Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Muhammad Abdul Rehman Rashid | -- | 2007 | 2023-10-16 14:06:55 | | | |
| 2 | Rita Xu | Meta information modification | 2007 | 2023-10-17 03:28:54 | | | | |
| 3 | Rita Xu | + 3 word(s) | 2010 | 2023-10-18 09:56:54 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Hao, Y.; Pan, Y.; Chen, W.; Rashid, M.A.R.; Li, M.; Che, N.; Duan, X.; Zhao, Y. Nucleotide-Binding Leucine-Rich Repeat Gene in Wheat. Encyclopedia. Available online: https://encyclopedia.pub/entry/50358 (accessed on 07 February 2026).
Hao Y, Pan Y, Chen W, Rashid MAR, Li M, Che N, et al. Nucleotide-Binding Leucine-Rich Repeat Gene in Wheat. Encyclopedia. Available at: https://encyclopedia.pub/entry/50358. Accessed February 07, 2026.
Hao, Yongchao, Yinghua Pan, Wuying Chen, Muhammad Abdul Rehman Rashid, Mengyao Li, Naixiu Che, Xu Duan, Yan Zhao. "Nucleotide-Binding Leucine-Rich Repeat Gene in Wheat" Encyclopedia, https://encyclopedia.pub/entry/50358 (accessed February 07, 2026).
Hao, Y., Pan, Y., Chen, W., Rashid, M.A.R., Li, M., Che, N., Duan, X., & Zhao, Y. (2023, October 16). Nucleotide-Binding Leucine-Rich Repeat Gene in Wheat. In Encyclopedia. https://encyclopedia.pub/entry/50358
Hao, Yongchao, et al. "Nucleotide-Binding Leucine-Rich Repeat Gene in Wheat." Encyclopedia. Web. 16 October, 2023.
Copy Citation
Wheat has a large and diverse repertoire of nucleotide-binding leucine-rich repeats (NLRs) involved in disease resistance, with over 1500 NLRs detected in some studies. These NLR genes occur as singletons or clusters containing copies of NLRs from different phylogenetic clades. The number of NLRs and cluster size can differ drastically among ecotypes and cultivars.
wheat
nucleotide-binding leucine-rich repeat
NLR genes
1. Introduction
Polyploid crops, such as wheat, triticale, oats, sweet potato, and peanuts, play a vital role in ensuring food security. Among these, the tetraploid durum wheat (pasta wheat, Triticum durum; 2n = 4x = 28) and the hexaploid bread wheat (Triticum aestivum; 2n = 6x = 42) are adaptable to various environmental conditions and, therefore, the most widely cultivated [1]. The production of wheat worldwide in 2021 was recorded to be as high as 781 million tons (FaoStat; https://www.fao.org/faostat/en/#data (accessed on 20 June 2023)). However, in spite of the high production, wheat and other crops are often threatened by abiotic and biotic stressors that significantly reduce their yield and quality. Specifically, diseases caused by pathogenic bacteria, viruses, fungi, and oomycetes have resulted in significant crop losses, affecting food security worldwide [2]. As in other plants, wheat resists pathogens via their multilayered innate immune responses: one mediated by pattern recognition receptors (PRRs) on the cell surface and the other mediated by nucleotide-binding leucine-rich repeats (NLRs) intracellularly [3][4]. Typically, plants sense pathogens via the immune receptors that detect the pathogen-derived molecules and initiate diverse defense responses interconnected to form a signaling network [5]. These PRRs, which are in the form of receptor-like proteins (RLPs) or receptor-like kinases (RLKs), detect conserved pathogen-associated molecular patterns (PAMPs) and activate pattern-triggered immunity (PTI) to instigate defensive responses against non-adapted pathogens. Additionally, the residual PTI provides basal resistance to the adapted pathogens [4]. Therefore, PTI is categorized as a non-race-specific resistance [4].
In order to overcome the impenetrable blockade of the first layer of the immune system, pathogenic organisms secrete a class of small molecules into plant cells called effectors [6]. Effectors of these pathogens subsequently interfere with PTI by hindering PRR translation, inhibiting the activity of PRRs and their complexes, affecting the transmission of MAPK and its downstream signals, and impacting vesicle transport and callose deposition [7][8][9]. In this way, pathogenic bacteria can successfully infect. However, plant cells will not surrender easily. Faced with this situation, plants will urgently activate the next immune layer. The NLR receptor within the cell can indirectly or directly spot the effector, causing the plant to initiate a second immune response, which is called effector-triggered immunity (ETI), and the effector recognized by the NLR protein is called avirulent (Avr) protein [10]. The ETI is usually accompanied by programmed cell death (PCD); this hypersensitive response (HR) is crucial for plant resistance to biotrophic pathogens [11]. In addition, the downstream immune response activated by ETI is similar to PTI but with greater intensity and longer duration [4].
