Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Chenzhong Li | -- | 2498 | 2023-10-12 15:09:44 | | | |
| 2 | Rita Xu | Meta information modification | 2498 | 2023-10-13 03:40:53 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Zhu, W.; Wang, J.; Luo, H.; Luo, B.; Li, X.; Liu, S.; Li, C. Single-Cell Analysis and Application. Encyclopedia. Available online: https://encyclopedia.pub/entry/50208 (accessed on 07 February 2026).
Zhu W, Wang J, Luo H, Luo B, Li X, Liu S, et al. Single-Cell Analysis and Application. Encyclopedia. Available at: https://encyclopedia.pub/entry/50208. Accessed February 07, 2026.
Zhu, Weitao, Jiaao Wang, Hongzhi Luo, Binwen Luo, Xue Li, Shan Liu, Chenzhong Li. "Single-Cell Analysis and Application" Encyclopedia, https://encyclopedia.pub/entry/50208 (accessed February 07, 2026).
Zhu, W., Wang, J., Luo, H., Luo, B., Li, X., Liu, S., & Li, C. (2023, October 12). Single-Cell Analysis and Application. In Encyclopedia. https://encyclopedia.pub/entry/50208
Zhu, Weitao, et al. "Single-Cell Analysis and Application." Encyclopedia. Web. 12 October, 2023.
Copy Citation
Biological parameters extracted from electrical signals from various body parts have been used for many years to analyze the human body and its behavior. In addition, electrical signals from cancer cell lines, normal cells, and viruses, among others, have been widely used for the detection of various diseases. Single-cell parameters such as cell and cytoplasmic conductivity, relaxation frequency, and membrane capacitance are important.
single-cell analysis
electrical manipulation
single-cell application
1. Introduction
Several decades ago, the introduction of the membrane-clamp technique revolutionized the study of cell physiology. It observes the function of individual ion channels in a variety of cells through a high-resolution electrochemical method. At the cellular level, the membrane-clamp technique allows the study of physiological processes, such as cellular signal transduction and synaptic transmission. These studies have contributed to a new understanding of the mechanism of certain disease processes and provided new ideas for the treatment of these diseases. Biological parameters extracted from electrical signals from the nervous system, muscles, heart, and other parts of the body have been used for years to analyze the human body and its behavior. In addition, electrical signals from cancer cell lines, normal cells, and viruses, among others, have been widely used for the detection of various diseases. Single-cell parameters such as cellular and cytoplasmic conductivity, relaxation frequency, and membrane capacitance are important. The physiological state of cells can change in response to stimulation, and therefore, the health state of cells can be characterized by measuring some specific cell markers [1]. Electrical parameters are original cell markers, which can identify diseases, enabling early diagnosis and prevention. The electrical properties of cells are often used to identify cells and describe their viability and growth, and these properties are closely related to cell structure and chemical composition [2]. Therefore, it is possible to obtain the dielectric properties of cells by quantitative analysis of their electrical parameters. The electrical properties of cells can provide key data that can provide insight into their complex physiological state. Electrical properties are important in cell quantification, isolation, and capture, and in single-cell characterization studies. For example, based on the deflection properties of a charged droplet in an electric field, the charging electrode charges the droplets, including the cells within, as the sample is ejected. The electrostatic properties of the cells determine the amount of charge they carry. They will be deflected from the mainstream in the electric field and move toward different electrode sheets, which achieves the purpose of sorting, separating, and capturing the cells [3].
Single-cell electrical manipulation can safely manipulate cells without causing harm, and analytical and numerical polarization models resulting from electric fields can be used to describe and characterize the dielectric electrophoretic behavior of cells [4]. Single-cell electrical manipulation enables the separation of individual cells to specific spatial regions for single-cell analysis, as well as the ability to isolate and characterize rare cells. In addition to the detection of membrane, cytoplasm, and nuclear antigens, single-cell analysis can also detect whole cells and cellular components, such as organelles, nuclei, DNA, RNA, chromosomes, cytokines, hormones, and protein content, which can also be studied by single-cell manipulation. Single-cell analysis also enables further exploration of cell proliferation and cell cycles [3].
There are many techniques that can be used to characterize biomaterials, such as nanotechnology, microstrip cavity resonance measurement, etc. Cellular techniques can be divided into single-cell analysis and cell identification. Single-cell analysis uses patch clamps and nanoprobes, and the latest technologies include microfluidic and micro-electrical impedance spectroscopy.
