
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Richard Zimmermann | -- | 2692 | 2023-09-27 15:36:24 | | | |
| 2 | Sirius Huang | Meta information modification | 2692 | 2023-09-28 03:21:14 | | | | |
| 3 | Richard Zimmermann | + 1149 word(s) | 3841 | 2025-09-11 20:06:15 | | |
Video Upload Options
The protein import into the organelle termed the endoplasmic reticulum (ER) is the first step in the biogenesis of about one-third of the different soluble and membrane proteins (MPs) of human cells and, therefore, represents a central cell biological research topic.
1. Introduction
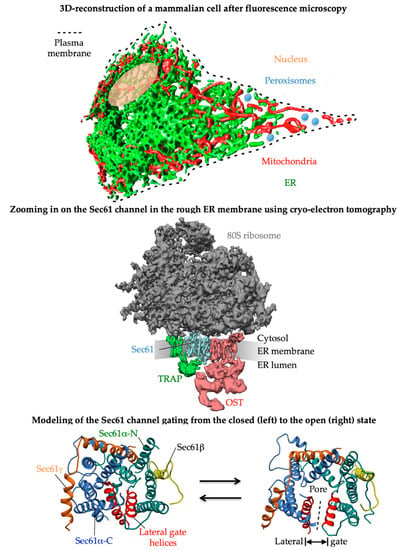
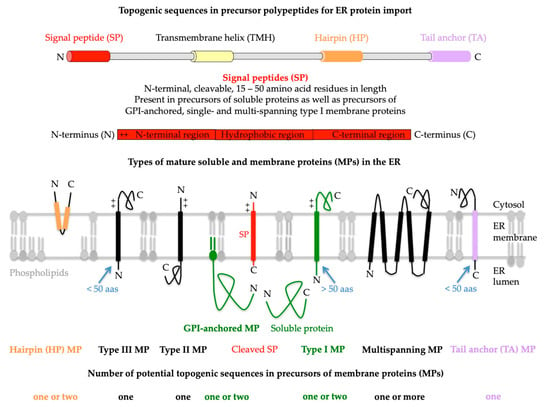
2. Proteins of the ER Membrane
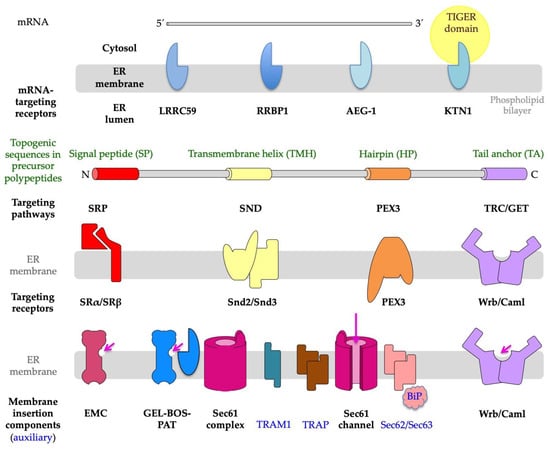
3. Targeting of Precursor Polypeptides to the ER Membrane
4. Insertion of Precursor Polypeptides into the ER Membrane
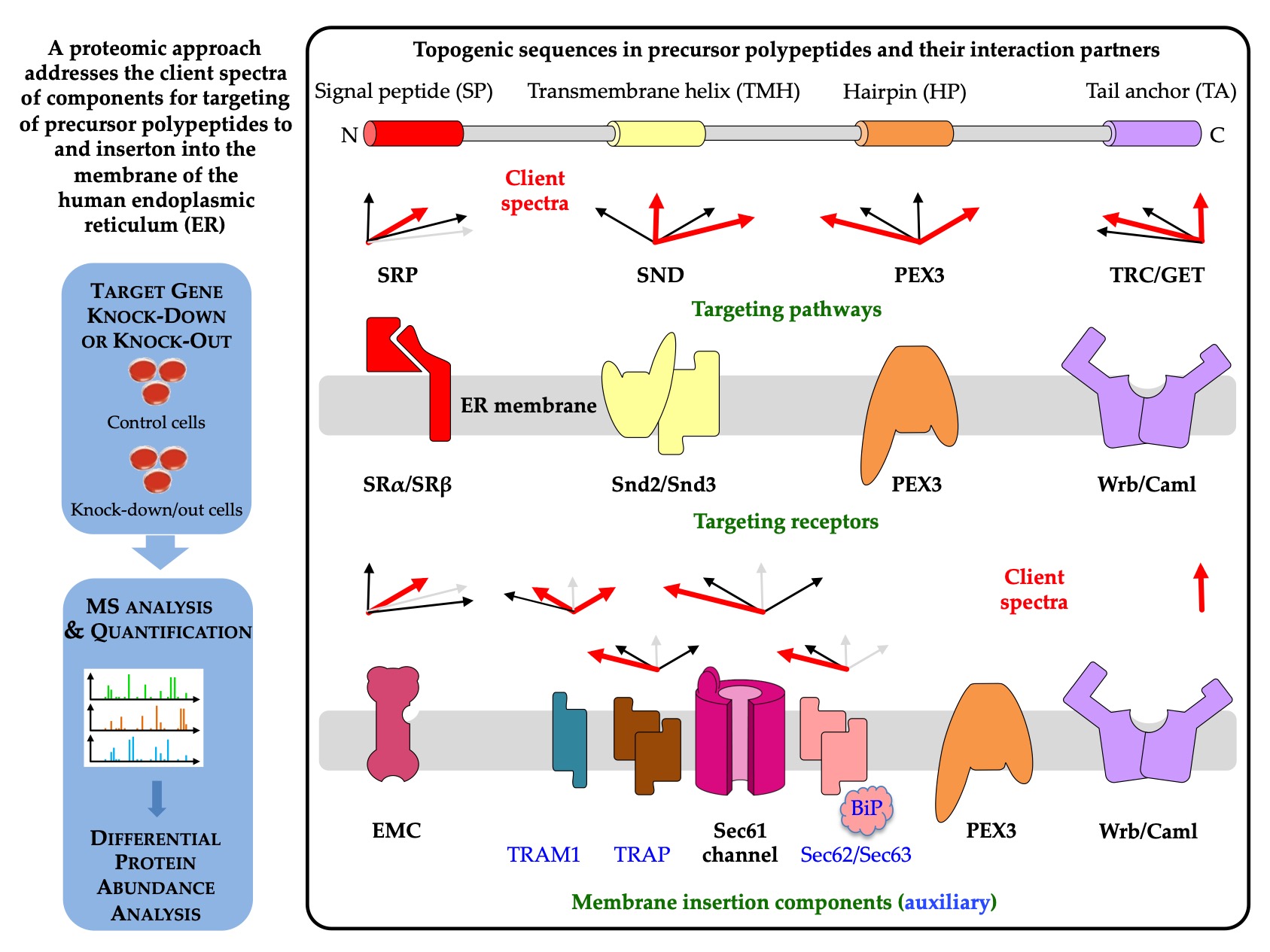
5. A Single Proteomic Approach to Address the Client Spectra of Various Components for Targeting of Precursor Polypeptides to and Insertion into the Human ER Membrane
6. Insertion of Precursor Polypeptides into the ER Membrane: An Update
In the original entry, based on the review by Jung and Zimmermann, the focus was on global lessons from the quantitative proteomics approach that addressed the question of which precursors use a certain targeting pathway or membrane insertion or translocation component in intact cells. In the last update of the original review[140], the focus was on the limitations of the proteomic approach in answering this cell biological question as well as on additional aspects, such as N-terminal methionine excision and N-terminal acetylation of membrane proteins.
