
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Natalie Krahn | -- | 1757 | 2023-10-31 16:29:32 | | | |
| 2 | Fanny Huang | Meta information modification | 1757 | 2023-11-01 04:23:18 | | |
Video Upload Options
Selenocysteine (Sec), the 21st amino acid, is structurally similar to cysteine but with a sulfur to selenium replacement. This single change retains many of the chemical properties of cysteine but often with enhanced catalytic and redox activity. Incorporation of Sec into proteins is unique, requiring additional translation factors and multiple steps to insert Sec at stop (UGA) codons. These Sec-containing proteins (selenoproteins) are found in all three domains of life where they often are involved in cellular homeostasis (e.g., reducing reactive oxygen species). The essential role of selenoproteins in humans requires us to maintain appropriate levels of selenium, the precursor for Sec, in our diet. Too much selenium is also problematic due to its toxic effects. Deciphering the role of Sec in selenoproteins is challenging for many reasons, one of which is due to their complicated biosynthesis pathway. However, clever strategies are surfacing to overcome this and facilitate production of selenoproteins.
1. Introduction
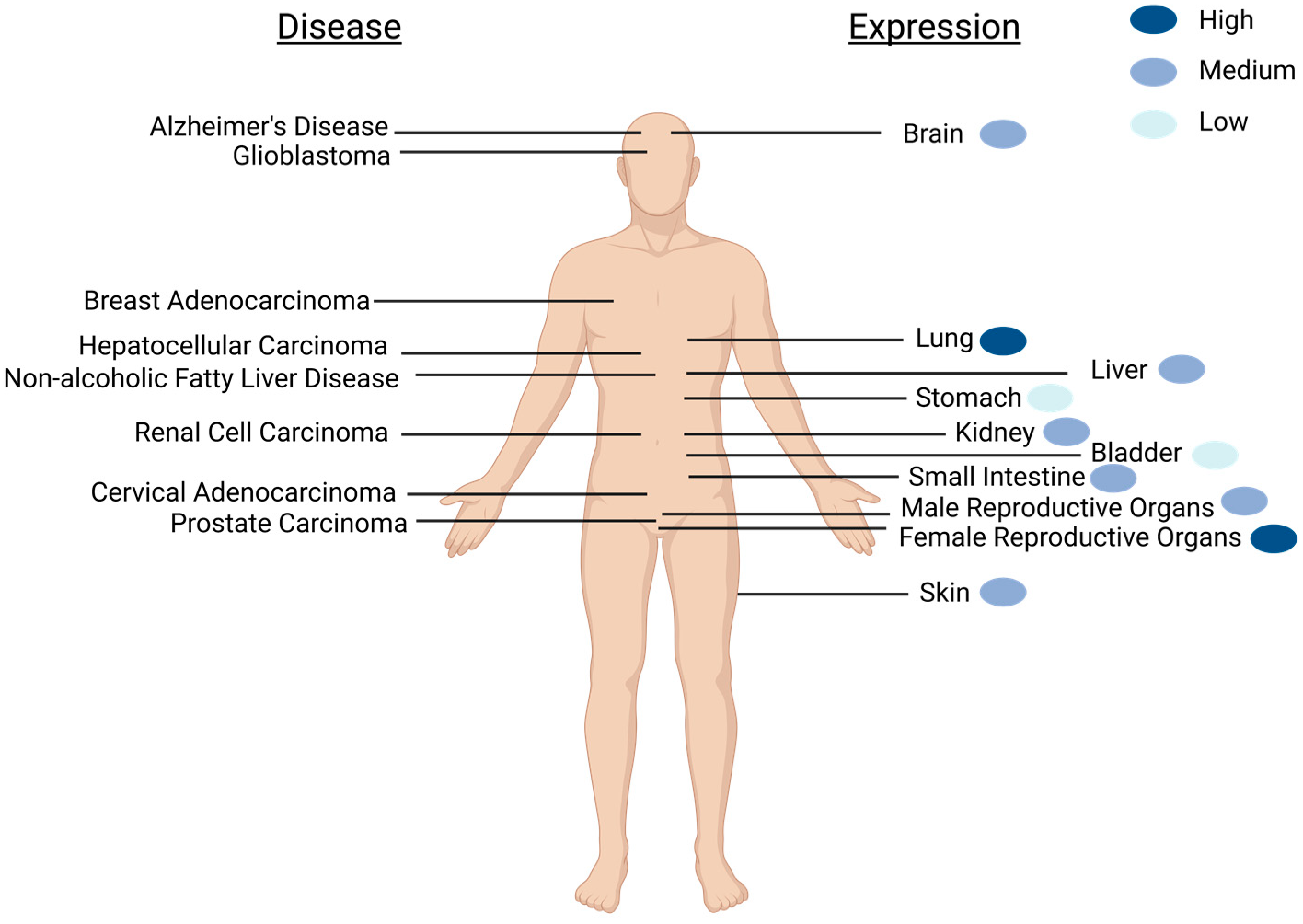
2. Expression of SELENOM Is Widespread
3. Elucidating SELENOM Function from Structure
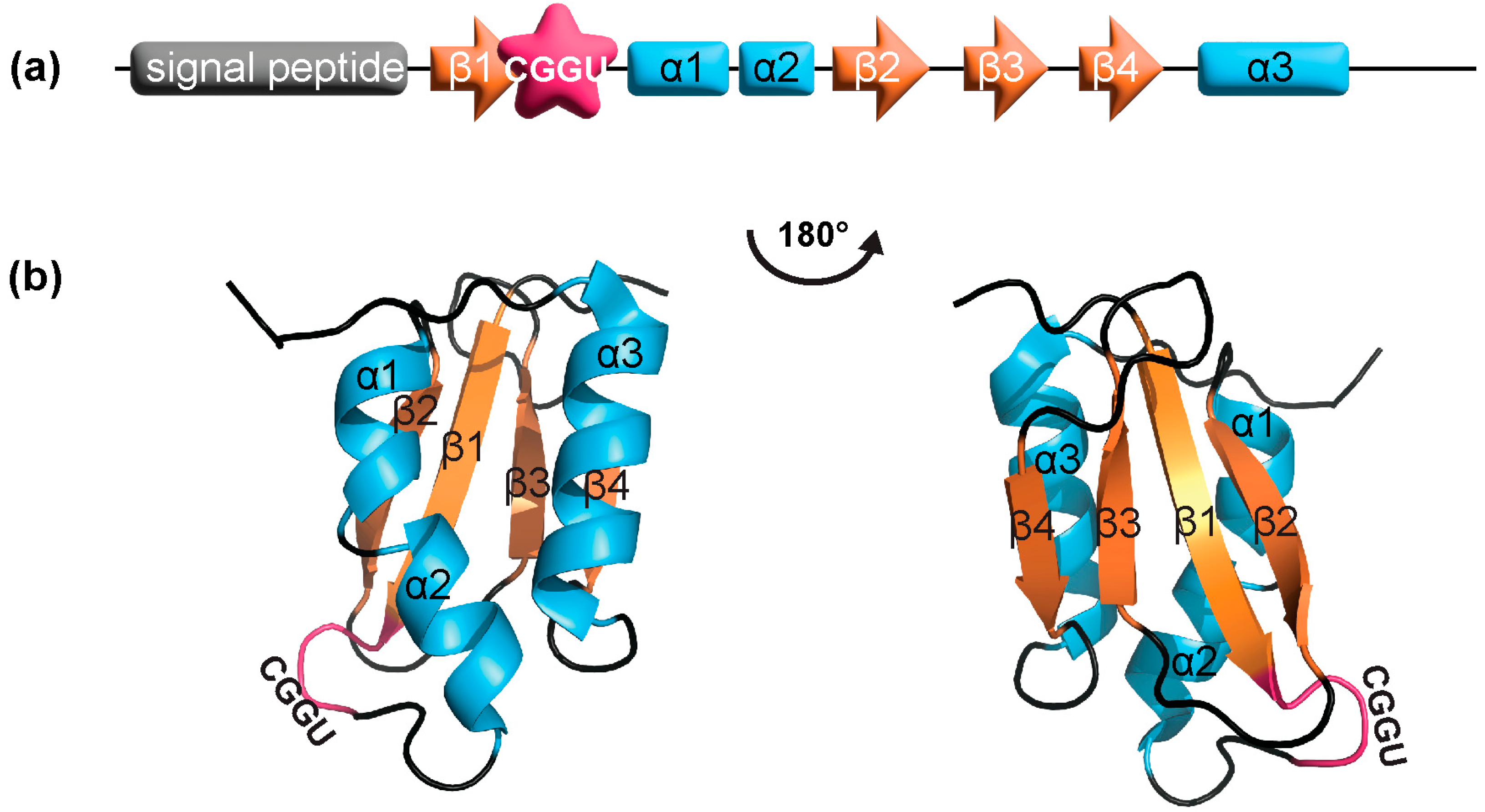
3.1. SELENOM Is Structurally Close to Thioredoxin
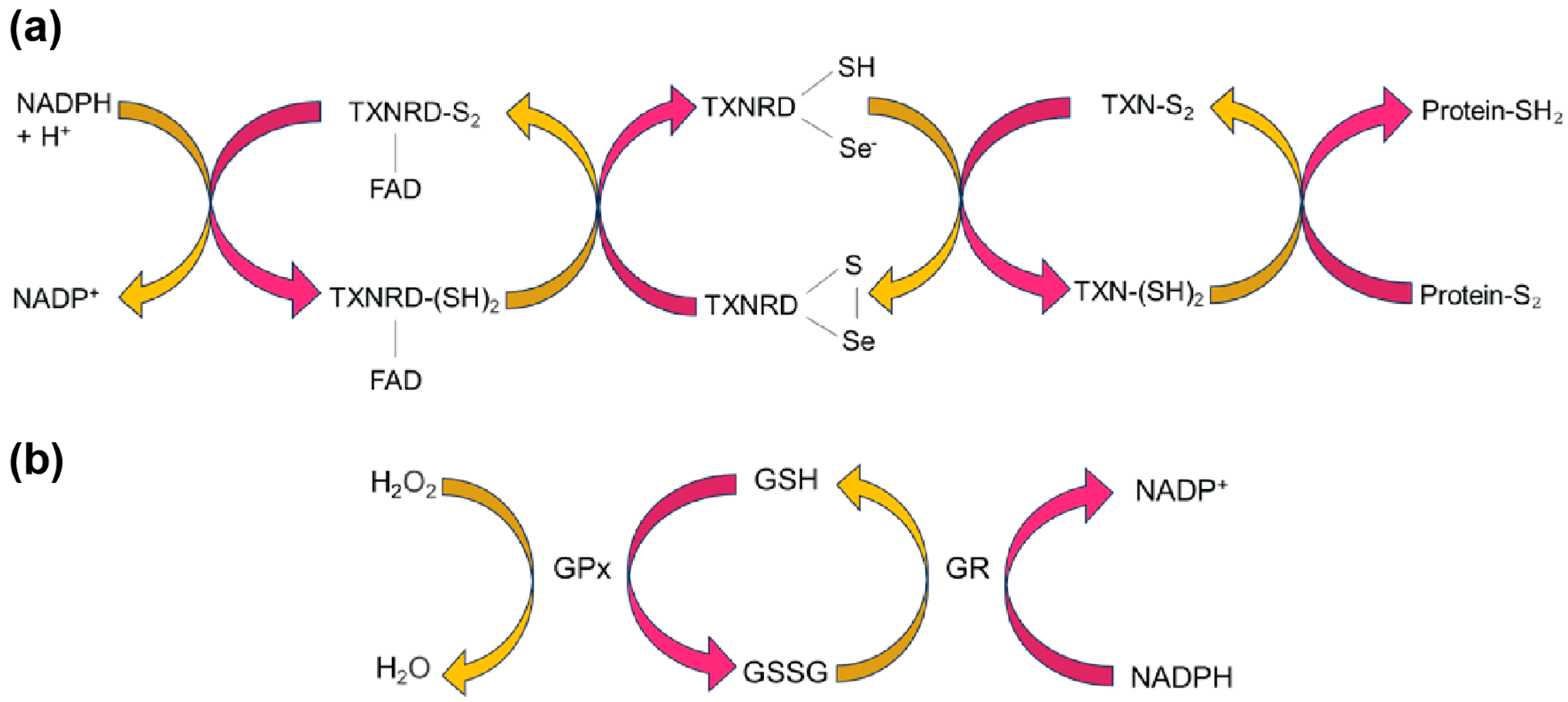
3.2. SELENOM Defines a New Thioredoxin Family
3.3. SELENOM Does Not Bind UGGT
References
- Rayman, M.P. Selenium intake, status, and health: A complex relationship. Hormones 2020, 19, 9–14.