2. Single-Cell Analysis and Application
In general, living cells are usually of greater research interest due to their more significant biological properties and applications. Therefore, whether analysis can be performed while maintaining the activity of the target cells is a hot issue in single-cell technology. Most single-cell analysis techniques have the advantage of not damaging the cell cytosol. For example, single-cell sequencing is a non-invasive method for tumor diagnosis and prognosis prediction. Single-cell sequencing can obtain comprehensive information, assist in early diagnosis of tumors, select the best therapeutic drugs, and monitor recurrence. The integrated analysis of single-cell sequencing with other omics can also provide valuable information that can promote the development of precision medicine in cancer. In addition, some researchers have combined on-chip oxygen control with a single IFC chip for sickle cell disease diagnosis and monitoring, and the living single-cell-based analysis results proved to be accurate and detailed (Figure 1) [5].
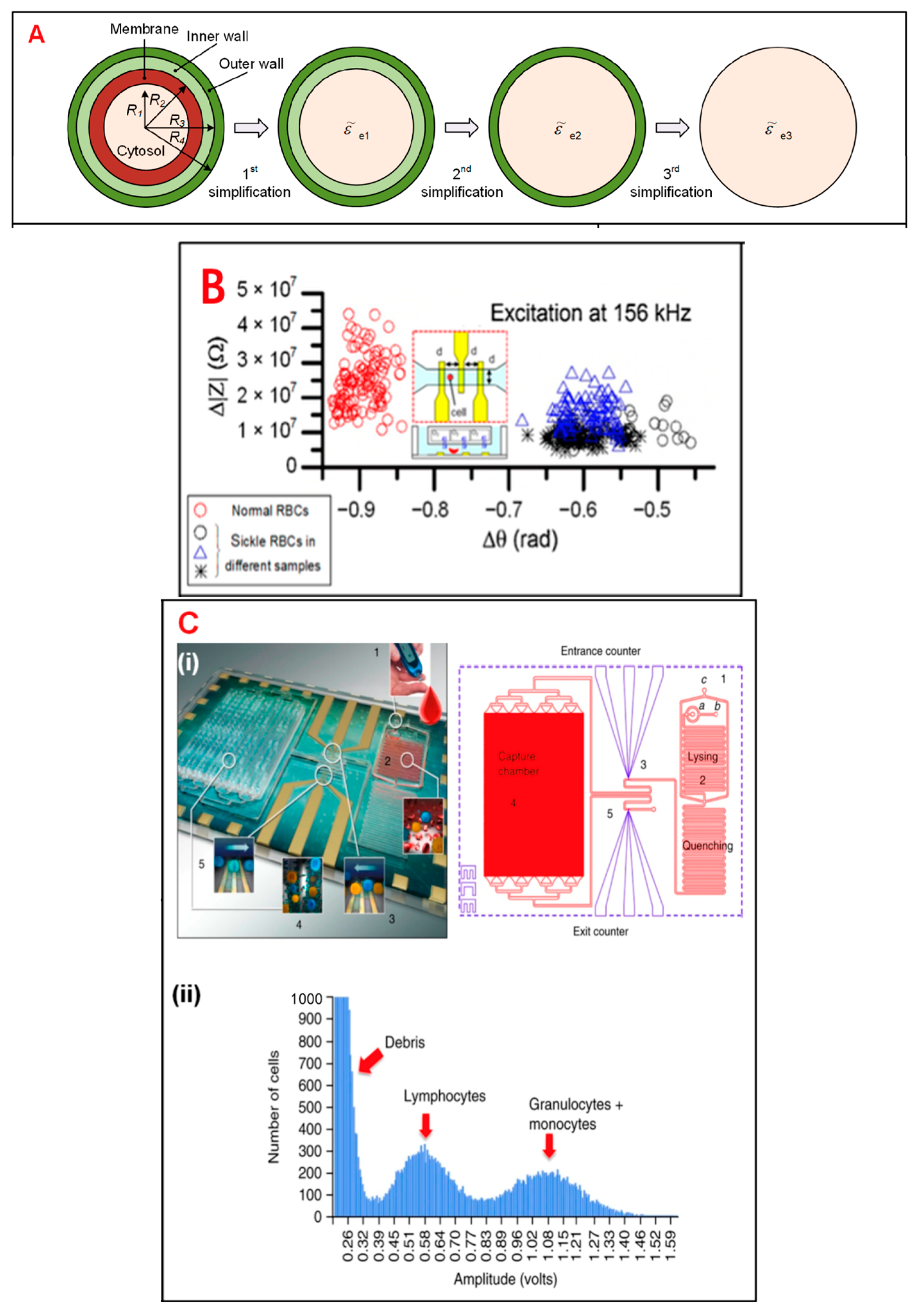
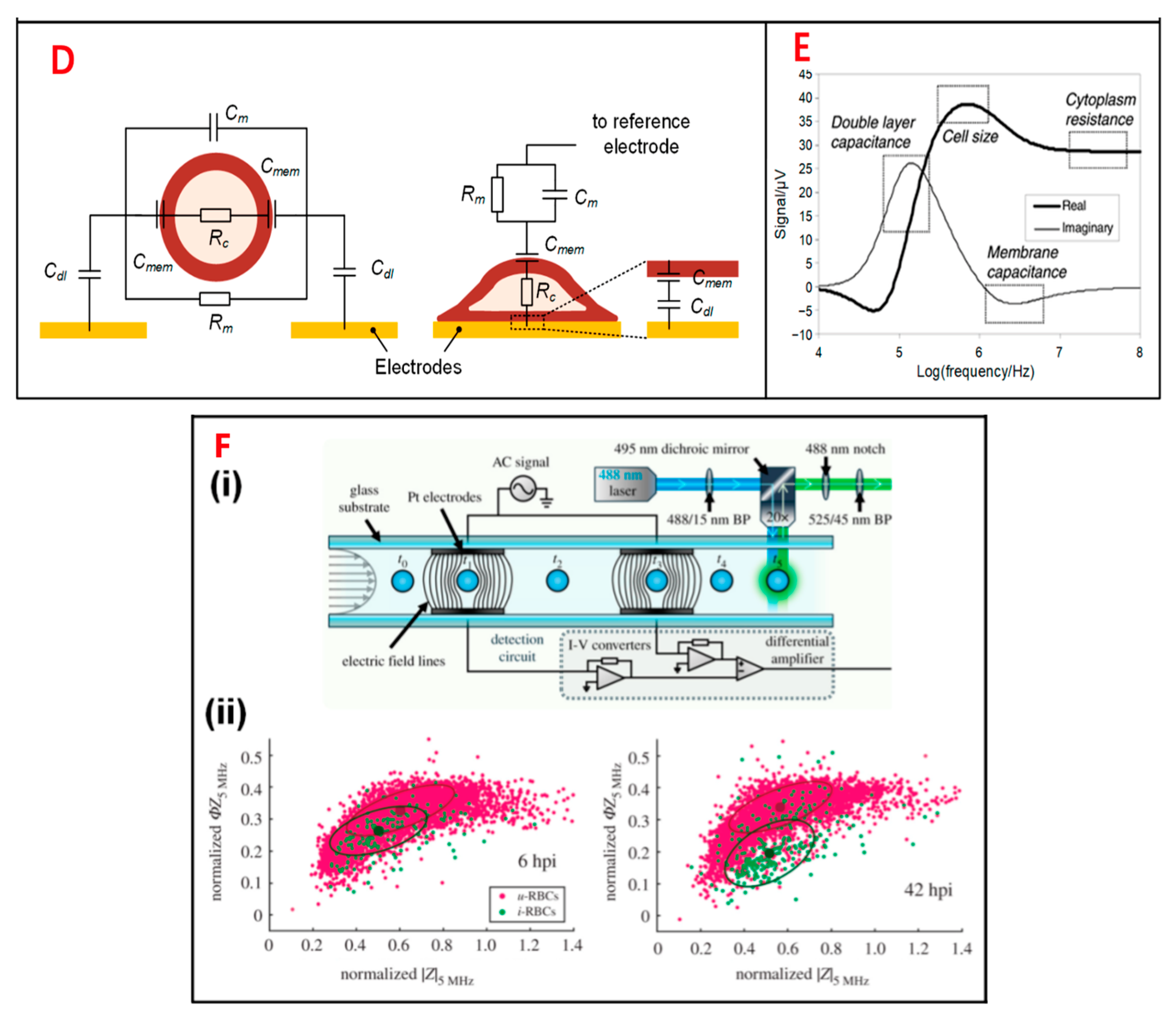
Figure 1. Electrical impedance sensing technique for single-cell analysis. (A) Electrical model and equivalent circuit models (ECMs) of a single cell. The mode is simplified to a homogeneous sphere. (B) Measurement of electrical impedance for normal and sickle RBCs at 156 kHz. (C) (i) Pictures of immunocapture biochip. (ii) Pulse amplitudes of impedance signals which show the distribution of cells by size. (D) The cell mode is suspended between a pair of sensors and adhered on a sensor. (E) Simulation results of an ECM model, which show different frequency domains related to cell parameters. (F) (i) Schematic of an IFC device. (ii) Measurement of impedance of infected and uninfected RBCs after 6 and 42 h.
