.; O’connell, D.; Bischof, A.G.; Maas, R.; Weitz, D.A.; Mahadevan, L.; Ingber, D.E. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods 2014, 11, 183–189.
- Scarcelli, G.; Yun, S.H. Confocal Brillouin microscopy for three-dimensional mechanical imaging. Nat. Photonics 2008, 2, 39–43.
- Prevedel, R.; Diz-Muñoz, A.; Ruocco, G.; Antonacci, G. Brillouin microscopy: An emerging tool for mechanobiology. Nat. Methods 2019, 16, 969–977.
- Antonacci, G.; Braakman, S. Biomechanics of subcellular structures by non-invasive Brillouin microscopy. Sci. Rep. 2016, 6, 37217.
- Kronenberg, N.M.; Liehm, P.; Steude, A.; Knipper, J.A.; Borger, J.G.; Scarcelli, G.; Franze, K.; Powis, S.J.; Gather, M.C. Long-term imaging of cellular forces with high precision by elastic resonator interference stress microscopy. Nat. Cell Biol. 2017, 19, 864–872.
More
Information
Subjects:
Biophysics; Optics
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
693
Revisions:
2 times
(View History)
Update Date:
03 Nov 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