- Marschall, T.A.; Bornhorst, J.; Kuehnelt, D.; Schwerdtle, T. Differing cytotoxicity and bioavailability of selenite, methylselenocysteine, selenomethionine, selenosugar 1 and trimethylselenonium ion and their underlying metabolic transformations in human cells. Mol. Nutr. Food Res. 2016, 60, 2622–2632.
- Lazard, M.; Dauplais, M.; Blanquet, S.; Plateau, P. Recent advances in the mechanism of selenoamino acids toxicity in eukaryotic cells. Biomol. Concepts 2017, 8, 93–104.
- Speckmann, B.; Steinbrenner, H. Selenium and selenoproteins in inflammatory bowel diseases and experimental colitis. Inflamm. Bowel Dis. 2014, 20, 1110–1119.
- Avery, J.C.; Hoffmann, P.R. Selenium, selenoproteins, and immunity. Nutrients 2018, 10, 1203.
- Fairweather-Tait, S.J.; Filippini, T.; Vinceti, M. Selenium status and immunity. Proc. Nutr. Soc. 2022, 82, 32–38.
- Vinceti, M.; Filippini, T.; Jablonska, E.; Saito, Y.; Wise, L.A. Safety of selenium exposure and limitations of selenoprotein maximization: Molecular and epidemiologic perspectives. Environ. Res. 2022, 211, 113092.
- Urbano, T.; Filippini, T.; Lasagni, D.; De Luca, T.; Sucato, S.; Polledri, E.; Bruzziches, F.; Malavolti, M.; Baraldi, C.; Santachiara, A.; et al. Associations between urinary and dietary selenium and blood metabolic parameters in a healthy northern italy population. Antioxidants 2021, 10, 1193.
- Urbano, T.; Vinceti, M.; Mandrioli, J.; Chiari, A.; Filippini, T.; Bedin, R.; Tondelli, M.; Simonini, C.; Zamboni, G.; Shimizu, M.; et al. Selenoprotein P concentrations in the cerebrospinal fluid and serum of individuals affected by amyotrophic lateral sclerosis, mild cognitive impairment and Alzheimer’s Dementia. Int. J. Mol. Sci. 2022, 23, 9865.
- Huang, Y.C.; Combs, G.F., Jr.; Wu, T.L.; Zeng, H.; Cheng, W.H. Selenium status and type 2 diabetes risk. Arch. Biochem. Biophys. 2022, 730, 109400.
- Steinbrenner, H.; Duntas, L.H.; Rayman, M.P. The role of selenium in type-2 diabetes mellitus and its metabolic comorbidities. Redox Biol. 2022, 50, 102236.
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268.
- Chung, C.Z.; Krahn, N. The selenocysteine toolbox: A guide to studying the 21st amino acid. Arch. Biochem. Biophys. 2022, 730, 109421.
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777.
- Krahn, N.; Fischer, J.T.; Söll, D. Naturally occurring tRNAs with non-canonical structures. Front. Microbiol. 2020, 11, 596914.
- Serrão, V.H.B.; Silva, I.R.; da Silva, M.T.A.; Scortecci, J.F.; de Freitas Fernandes, A.; Thiemann, O.H. The unique tRNASec and its role in selenocysteine biosynthesis. Amino Acids 2018, 50, 1145–1167.