Single-cell technology can also be used in biological tissue biopsies. Circulating tumor cells (CTCs) are tumor cells that originate from epithelial primary or metastatic tumors and enter the bloodstream with high viability and potential for metastasis. Liquid biopsy of CTCs can monitor tumor progression in real time [6]. Tumor metastasis is one of the main causes of death in cancer patients. CTCs can provide early warning of tumor heterogeneity and drug resistance and identify the mechanism of tumor occurrence and development [7]. Current methods for detecting tumor metastasis are mainly based on imaging, but early tumor metastasis is difficult to detect at the cellular level, even when viewed under high-resolution images. However, through high-resolution imaging combined with dynamic monitoring of the number and nature of CTCs, potential clues of tumor lesion metastasis can be found, providing potential for early targeted therapy [8]. Often, high-throughput sequencing analyses of tumor tissues ignore their heterogeneity because they target mixed samples of millions of cells that only reflect the overall genomic characteristics of the cells, while ignoring genetic material from low-abundance but functionally important cells, such as CTCs and cancer stem cells [9]. The separation of CTCs can be divided into two steps: enrichment and capture. The technology is mainly based on cell surface labeling and microfluidic chips. Enrichment based on cell surface markers generally includes positive and negative selection. This method utilizes antibodies, such as anti-epithelial cell adhesion molecules (EpCAM) and cytokeratin (CK), to capture and enrich tumor cells from the epithelium, while leucocyte-derived antibodies are utilized to eliminate white blood cells [6]. Based on the enrichment of microfluidic chips, peripheral blood mononuclear cells (PBMCs) of tumor patients were first isolated according to the biological and physical characteristics of tumor cells. The PBMCs were then slowly passed through a microfluidic chip coated with EpCAM antibodies under slow laminar flow control. EpCAM+ cells were captured and bound to the bottom of the chip, while other lymphocytes flowed out with the fluid [10].
In fact, due to the small amount of PBMCs in peripheral blood, PBMCs can only reflect the occurrence of disease to a certain extent. In contrast, whole genome RNA expression profiling can reflect the occurrence of disease more comprehensively. Some researchers have taken bacterial meningitis as an example. The researchers extracted whole blood RNA from a group of patients for gene expression profiling and confirmed the expression levels of 10 functionally related genes (CD177, IL1R2, IL18R1, IL18RAP, OLFM4, TLR5, CPA3, FCER1A, IL5RA, and IL7R) with a high degree of statistical significance by real-time quantitative PCR (qRT). The researchers ultimately demonstrated that whole blood RNA analysis could better reflect the occurrence and course of the disease, proving that the transcript levels of certain immune genes are associated with pathogens with some specificity [11]. The authors also suggested that whole blood RNA analysis could also reveal some specific cellular activations [11]. Combining whole blood RNA analysis with single-cell gene analysis can better assist the diagnosis and treatment of diseases, which is a promising direction.