6.1. N-terminal methionine excision and acetylation of membrane proteins
Recent work has shown that the nascent chain-associated complex (NAC) coordinates N-terminal methionine excision and N-terminal acetylation of the majority of nascent polypeptides in the cytosol[141,142,143]. Based on protein data bases, an estimated 80% of all cellular proteins are enzymatically modified in this way, including MPs. In short, the heterodimeric chaperone NAC, which is equimolar to ribosomes in the cytosol and is typically bound to them, binds first to the nascent chains during polypeptide synthesis as they emerge from the ribosomal tunnel exit. It then recruits one of the two methionine aminopeptidases (METAPs) and subsequently one of several N-acetyltransferases (NATs). In both cases, substrate specificity is determined by the amino acid residue following the initiating methionine. Alternatively, the initiating methionine is retained and becomes N-acetylated. Apparently, SRP can antagonize NAC binding in the case of SP-containing precursor polypeptides of secretory and membrane proteins, thus inhibiting cotranslational N-terminal methionine excision and N-terminal acetylation. Hydrophobic SPs trigger NAC release through direct interaction. Notably, in yeast, the artificial enzymatic modification of an SP was observed to inhibit ER import of the polypeptide. However, many TMHs of MPs are not N-terminal, raising the question of whether or not these MPs can undergo these particular enzymatic modifications.