- Hilal, T.; Killam, B.Y.; Grozdanović, M.; Dobosz-Bartoszek, M.; Loerke, J.; Bürger, J.; Mielke, T.; Copeland, P.R.; Simonović, M.; Spahn, C.M.T. Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon. Science 2022, 376, 1338–1343.
- Peng, J.J.; Yue, S.Y.; Fang, Y.H.; Liu, X.L.; Wang, C.H. Mechanisms affecting the biosynthesis and incorporation rate of selenocysteine. Molecules 2021, 26, 7120.
- Korotkov, K.V.; Novoselov, S.V.; Hatfield, D.L.; Gladyshev, V.N. Mammalian selenoprotein in which selenocysteine (Sec) incorporation is supported by a new form of Sec insertion sequence element. Mol. Cell. Biol. 2002, 22, 1402–1411.
- Zhang, Y.; Zhou, Y.; Schweizer, U.; Savaskan, N.E.; Hua, D.; Kipnis, J.; Hatfield, D.L.; Gladyshev, V.N. Comparative analysis of selenocysteine machinery and selenoproteome gene expression in mouse brain identifies neurons as key functional sites of selenium in mammals. J. Biol. Chem. 2008, 283, 2427–2438.
- Pitts, M.W.; Reeves, M.A.; Hashimoto, A.C.; Ogawa, A.; Kremer, P.; Seale, L.A.; Berry, M.J. Deletion of selenoprotein M leads to obesity without cognitive deficits. J. Biol. Chem. 2013, 288, 26121–26134.
- Gong, T.; Hashimoto, A.C.; Sasuclark, A.R.; Khadka, V.S.; Gurary, A.; Pitts, M.W. Selenoprotein M promotes hypothalamic leptin signaling and thioredoxin antioxidant activity. Antioxid. Redox Signal. 2021, 35, 775–787.
- Sasuclark, A.R.; Khadka, V.S.; Pitts, M.W. Cell-type specific analysis of selenium-related genes in brain. Antioxidants 2019, 8, 120.
- Ferguson, A.D.; Labunskyy, V.M.; Fomenko, D.E.; Araç, D.; Chelliah, Y.; Amezcua, C.A.; Rizo, J.; Gladyshev, V.N.; Deisenhofer, J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J. Biol. Chem. 2006, 281, 3536–3543.
- Oberacker, T.; Kraft, L.; Schanz, M.; Latus, J.; Schricker, S. The importance of thioredoxin-1 in health and disease. Antioxidants 2023, 12, 1078.
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87.
- Muri, J.; Kopf, M. The thioredoxin system: Balancing redox responses in immune cells and tumors. Eur. J. Immunol. 2023, 53, e2249948.
- Alanen, H.I.; Williamson, R.A.; Howard, M.J.; Lappi, A.K.; Jantti, H.P.; Rautio, S.M.; Kellokumpu, S.; Ruddock, L.W. Functional characterization of ERp18, a new endoplasmic reticulum-located thioredoxin superfamily member. J. Biol. Chem. 2003, 278, 28912–28920.
- Anelli, T.; Alessio, M.; Mezghrani, A.; Simmen, T.; Talamo, F.; Bachi, A.; Sitia, R. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002, 21, 835–844.
- Flowers, B.; Bochnacka, O.; Poles, A.; Diamond, A.M.; Kastrati, I. Distinct roles of SELENOF in different human cancers. Biomolecules 2023, 13, 486.
- Korotkov, K.V.; Kumaraswamy, E.; Zhou, Y.; Hatfield, D.L.; Gladyshev, V.N. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 2001, 276, 15330–15336.
- Qi, Y.; Grishin, N.V. Structural classification of thioredoxin-like fold proteins. Proteins 2005, 58, 376–388.
- Labunskyy, V.M.; Ferguson, A.D.; Fomenko, D.E.; Chelliah, Y.; Hatfield, D.L.; Gladyshev, V.N. A novel cysteine-rich domain of Sep15 mediates the interaction with UDP-glucose:glycoprotein glucosyltransferase. J. Biol. Chem. 2005, 280, 37839–37845.





