
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tigran Harutyunyan | -- | 1125 | 2023-09-29 11:04:43 | | | |
| 2 | Wendy Huang | Meta information modification | 1125 | 2023-10-07 09:34:00 | | |
Video Upload Options
The term cell-free DNA (cfDNA) encompasses all kinds of extracellular DNA molecules found in serum or plasma and other body fluids, and includes genomic DNA (gDNA) and mitochondrial DNA (mtDNA), as well as DNA of bacterial or viral origin. Cell-free DNA (cfDNA), freely circulating in the bloodstream, urine, and other fluids (or encapsulated in vesicles) may be derived from both normal and diseased cells. cfDNA is extremely dynamic and responsive, providing sensitive indicators of changes that are not detectable by standard clinical tests. It can be used as a reliable, safe, and objective tool to reflect disease progression and supplement clinical data in a particular patient, and thus, represents a new path in personalized medicine.
1. Introduction
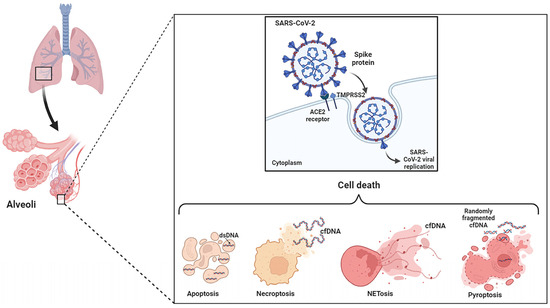
2. Apoptosis, Necrosis and NETosis
3. Extracellular Vesicles
4. Chromosomal Instability and Micronucleus Formation
5. Erythroblast Enucleation
References
- Tan, E.M.; Kunkel, H.G. Characteristics of a Soluble Nuclear Antigen Precipitating with Sera of Patients with Systemic Lupus Erythematosus. J. Immunol. 1966, 96, 464–471.
- Leon, S.A.; Ehrlich, G.E.; Shapiro, B.; Labbate, V.A. Free DNA in the Serum of Rheumatoid Arthritis Patients. J. Rheumatol. 1977, 4, 139–143.
- Dennis Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.G.; Wainscoat, J.S. Presence of Fetal DNA in Maternal Plasma and Serum. Lancet 1997, 350, 485–487.
- Zhong, S.; Ng, M.C.Y.; Lo, Y.M.D.; Chan, J.C.N.; Johnson, P.J. Presence of Mitochondrial TRNA(Leu(UUR)) A to G 3243 Mutation in DNA Extracted from Serum and Plasma of Patients with Type 2 Diabetes Mellitus. J. Clin. Pathol. 2000, 53, 466–469.
- Ha, T.T.N.; Huy, N.T.; Murao, L.A.; Lan, N.T.P.; Thuy, T.T.; Tuan, H.M.; Nga, C.T.P.; van Tuong, V.; van Dat, T.; Kikuchi, M.; et al. Elevated Levels of Cell-Free Circulating DNA in Patients with Acute Dengue Virus Infection. PLoS ONE 2011, 6, e25969.
- Phuong, N.T.N.; Manh, D.H.; Dumre, S.P.; Mizukami, S.; Weiss, L.N.; van Thuong, N.; Ha, T.T.N.; Phuc, L.H.; van An, T.; Tieu, T.M.; et al. Plasma Cell-Free DNA: A Potential Biomarker for Early Prediction of Severe Dengue. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 10.
- Outinen, T.K.; Kuparinen, T.; Jylhävä, J.; Leppänen, S.; Mustonen, J.; Mäkelä, S.; Pörsti, I.; Syrjänen, J.; Vaheri, A.; Hurme, M. Plasma Cell-Free DNA Levels Are Elevated in Acute Puumala Hantavirus Infection. PLoS ONE 2012, 7, e31455.
- Yi, J.; Zhang, Y.; Zhang, Y.; Ma, Y.; Zhang, C.; Li, Q.; Liu, B.; Liu, Z.; Liu, J.; Zhang, X.; et al. Increased Plasma Cell-Free DNA Level during HTNV Infection: Correlation with Disease Severity and Virus Load. Viruses 2014, 6, 2723–2734.
- Bakir, M.; Engin, A.; Kuskucu, M.A.; Bakir, S.; Gündag, O.; Midilli, K. Relationship of Plasma Cell-Free DNA Level with Mortality and Prognosis in Patients with Crimean-Congo Hemorrhagic Fever. J. Med. Virol. 2016, 88, 1152–1158.
- Stawski, R.; Nowak, D.; Perdas, E. Cell-Free DNA: Potential Application in COVID-19 Diagnostics and Management. Viruses 2022, 14, 321.
- Teo, Y.V.; Capri, M.; Morsiani, C.; Pizza, G.; Faria, A.M.C.; Franceschi, C.; Neretti, N. Cell-free DNA as a biomarker of aging. Aging Cell 2019, 18, e12890.
- Bai, Y.; Zheng, F.; Zhang, T.; Luo, Q.; Luo, Y.; Zhou, R.; Jin, Y.; Shan, Y.; Cheng, J.; Yang, Z.; et al. Integrating Plasma Cell-free DNA with Clinical Laboratory Results Enhances the Prediction of Critically Ill Patients with COVID-19 at Hospital Admission. Clin. Transl. Med. 2022, 12, e966.