CTCs are mainly used in the following aspects: to reduce the interference of tumor heterogeneity, and to compare the differences between single-cell genomes, transcriptomes, and epigenomes between the tumor primary site and metastatic site and between peripheral blood CTCs and metastatic lymph nodes [12]. Single-cell transcriptome sequencing analysis of CTCs can be used to understand the therapeutic effect of tumors, as well as the genetic heterogeneity, evolution, and drug resistance of tumor cells (Figure 2). Surface markers and cell sizes of CTCs vary from tumor to tumor. Most CTCs are aggregated, which is also a significant feature of tumor stem cells, and the presence of CTCs in the peripheral blood of tumor patients can indicate the progression of the tumor [13]. For rare peripheral blood CTC detection, traditional single-cell sorting methods, such as fluorescence activated cell sorting (FACS), are no longer suitable. Instead, micromanipulation, microfluidic technology, and cell selection systems are used.
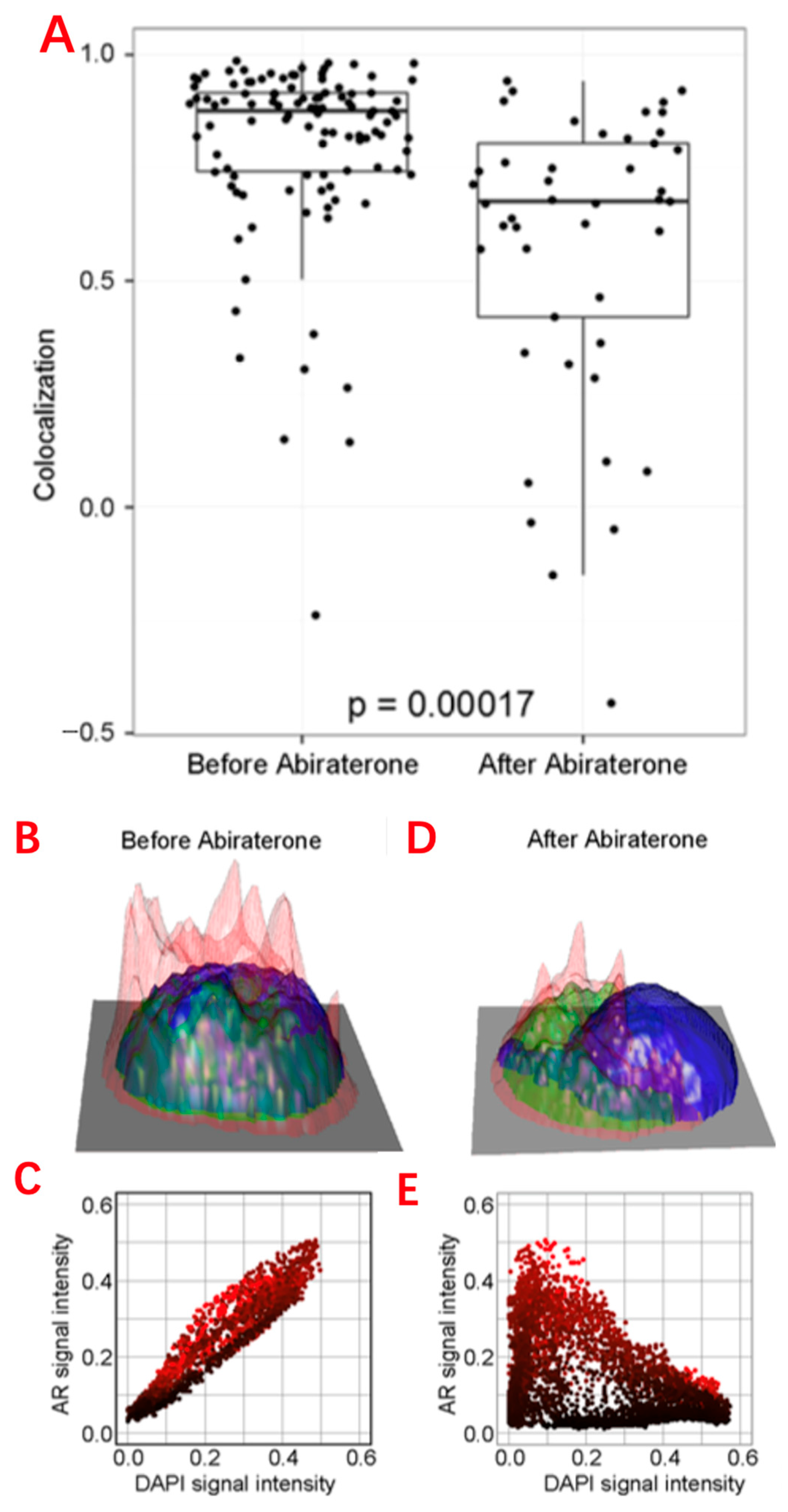
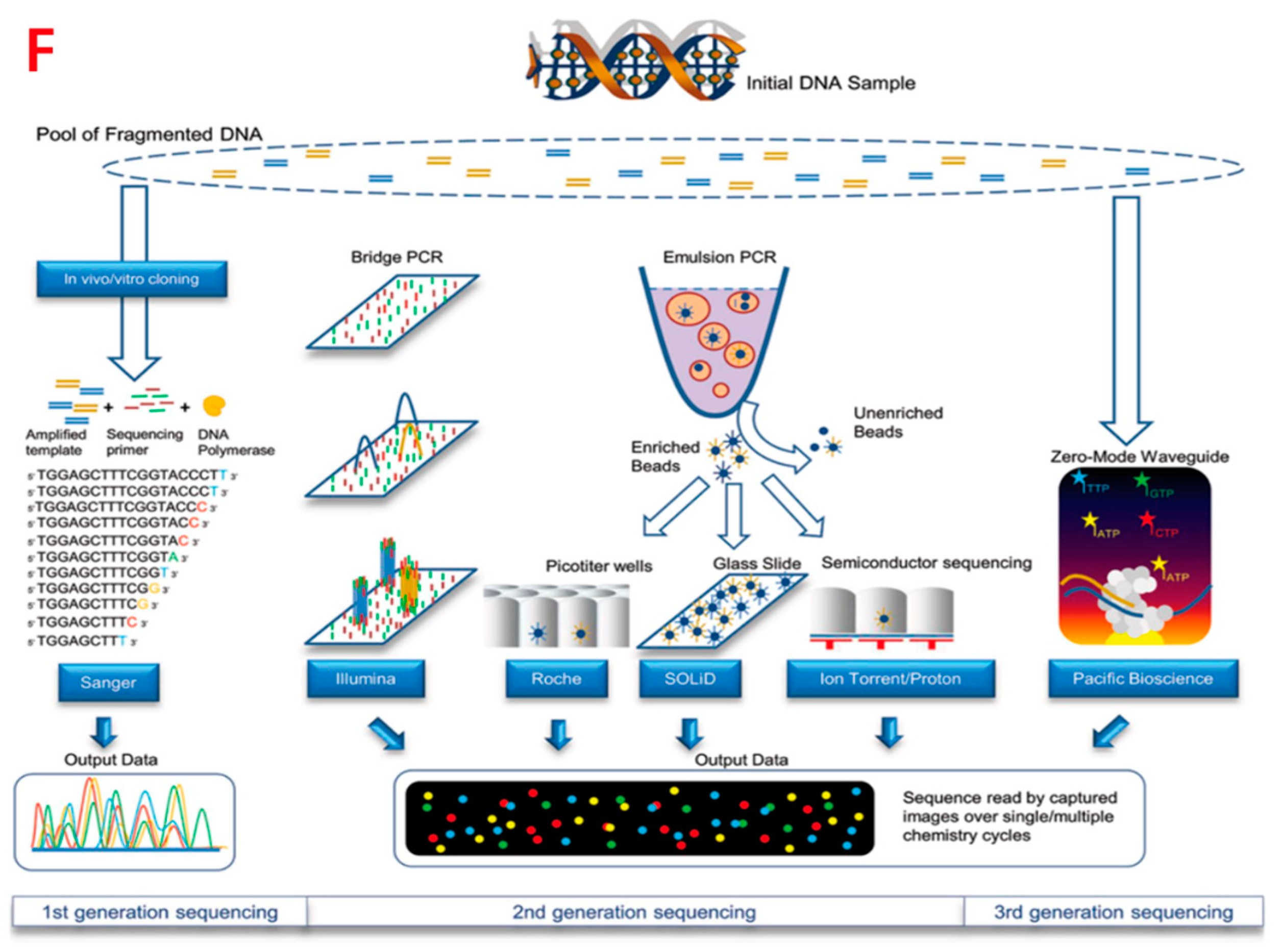
Figure 2. Single-cell analysis applications in CTCs. (A) Comparison of the AR subcellular localization in each CTC identified in the blood before and after 9 weeks of abiraterone treatment, which reveals the intracellular colocalization relationship between the AR and DAPI signals of CTCs. (B,D) Height maps of AR signal intensity of a single CTC, before and after 9 weeks of treatment. (C,E) AR versus DAPI signal intensities for each pixel inside the single CTC, revealing the relationship of nuclear exclusion as a negative correlation and nuclear localization as a positive correlation. (F) Different sequencing approaches. Technical features include sample preparation, sequencing chemistries, and data output formats.
Single-cell technology can also be combined with viral recombinant technology for the precise treatment of human diseases. Studies have demonstrated gene therapy at the single-cell level, presented experimental and computational methods for parallel characterization of recombinant adeno-associated virus (rAAVs) tropism, and enabled safe and precise gene delivery vectors. Recombinant AAVs (rAAVs) are the gene delivery vector of choice for many studies because of their broad viral tropism, ability to transect dividing and nondividing cells, and their ability to ensure long-term transgene expression with stability and persistence [14]. However, rAAV has poor target specificity and a relatively low