2. NLR Gene Structure and Categories
Typically, the structure of NLR consists of three parts, including a variable N-terminal domain, a conserved, central nucleotide-binding adapter (NB-ARC) domain, and a C-terminal leucine-rich repeat (LRR) domain [12][13][14]. The variable N-terminal domain is divided into two categories based on the protein sequence and similarity to known proteins: Toll interleukin-1 receiver (TIR) and coiled coil (CC) [15][16]. The NLR genes are classified into subgroups depending on their domain architecture, including NBS, TN, TNL, CN, NL, and CNL (Figure 1) [17]. Some studies have indicated that NLRs with a deletion of the domain at the N-terminus cannot elicit plant immunity, while their expression alone is sufficient to induce HR responses [18][19]. Therefore, this domain of NLR genes is believed to stimulate and transmit immune signals downstream. In animal NLRs, when the effector is recognized, the entire NLR protein can oligomerize and form high molecular weight complexes known as inflammasomes to stimulate immune signaling [20]. In plants, several studies have reported that NLRs self-associate upon specific recognition of their corresponding effectors, including TIR-NLRs (tobacco N and Arabidopsis RPP1) and a CC-NLR (Arabidopsis ZAR1) [21][22][23]. The N-terminal TIR and CC domains may be crucial for NLR oligomerization and activation [24][25].
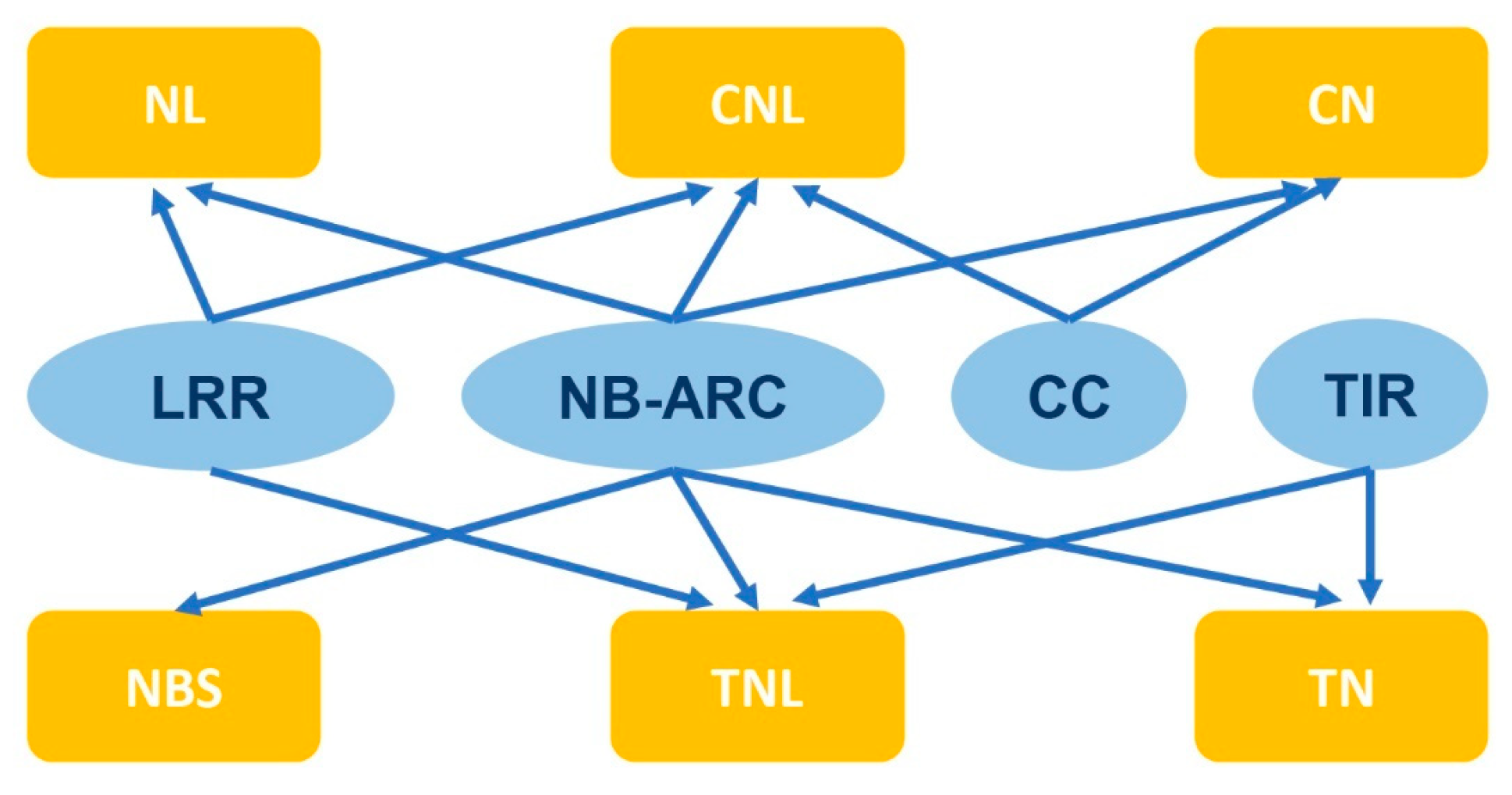
Figure 1. Subgroups of NLR genes based on domain structures.