- Danthi, P. Viruses and the Diversity of Cell Death. Annu. Rev. Virol. 2016, 3, 533–553.
- Rex, D.A.B.; Prasad, T.S.K.; Kandasamy, R.K. Revisiting Regulated Cell Death Responses in Viral Infections. Int. J. Mol. Sci. 2022, 23, 7023.
- Thiam, H.R.; Wong, S.L.; Wagner, D.D.; Waterman, C.M. Cellular Mechanisms of NETosis. Annu. Rev. Cell Dev. Biol. 2020, 36, 191–218.
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532–1535.
- Keshari, R.S.; Jyoti, A.; Kumar, S.; Dubey, M.; Verma, A.; Srinag, B.S.; Krishnamurthy, H.; Barthwal, M.K.; Dikshit, M. Neutrophil Extracellular Traps Contain Mitochondrial as Well as Nuclear DNA and Exhibit Inflammatory Potential. Cytom. Part A 2012, 81 A, 238–247.
- Liu, Y.; Garron, T.M.; Chang, Q.; Su, Z.; Zhou, C.; Qiu, Y.; Gong, E.C.; Zheng, J.; Whitney Yin, Y.; Ksiazek, T.; et al. Cell-Type Apoptosis in Lung during Sars-Cov-2 Infection. Pathogens 2021, 10, 509.
- André, S.; Picard, M.; Cezar, R.; Roux-Dalvai, F.; Alleaume-Butaux, A.; Soundaramourty, C.; Cruz, A.S.; Mendes-Frias, A.; Gotti, C.; Leclercq, M.; et al. T Cell Apoptosis Characterizes Severe COVID-19 Disease. Cell Death Differ. 2022, 29, 1486–1499.
- Sun, C.; Han, Y.; Zhang, R.; Liu, S.; Wang, J.; Zhang, Y.; Chen, X.; Jiang, C.; Wang, J.; Fan, X.; et al. Regulated Necrosis in COVID-19: A Double-Edged Sword. Front. Immunol. 2022, 13, 917141.
- Bader, S.M.; Cooney, J.P.; Pellegrini, M.; Doerflinger, M. Programmed Cell Death: The Pathways to Severe COVID-19? Biochem. J. 2022, 479, 609–628.
- Da Silva, M.M.; de Lucena, A.S.L.; Júnior, S.d.S.L.P.; de Carvalho, V.M.F.; de Oliveira, P.S.S.; da Rosa, M.M.; Rego, M.J.B.d.M.; Pitta, M.G.d.R.; Pereira, M.C. Cell Death Mechanisms Involved in Cell Injury Caused by SARS-CoV-2. Rev. Med. Virol. 2022, 32, 3.
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727.
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell Res. 2014, 24, 766–769.
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rodriguez, I.R.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int. J. Mol. Sci. 2020, 21, 8062.
- Lázaro-Ibáñez, E.; Lässer, C.; Shelke, G.V.; Crescitelli, R.; Jang, S.C.; Cvjetkovic, A.; García-Rodríguez, A.; Lötvall, J. DNA Analysis of Low- and High-Density Fractions Defines Heterogeneous Subpopulations of Small Extracellular Vesicles Based on Their DNA Cargo and Topology. J. Extracell. Vesicles 2019, 8, 1656993.
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New Evidence That a Large Proportion of Human Blood Plasma Cell-Free DNA Is Localized in Exosomes. PLoS ONE 2017, 12, e0183915.
- Mondelo-Macía, P.; Castro-Santos, P.; Castillo-García, A.; Muinelo-Romay, L.; Diaz-Peña, R. Circulating Free DNA and Its Emerging Role in Autoimmune Diseases. J. Pers. Med. 2021, 11, 151.
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes Maintain Cellular Homeostasis by Excreting Harmful DNA from Cells. Nat. Commun. 2017, 8, 9.
- Cai, J.; Han, Y.; Ren, H.; Chen, C.; He, D.; Zhou, L.; Eisner, G.M.; Asico, L.D.; Jose, P.A.; Zeng, C. Extracellular Vesicle-Mediated Transfer of Donor Genomic DNA to Recipient Cells Is a Novel Mechanism for Genetic Influence between Cells. J. Mol. Cell Biol. 2013, 5, 227–238.
- Fenech, M.; Knasmueller, S.; Bolognesi, C.; Holland, N.; Bonassi, S.; Kirsch-Volders, M. Micronuclei as Biomarkers of DNA Damage, Aneuploidy, Inducers of Chromosomal Hypermutation and as Sources of pro-Inflammatory DNA in Humans. Mutat. Res.-Rev. Mutat. Res. 2020, 786, 108342.
- MacKenzie, K.J.; Carroll, P.; Martin, C.A.; Murina, O.; Fluteau, A.; Simpson, D.J.; Olova, N.; Sutcliffe, H.; Rainger, J.K.; Leitch, A.; et al. CGAS Surveillance of Micronuclei Links Genome Instability to Innate Immunity. Nature 2017, 548, 461–465.