therapeutic index for systemic gene therapy [15], and optimized AAV gene delivery vectors that can be used for cell-type-specific delivery are urgently needed [16]. Single-cell RNA sequencing (scRNA-seq) enables comprehensive analysis of the transcriptome at the entire-cell-type level. High-throughput single-cell transcriptome analysis can aid in further understanding AVV. Studies provided tropism information beyond several AAV variants and beyond the predominant cell types.
Additionally, the use of high-throughput RNA Seq on samples with the rRNA removed allows for the detection of almost all coding and non-coding RNA species in a given sample [17]. Such studies provide a large amount of data to reference for single-cell RNA sequencing analysis. Combining the two and carrying out single-cell sequencing analysis based on sequencing of mass samples is also a promising research direction.
Outside of disease research, single-cell sequencing technology can also be used to reveal the development process of various human tissues. For example, single-cell sequencing has been applied to the research of cortical cells of the nervous system, which assists the analysis of single cells in the developing human prefrontal cortex from 8 to 26 weeks of gestation, identifying cell types and subtypes within major categories [18].
The IFC device mentioned above is one of the most widely used single-cell analysis techniques. IFC has been used to classify two tumor cell lines, A549 and H1299 (human lung alveolar-like cell line, commonly used in non-small-cell lung cancer respiratory research), according to differences in cell membrane capacitance and cytoplasmic conductivity [19]. Other researchers have used IFC to isolate lung cancer DTCs (LC-DTCs) from red blood cells, peripheral blood mononuclear cells (PBMCs), and normal lung cells based on impedance amplitude [20]. Another investigator developed an IFC device to isolate individual PDAC tumor cells against a xenograft [21]. They found that the phases of impedance signals in six PDAC cells were correlated with specific gene expressions. The combination of individual intrinsic bioelectrical markers, such as membrane capacitance, cytoplasmic conductivity, and cell diameter, can significantly improve the classification accuracy of both A459 and HEPG2 tumor cells. In addition, some researchers used a commercial IFC device and found that necrotic and surviving U937 human lymphoma cells could be clearly distinguished based on the phase of the impedance signal [22]. In addition to its application to research directly related to humans, IFC equipment has been widely used for the detection, isolation, and activity analysis of unicellular microorganisms. Studies have demonstrated that IFC devices can detect bacteria based on cell size [23], can accurately measure the diameter of different bacteria [18], and can achieve higher sensitivity and detection throughput of bacterial size [24].