NB-ARC, the relatively large domain of NLRs, includes three subdomains: the NB, the ARC1, and the ARC2 subdomains. Among these, NB represents nucleotide binding, and ARC is so named because this domain appears in Apaf-1, R, and CED-4 proteins [26][27]. The NB-ARC domain is considered an ATPase domain due to its ability to bind with, exchange, or hydrolyze ADP or ATP nucleotides [28]. Generally, the NLR protein combined with ADP is in a closed or inactive state, and the combination with ATP promotes a conformational change in the NLR protein, turning it into an open or activated state. After NLR activation, it will transmit signals downstream to activate the plant’s immune system. Effector recognition by NLRs (directly or indirectly) will prompt NB-ARC to release ADP and bind ATP [29]. Additionally, this binding feature acts as a “molecular switch” model of NLRs in plant immunity [30]. The properties of the NB-ARC domain and its binding with different nucleotides form the structural basis of this “molecular switch” model [31]. LRR is named for the presence of multiple tandem leucine-rich repeats in this domain, and LRR domains regulate the activity of NLRs through intramolecular or intermolecular interactions [13][32]. Research has demonstrated that LRR domains negatively influence the activity of NLRs [33][34], and the LRR domain physically associates with the NB-ARC domain [35][36][37]. In addition, deletion of the LRR domain results in auto-activation [37][38]. However, other studies have shown that LRRs can also positively regulate the activity of NLRs, and self-activating mutations in potato Rx led to HR responses without pathogenic bacteria, whereas deletion of its LRRs suppressed HR responses [35].
3. Gene Duplication in Wheat
The genome of bread wheat is one of the largest and most complex of all cultivated plant species. This complexity originated as a result of two rounds of historical whole genome duplication events [39] and recent small-scale duplications [40]. Gene duplication provides “raw genetic material” for the evolution of genes and gene functions to improve crops; it is also a key factor driving the evolution, domestication, and diversification of species [41]. Scientists have deciphered numerous large, complex, and highly repetitive genomes of various species of the Triticeae tribe [1][42][43][44][45][46][47]. Comparing the available genome sequences and functional genomic data of plants, researchers have gained immense knowledge of how genes are duplicated, how these duplicated genes gain novel roles, and their ultimate impact on genome evolution [48][49]. Among the various events or mechanisms, the duplication of the whole genome or an entire chromosome is the most extensive form of gene duplication [49]. Whole genome duplication (WGD or polyploidization) is an extreme mechanism that has benefited wheat’s complex evolutionary history; the evolution of hexaploid wheat through two hybridization and polyploidization events formed a new species with a huge genome and abundant gene set [50][51]. Then, hexaploid wheat spread worldwide (Figure 2A). About 55% of the bread wheat homologous genes exhibit 1:1:1 correspondence across the three homologous subgenomes, and another 15% possess a minimum of one gene copy in at least one of the subgenomes [8]. Tandem duplication or recombination is the event when two or more genes after duplication are positioned adjacent to each other on the same chromosome [49][52][53]. It is likely a major mechanism of NLR gene expansion to form NLR gene clusters in wheat (Figure 2B). Additionally, researchers found a recent burst of gene duplications in all sequenced species of the Triticeae tribe, and detailed analysis of the features of the gene duplication and their flanking sequences suggested the role of transposable elements (TEs) in causing this recent event [40]. In contrast to local tandem duplication, replicative transposition by TEs forms dispersed duplicates [54] (Figure 2C).
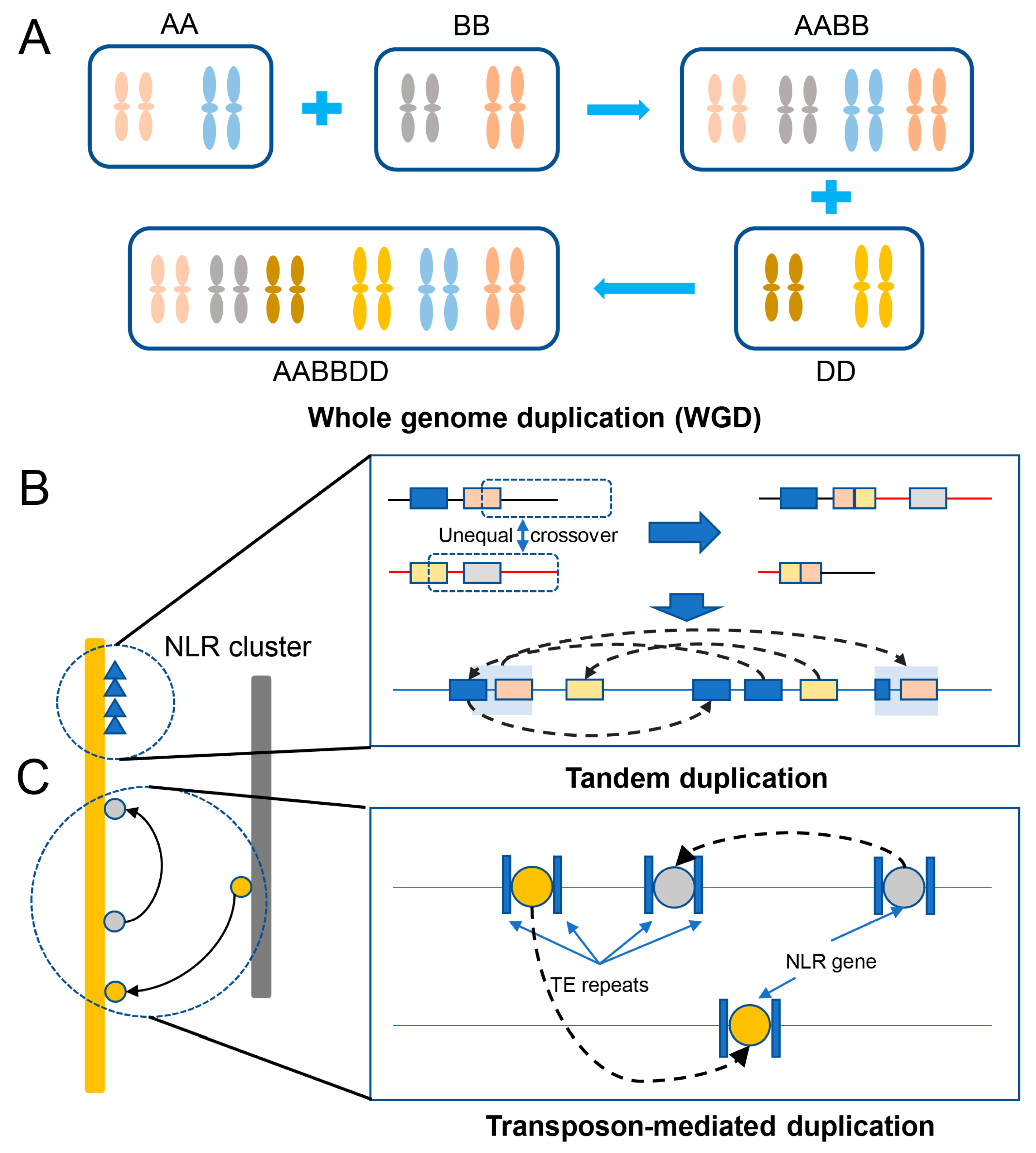
Figure 2. NLR duplication in wheat. (A) Whole genome duplication (WGD) through an increase in ploidy. (B) Tandem duplication through unequal recombination between similar alleles to form a gene cluster. (C) Transposon-mediated duplication of a gene associated with transposable elements (TE repeats) via replicative transposition to form dispersed duplicates.
4. Diversity and Divergence of NLR Genes for Disease Resistance
Earlier research suggested great variation in the number of NLRs among closely associated species [55], and the number is not related to the size of the genome or the level of ploidy [56]. However, more than 1500 NLRs have been detected by analyzing the transcriptional and physical organization of the intracellular immune receptor repertoire in bread wheat [57]. More than 2000 NLRs have been identified using the fully annotated reference genome of bread wheat [45]. A pan-genome provides an opportunity to further study the numerical variation of wheat NLRs, and researchers used multi-genome comparisons (11 wheat accessions) for characterizing NLRs and identified around 2500 loci with NLR signatures in each accession, while only 31–34% of the NLR signatures are shared across all genomes. They identified 5905 (98% identity) to 7780 (100% identity) unique NLRs in all wheat genomes, emphasizing the complexity and size of the immune receptor repertoire guiding disease resistance [1]. This may also demonstrate the relationship between NLR number and genome size and between NLR number and ploidy level. A recent study used a panel of 907 winter wheat accessions to construct a resistance gene atlas, and the majority of NLRs (96%) were grouped into 39,073 orthogroups, with the remaining being unique. Most orthogroups contained NLRs from at least 20 different accessions, but very few contained members from more than 500 accessions, which showed a different pattern of NLR diversity compared to the model plant A. thaliana [58].