On the other hand, single-cell analysis techniques are of interest in the field of metabolism and help to gain insight into the relationship between the physiological behavior of single cells and their chemical components. As the ultimate products of cellular biochemical reactions, metabolites can accurately and in a real-time manner reflect the biochemical phenotypes of cells. Moreover, metabolite features can be converted into signals, such as electrochemical, optical, or mass-to-charge ratio. They can be measured relatively easily by electrochemical, optical, and mass spectrometry detection methods [25]. Therefore, they are generally regarded as the research objects of cellular metabolic processes. As a result of the heterogeneity of cells and the extremely fast turnover rate of some metabolites, traditional methods for studying the state of a large number of cells struggle to accurately reflect the metabolic processes of individual cells [26]. The introduction of single-cell analysis techniques has solved this problem well. Based on the traditional nanoelectrospray ionization mass spectrometry (nano-ESI-MS) technique, some researchers have further designed induced nano-ESI-MS. The new technique, which uses high-voltage alternating current to generate an inductive electrospray charge, targets the metabolite molecules, then measures them by the current, resistance, and voltage changes in the electrodes. It was used to study the metabolism of the internal metabolites of single neurons and revealed a novel pathway for brain glutamate biosynthesis [27]. Another technique, pulsed direct-current electrospray ionization mass spectrometry (pulsed-dc-ESI-MS), is often combined with microwell-based droplet microextraction and has been applied to the study of phospholipids and glucose phosphates in single cells [28]. However, several of the MS methods mentioned above use manual capillary probes for sampling, which makes their analytical efficiency more limited. To solve this problem, researchers developed a novel asymmetric serpentine channel microfluidic chip and combined it with pulsed electric-field-induced electrospray ionization high-resolution mass spectrometry (chip-PEF-ESI-HRMS). This method suspends cells in physiological saline droplets and uses a microfluidic device for continuous cell separation and inertial focusing operations. In this method, the analysis is performed in a near-physiological environment, and the efficiency is up to 80 cells per minute, it can be stable for more than 3 h, and 3000 cells can be analyzed consecutively in a single experiment. The researchers used this method to label 120 cancer cell metabolites and successfully distinguished two different cancer cells (MCF7 and HepG2 cells) based on single-cell metabolic profiles. This method can efficiently obtain metabolomic information of various single cells under near-physiological conditions, which has wide application prospects [26].
References
- Zhang, L.; Wan, S.; Jiang, Y.; Wang, Y.; Fu, T.; Liu, Q.; Cao, Z.; Qiu, L.; Tan, W. Molecular Elucidation of Disease Biomarkers at the Interface of Chemistry and Biology. J. Am. Chem. Soc. 2017, 139, 2532–2540.
- Schwarz, M.; Jendrusch, M.; Constantinou, I. Spatially resolved electrical impedance methods for cell and particle characterization. Electrophoresis 2020, 41, 65–80.
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176.
- Gagnon, Z.R. Cellular dielectrophoresis: Applications to the characterization, manipulation, separation and patterning of cells. Electrophoresis 2011, 32, 2466–2487.
- Liu, J.; Qiang, Y.; Alvarez, O.; Du, E. Electrical impedance microflow cytometry with oxygen control for detection of sickle cells. Sens. Actuators B Chem. 2018, 255 (Pt 2), 2392–2398.
- Lee, J.; Kwak, B. Simultaneous on-chip isolation and characterization of circulating tumor cell sub-populations. Biosens. Bioelectron. 2020, 168, 112564.
- Nagano, T.; Lubling, Y.; Stevens, T.J.; Schoenfelder, S.; Yaffe, E.; Dean, W.; Laue, E.D.; Tanay, A.; Fraser, P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013, 502, 59–64.
- Xu, J.; Liao, K.; Yang, X.; Wu, C.; Wu, W. Correction to: Using single-cell sequencing technology to detect circulating tumor cells in solid tumors. Mol. Cancer 2022, 21, 100.
- Lim, S.B.; Lim, C.T.; Lim, W.T. Single-Cell Analysis of Circulating Tumor Cells: Why Heterogeneity Matters. Cancers 2019, 11, 1595.
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239.