Due to the diversity and abundance of pathogens in different environments, plants face dynamic selection pressure from habitat pathogens during their evolution, which results in the inability to maintain a stable distribution of NLR genes between and even within species [59]. It has been found in research on different angiosperms that significant pathogen infection pressure has driven the expansion of NLR genes [60][61][62]. In the study of wild emmer wheat, it was found that changes in the NLR gene appeared to be rapid within the species [63]. Due to differences in the abundance of pathogenic bacteria in habitats, the population of wild wheat differentiated, with wild wheat growing in areas where powdery mildew was prevalent, and then evolving resistance to powdery mildew [36]. As stated earlier, the clusters (medium and large) may vary greatly in their size among the ecotypes and cultivated varieties, indicating potential local adaptability [64][65]. For clusters containing highly homologous NLRs from an ancestral gene with few inversions, direct duplication of genes probably resulted in the original clustering; subsequently, increased rates of unequal crossing-over (UCO) during meiosis probably provided the material for rapid evolution and increased diversity in immune sensors, thereby expanding the cluster [66]. Under different pressures, these clusters rapidly contract or expand, which explains the large variations in cluster patterns between ecotypes [1][67]. However, the abundance of NLR genes is not solely beneficial; the more NLR genes plants maintain, the more health costs they incur [68]. Under no or less pathogen selection pressure, various plant species have exhibited contraction of NLR genes [69][70], leading to significant variations in the number and diversity of NLR genes between or within species.
Gene recombination is crucial for NLR diversification. Current evidence shows that the unconventional recombination (illegitimate recombination) between NLR genes is the main way to change the number of repeats of the LRR domain, which can cause a rapid increase or decrease in the repeat number of the LRR domain, further increasing the potential of NLRs to recognize different effectors [44]. In addition, if the NLR gene recombines with other genes (such as WRKY, NAC), it can cause the fusion of the domains of other genes to NLRs, further enriching the diversity of NLR structure and function (Figure 2B). For example, a WRKY domain is attached to the Arabidopsis NLR gene RRS1 at the C-terminus [71][72]. Further research found that this gene fusion phenomenon is very common in plant NLR genes [73]. In addition, some gene recombination events can also cause the loss of the NLR domain, resulting in truncated NLRs. Truncated NLRs are also ubiquitous in various plants and also play very important roles in plant immunity [19][74]. Interestingly, although the domains of truncated NLRs are incomplete, they can also perform similar functions to intact NLRs: they can directly or indirectly recognize effectors and induce plant immunity [40][74]. Taken together, gene duplication of NLRs provided diversity to cope with the infection of thousands of evolving pathogenic bacteria.
References
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283.
- Keller, B.; Wicker, T.; Krattinger, S.G. Advances in Wheat and Pathogen Genomics: Implications for Disease Control. Annu. Rev. Phytopathol. 2018, 56, 67–87.
- Lacaze, A.; Joly, D.L. Structural specificity in plant-filamentous pathogen interactions. Mol. Plant Pathol. 2020, 21, 1513–1525.
- Ngou, B.P.M.; Ding, P.; Jones, J.D.G. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 34, 1447–1478.
- Lu, Y.; Tsuda, K. Intimate Association of PRR- and NLR-Mediated Signaling in Plant Immunity. Mol. Plant-Microbe Interact. 2021, 34, 3–14.
- de Jonge, R.; Bolton, M.D.; Thomma, B.P. How filamentous pathogens co-opt plants: The ins and outs of fungal effectors. Curr. Opin. Plant Biol. 2011, 14, 400–406.
- Yahiaoui, N.; Brunner, S.; Keller, B. Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J. 2006, 47, 85–98.
- Toruno, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441.
- Yu, X.; Feng, B.; He, P.; Shan, L. From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137.
- Plissonneau, C.; Benevenuto, J.; Mohd-Assaad, N.; Fouche, S.; Hartmann, F.E.; Croll, D. Using Population and Comparative Genomics to Understand the Genetic Basis of Effector-Driven Fungal Pathogen Evolution. Front. Plant Sci. 2017, 8, 119.
- Pruitt, R.N.; Gust, A.A.; Nurnberger, T. Plant immunity unified. Nat. Plants 2021, 7, 382–383.
- Cesari, S.; Bernoux, M.; Moncuquet, P.; Kroj, T.; Dodds, P.N. A novel conserved mechanism for plant NLR protein pairs: The “integrated decoy” hypothesis. Front. Plant Sci. 2014, 5, 606.
- Maruta, N.; Burdett, H.; Lim, B.Y.J.; Hu, X.; Desa, S.; Manik, M.K.; Kobe, B. Structural basis of NLR activation and innate immune signalling in plants. Immunogenetics 2022, 74, 5–26.
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337.
- Shaw, M.H.; Reimer, T.; Kim, Y.G.; Nunez, G. NOD-like receptors (NLRs): Bona fide intracellular microbial sensors. Curr. Opin. Immunol. 2008, 20, 377–382.
- Franchi, L.; Warner, N.; Viani, K.; Nunez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128.
- Sanchez-Martin, J.; Keller, B. NLR immune receptors and diverse types of non-NLR proteins control race-specific resistance in Triticeae. Curr. Opin. Plant Biol. 2021, 62, 102053.
- Wu, Z.; Tian, L.; Liu, X.; Huang, W.; Zhang, Y.; Li, X. The N-terminally truncated helper NLR NRG1C antagonizes immunity mediated by its full-length neighbors NRG1A and NRG1B. Plant Cell 2022, 34, 1621–1640.
- Cox, K.L. Unexpected help: Role of an N-terminally truncated helper NLR in plant immunity. Plant Cell 2022, 34, 1427–1428.
- Lamkanfi, M.; Dixit, V.M. Mechanisms and functions of inflammasomes. Cell 2014, 157, 1013–1022.
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, eaav5870.
- Schreiber, K.J.; Bentham, A.; Williams, S.J.; Kobe, B.; Staskawicz, B.J. Multiple Domain Associations within the Arabidopsis Immune Receptor RPP1 Regulate the Activation of Programmed Cell Death. PLoS Pathog. 2016, 12, e1005769.
- Mestre, P.; Baulcombe, D.C. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 2006, 18, 491–501.
- Wang, J.; Chen, T.; Han, M.; Qian, L.; Li, J.; Wu, M.; Han, T.; Cao, J.; Nagalakshmi, U.; Rathjen, J.P.; et al. Plant NLR immune receptor Tm-22 activation requires NB-ARC domain-mediated self-association of CC domain. PLoS Pathog. 2020, 16, e1008475.
- Ve, T.; Williams, S.J.; Kobe, B. Structure and function of Toll/interleukin-1 receptor/resistance protein (TIR) domains. Apoptosis 2015, 20, 250–261.
- van der Biezen, E.A.; Jones, J.D.G. The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 1998, 8, R226–R228.
- van Ooijen, G.; Mayr, G.; Kasiem, M.M.A.; Albrecht, M.; Cornelissen, B.J.C.; Takken, F.L.W. Structure–function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 2008, 59, 1383–1397.
- Danot, O.; Marquenet, E.; Vidal-Ingigliardi, D.; Richet, E. Wheel of Life, Wheel of Death: A Mechanistic Insight into Signaling by STAND Proteins. Structure 2009, 17, 172–182.
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109.
- Bonardi, V.; Cherkis, K.; Nishimura, M.T.; Dangl, J.L. A new eye on NLR proteins: Focused on clarity or diffused by complexity? Curr. Opin. Immunol. 2012, 24, 41–50.
- Takken, F.L.; Goverse, A. How to build a pathogen detector: Structural basis of NB-LRR function. Curr. Opin. Plant Biol. 2012, 15, 375–384.
- Fick, A.; Swart, V.; van den Berg, N. The Ups and Downs of Plant NLR Expression During Pathogen Infection. Front. Plant Sci. 2022, 13, 921148.
- Hu, Z.; Yan, C.; Liu, P.; Huang, Z.; Ma, R.; Zhang, C.; Wang, R.; Zhang, Y.; Martinon, F.; Miao, D.; et al. Crystal Structure of NLRC4 Reveals Its Autoinhibition Mechanism. Science 2013, 341, 172–175.
- Wang, J.; Wang, J.; Hu, M.; Wu, S.; Qi, J.; Wang, G.; Han, Z.; Qi, Y.; Gao, N.; Wang, H.W.; et al. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 2019, 364, eaav5868.
- Moffett, P.; Farnham, G.; Peart, J.; Baulcombe, D.C. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 2002, 21, 4511–4519.
- Leister, R.T.; Dahlbeck, D.; Day, B.; Li, Y.; Chesnokova, O.; Staskawicz, B.J. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 2005, 17, 1268–1278.