- Lill, M.; Kõks, S.; Soomets, U.; Schalkwyk, L.C.; Fernandes, C.; Lutsar, I.; Taba, P. Peripheral blood RNA gene expression profiling in patients with bacterial meningitis. Front. Neurosci. 2013, 7, 33.
- Fernandez, S.V.; Bingham, C.; Fittipaldi, P.; Austin, L.; Palazzo, J.; Palmer, G.; Alpaugh, K.; Cristofanilli, M. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast. Cancer Res. 2014, 16, 445.
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122.
- Bedbrook, C.N.; Deverman, B.E.; Gradinaru, V. Viral Strategies for Targeting the Central and Peripheral Nervous Systems. Annu. Rev. Neurosci. 2018, 41, 323–348.
- Mével, M.; Bouzelha, M.; Leray, A.; Pacouret, S.; Guilbaud, M.; Penaud-Budloo, M.; Alvarez-Dorta, D.; Dubreil, L.; Gouin, S.G.; Combal, J.P.; et al. Chemical modification of the adeno-associated virus capsid to improve gene delivery. Chem. Sci. 2019, 11, 1122–1131.
- Paulk, N. Gene Therapy: It Is Time to Talk About High-Dose AAV: The Deaths of Two Children With X-Linked Myotubular Myopathy in the ASPIRO Trial Prompts a Reexamination of Vector Safety. Genet. Eng. Biotechnol. News 2020, 40, 14–16.
- Reemann, P.; Reimann, E.; Ilmjärv, S.; Porosaar, O.; Silm, H.; Jaks, V.; Vasar, E.; Kingo, K.; Kõks, S. Melanocytes in the skin–comparative whole transcriptome analysis of main skin cell types. PLoS ONE 2014, 9, e115717.
- Choi, H.; Jeon, C.S.; Hwang, I.; Ko, J.; Lee, S.; Choo, J.; Boo, J.H.; Kim, H.C.; Chung, T.D. A flow cytometry-based submicron-sized bacterial detection system using a movable virtual wall. Lab Chip 2014, 14, 2327–2333.
- Zhao, Y.; Jiang, M.; Chen, D.; Zhao, X.; Xue, C.; Hao, R.; Yue, W.; Wang, J.; Chen, J. Single-Cell Electrical Phenotyping Enabling the Classification of Mouse Tumor Samples. Sci. Rep. 2016, 6, 19487.
- Desai, S.P.; Coston, A.; Berlin, A. Micro-electrical impedance spectroscopy and identification of patient-derived, dissociated tumor cells. IEEE Trans. Nanobiosci. 2019, 18, 369–372.
- McGrath, J.S.; Honrado, C.; Moore, J.H.; Adair, S.J.; Varhue, W.B.; Salahi, A.; Farmehini, V.; Goudreau, B.J.; Nagdas, S.; Blais, E.M.; et al. Electrophysiology-based stratification of pancreatic tumorigenicity by label-free single-cell impedance cytometry. Anal. Chim. Acta 2020, 1101, 90–98.
- Ostermann, M.; Sauter, A.; Xue, Y.; Birkeland, E.; Schoelermann, J.; Holst, B.; Cimpan, M.R. Label-free impedance flow cytometry for nanotoxicity screening. Sci. Rep. 2020, 10, 142.
- Bernabini, C.; Holmes, D.; Morgan, H. Micro-impedance cytometry for detection and analysis of micron-sized particles and bacteria. Lab Chip 2011, 11, 407–412.
- Guler, M.T.; Bilican, I. Capacitive detection of single bacterium from drinking water with a detailed investigation of electrical flow cytometry. Sens. Actuator A Phys. 2018, 269, 454–463.
- Yang, J.; Huang, L.; Qian, K. Nanomaterials-assisted metabolic analysis toward in vitro diagnostics. Exploration 2022, 2, 20210222.
- Feng, D.; Li, H.; Xu, T.; Zheng, F.; Hu, C.; Shi, X.; Xu, G. High-throughput single cell metabolomics and cellular heterogeneity exploration by inertial microfluidics coupled with pulsed electric field-induced electrospray ionization-high resolution mass spectrometry. Anal. Chim. Acta 2022, 1221, 340116.
- Flangea, C.; Serb, A.; Sisu, E.; Zamfir, A.D. Chip-based nanoelectrospray mass spectrometry of brain gangliosides. Biochim. Biophys. Acta 2011, 1811, 513–535.
- Wei, Z.; Xiong, X.; Guo, C.; Si, X.; Zhao, Y.; He, M.; Yang, C.; Xu, W.; Tang, F.; Fang, X.; et al. Pulsed Direct Current Electrospray: Enabling Systematic Analysis of Small Volume Sample by Boosting Sample Economy. Anal. Chem. 2015, 87, 11242–11248.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
538
Revisions:
2 times
(View History)
Update Date:
13 Oct 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