- Ade, J.; DeYoung, B.J.; Golstein, C.; Innes, R.W. Indirect activation of a plant nucleotide binding site–leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 2006, 104, 2531–2536.
- Tameling, W.I.; Nooijen, C.; Ludwig, N.; Boter, M.; Slootweg, E.; Goverse, A.; Shirasu, K.; Joosten, M.H. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 2010, 22, 4176–4194.
- Salamini, F.; Ozkan, H.; Brandolini, A.; Schafer-Pregl, R.; Martin, W. Genetics and geography of wild cereal domestication in the near east. Nat. Rev. Genet. 2002, 3, 429–441.
- Wang, X.; Yan, X.; Hu, Y.; Qin, L.; Wang, D.; Jia, J.; Jiao, Y. A recent burst of gene duplications in Triticeae. Plant Commun. 2022, 3, 100268.
- Juery, C.; Concia, L.; De Oliveira, R.; Papon, N.; Ramirez-Gonzalez, R.; Benhamed, M.; Uauy, C.; Choulet, F.; Paux, E. New insights into homoeologous copy number variations in the hexaploid wheat genome. Plant Genome 2021, 14, e20069.
- Luo, M.C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502.
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433.
- Zhao, G.; Zou, C.; Li, K.; Wang, K.; Li, T.; Gao, L.; Zhang, X.; Wang, H.; Yang, Z.; Liu, X.; et al. The Aegilops tauschii genome reveals multiple impacts of transposons. Nat. Plants 2017, 3, 946–955.
- International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191.
- Ling, H.Q.; Ma, B.; Shi, X.; Liu, H.; Dong, L.; Sun, H.; Cao, Y.; Gao, Q.; Zheng, S.; Li, Y.; et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 2018, 557, 424–428.
- Zhou, Y.; Bai, S.; Li, H.; Sun, G.; Zhang, D.; Ma, F.; Zhao, X.; Nie, F.; Li, J.; Chen, L.; et al. Introgressing the Aegilops tauschii genome into wheat as a basis for cereal improvement. Nat. Plants 2021, 7, 774–786.
- Bomblies, K. When everything changes at once: Finding a new normal after genome duplication. Proc. Biol. Sci. 2020, 287, 20202154.
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316.
- Matsuoka, Y. Evolution of polyploid triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011, 52, 750–764.
- Luo, M.C.; Yang, Z.L.; You, F.M.; Kawahara, T.; Waines, J.G.; Dvorak, J. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor. Appl. Genet. 2007, 114, 947–959.
- Hao, Y.; Xu, S.; lyu, Z.; Wang, H.; Kong, L.; Sun, S. Comparative Analysis of the Glutathione S-Transferase Gene Family of Four Triticeae Species and Transcriptome Analysis of GST Genes in Common Wheat Responding to Salt Stress. Int. J. Genom. 2021, 2021, 6289174.
- Hao, Y.; Hao, M.; Cui, Y.; Kong, L.; Wang, H. Genome-wide survey of the dehydrin genes in bread wheat (Triticum aestivum L.) and its relatives: Identification, evolution and expression profiling under various abiotic stresses. BMC Genom. 2022, 23, 73.
- Kapitonov, V.V.; Jurka, J. Helitrons on a roll: Eukaryotic rolling-circle transposons. Trends Genet. 2007, 23, 521–529.
- Seo, E.; Kim, S.; Yeom, S.I.; Choi, D. Genome-Wide Comparative Analyses Reveal the Dynamic Evolution of Nucleotide-Binding Leucine-Rich Repeat Gene Family among Solanaceae Plants. Front. Plant Sci. 2016, 7, 1205.
- Borrelli, G.M.; Mazzucotelli, E.; Marone, D.; Crosatti, C.; Michelotti, V.; Vale, G.; Mastrangelo, A.M. Regulation and Evolution of NLR Genes: A Close Interconnection for Plant Immunity. Int. J. Mol. Sci. 2018, 19, 1662.
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Harvey Millar, A.; Singh, K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009, 58, 53–68.
- Kale, S.M.; Schulthess, A.W.; Padmarasu, S.; Boeven, P.H.G.; Schacht, J.; Himmelbach, A.; Steuernagel, B.; Wulff, B.B.H.; Reif, J.C.; Stein, N.; et al. A catalogue of resistance gene homologs and a chromosome-scale reference sequence support resistance gene mapping in winter wheat. Plant Biotechnol. J. 2022, 20, 1730–1742.
- Casadevall, A. Fungal virulence, vertebrate endothermy, and dinosaur extinction: Is there a connection? Fungal. Genet. Biol. 2005, 42, 98–106.
- Shao, Z.Q.; Xue, J.Y.; Wu, P.; Zhang, Y.M.; Wu, Y.; Hang, Y.Y.; Wang, B.; Chen, J.Q. Large-Scale Analyses of Angiosperm Nucleotide-Binding Site-Leucine-Rich Repeat Genes Reveal Three Anciently Diverged Classes with Distinct Evolutionary Patterns. Plant Physiol. 2016, 170, 2095–2109.
- Shao, Z.Q.; Zhang, Y.M.; Hang, Y.Y.; Xue, J.Y.; Zhou, G.C.; Wu, P.; Wu, X.Y.; Wu, X.Z.; Wang, Q.; Wang, B.; et al. Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: Understanding gained from and beyond the legume family. Plant Physiol. 2014, 166, 217–234.
- Glover, N.M.; Daron, J.; Pingault, L.; Vandepoele, K.; Paux, E.; Feuillet, C.; Choulet, F. Small-scale gene duplications played a major role in the recent evolution of wheat chromosome 3B. Genome Biol. 2015, 16, 188.
- Wang, H.; Yin, H.; Jiao, C.; Fang, X.; Wang, G.; Li, G.; Ni, F.; Li, P.; Su, P.; Ge, W.; et al. Sympatric speciation of wild emmer wheat driven by ecology and chromosomal rearrangements. Proc. Natl. Acad. Sci. USA 2020, 117, 5955–5963.
- Bahram, M.; Netherway, T.; Frioux, C.; Ferretti, P.; Coelho, L.P.; Geisen, S.; Bork, P.; Hildebrand, F. Metagenomic assessment of the global diversity and distribution of bacteria and fungi. Environ. Microbiol. 2021, 23, 316–326.
- Liu, Y.; Zeng, Z.; Zhang, Y.M.; Li, Q.; Jiang, X.M.; Jiang, Z.; Tang, J.H.; Chen, D.; Wang, Q.; Chen, J.Q.; et al. An angiosperm NLR Atlas reveals that NLR gene reduction is associated with ecological specialization and signal transduction component deletion. Mol. Plant 2021, 14, 2015–2031.
- Van de Weyer, A.L.; Monteiro, F.; Furzer, O.J.; Nishimura, M.T.; Cevik, V.; Witek, K.; Jones, J.D.G.; Dangl, J.L.; Weigel, D.; Bemm, F. A Species-Wide Inventory of NLR Genes and Alleles in Arabidopsis thaliana. Cell 2019, 178, 1260–1272 e1214.
- van Wersch, S.; Li, X. Stronger When Together: Clustering of Plant NLR Disease resistance Genes. Trends Plant Sci. 2019, 24, 688–699.
- Karasov, T.L.; Kniskern, J.M.; Gao, L.; DeYoung, B.J.; Ding, J.; Dubiella, U.; Lastra, R.O.; Nallu, S.; Roux, F.; Innes, R.W.; et al. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 2014, 512, 436–440.
- Xue, J.Y.; Zhao, T.; Liu, Y.; Liu, Y.; Zhang, Y.X.; Zhang, G.Q.; Chen, H.; Zhou, G.C.; Zhang, S.Z.; Shao, Z.Q. Genome- Wide Analysis of the Nucleotide Binding Site Leucine-Rich Repeat Genes of Four Orchids Revealed Extremely Low Numbers of Disease Resistance Genes. Front. Genet. 2019, 10, 1286.
- Lin, X.; Zhang, Y.; Kuang, H.; Chen, J. Frequent loss of lineages and deficient duplications accounted for low copy number of disease resistance genes in Cucurbitaceae. BMC Genom. 2013, 14, 335.
- Birker, D.; Heidrich, K.; Takahara, H.; Narusaka, M.; Deslandes, L.; Narusaka, Y.; Reymond, M.; Parker, J.E.; O’Connell, R. A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J. 2009, 60, 602–613.
- Narusaka, M.; Shirasu, K.; Noutoshi, Y.; Kubo, Y.; Shiraishi, T.; Iwabuchi, M.; Narusaka, Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009, 60, 218–226.
- Sarris, P.F.; Cevik, V.; Dagdas, G.; Jones, J.D.; Krasileva, K.V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016, 14, 8.
- Chen, Y.; Zhong, G.; Cai, H.; Chen, R.; Liu, N.; Wang, W.; Tang, D. A Truncated TIR-NBS Protein TN10 Pairs with Two Clustered TIR-NBS-LRR Immune Receptors and Contributes to Plant Immunity in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 4004.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
817
Revisions:
3 times
(View History)
Update Date:
18 Oct 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