Therefore, clients of components involved in the biogenesis of MPs at the human ER were screened for N-terminal methionine excision and N-acetylation in the NCBI protein database. The results clearly demonstrated that the two cotranslational N-terminal modifications are compatible with ER targeting and membrane insertion of MP clients for almost all analyzed components in human cells, with the exception of TRAM1 with a comparatively low number of putative MP clients[140]. Starting from a modified EMC client with a TMH in the range from amino acid residues 29 to 49, the critical starting point of the TMH enabling the two modifcations is on average 52 amino acid residues, i.e. just downstream of the average SP with a typical length of 15 to 50 amino acid residues. Considering that at any given time of protein synthesis, approximatley 40 amino acid residues of a nascent polypeptide chain are buried in the ribosomal polypeptide tunnel, it appears that cotranslational N-terminal methionine excision and N-acetylation of MPs with TMH is possible in human cells, as long as the TMH is not located at the immediate N-terminus.
6.2. Limitations of the proteomic approach for the analysis of components and pathways that are involved in biogenesis of membrane proteins at the ER
The results from the in vivo-like approach confirmed and further consolidated many of the lessons from the classical in vitro experiments, but also added new ones, such as that not only the polarity or hydrophobicity of the SP or TMH matters, and that not only what is up-front, i.e., at the N-terminus, that counts. In addition, the glycine and proline content, i.e., the helix propensity of SP or TMH, as well as clusters of charged amino acid residues downstream of SP, play a role in substrate specificity and affinity.
An undisputable limitation of the present global analyses of MP biogenesis at the ER concerns the Sec61 complex. Since this complex contributes to MP biogenesis in two ways: it functions as an MP insertase and provides a ribosome binding site to the MP insertase that is termed PAT-GEL-BOS supercomplex, Sec61 clients cannot be distinguished between true clients of the Sec61 channel and clients of the heterotrimeric supercomplex within the MPT, i.e., the ribosome binding site of Sec61. This puzzle will have to be resolved in future research.
The interpretation of the proteomic data was weakest where the total number of putative MP clients was lowest, such as in the above-mentioned case of TRAM1. Typically, 30% of the proteome of a human cell comprises clients of ER protein import. However, even in the case of Sec61 depletion, only 197 proteins with SP plus 98 with TMH, i.e., about 300 of the 6000 quantified proteins or 5%, were negatively affected by the depletion. This raises the question of why we observed only such an abysmal effect. We envision several responsible factors under conditions of siRNA-mediated knockdown: The depletion efficiency and its duration, which were optimized for minimal effects on cell growth and viability, were not high enough to cause significant accumulation and degradation of the respective clients. The MS data suggested a depletion efficiency of approximately 90% for the respective component. The residual amount of the component may have been sufficient to sustain the physiological functions of depleted proteins over the duration of the experiment. A certain function in ER protein import in human cells is compensated for by other components or pathways. Except for the Sec61 complex, we expected that to be the case. Some client proteins may have remained largely unaffected due to either longer half-lives than the respective component or higher than average affinities for the component. Furthermore, some accumulating precursors may have stayed soluble in the cytosol, aggregated, or ended up in mitochondria, where they were out of reach for degradation by the ubiquitin/proteasome system. We propose that all these factors together may have contributed, possibly to a different extent for different precursors.
Therefore, the question arises whether CRISPR/Cas9-mediated knock-out cells would not have been the better choice to begin with. Indeed, the EMC clients were identified in this kind of cells, where the half-lives of clients should not matter. To address this question, the positively affected proteins after transient knock-down of Sec62 or Sec63 and after knock-out were re-interrogated[140]. Although the number of negatively affected proteins were higher in the Sec62 knock-out versus knock-down cells, as had to be expected, both the total numbers of positively affected proteins as well as the number of proteins that are related to the ubiquitin/proteasome system were lower in the Sec62 knock-out (13 and 5, respectively) versus knock-down cells (25 and 14, respectively). The impression from this comparison is that in the knock-out cells different adaptation processes must have occurred as compared to the knock-down cells. It appears that adaptation in the latter occurred at the level of the clients via increased chaperone and ubiquitin/proteasome activity and/or capacity. In contrast, knock-out cells may have adapted by globally lowering the expression of genes or mRNAs that code for clients. Notably, we originally refrained from using knock-out cells because we wanted to compare all depleted components with the essential Sec61α1 protein.
References
- Blobel, G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA 1980, 77, 1496–1500.
- Egea, P.F.; Stroud, R.M.; Walter, P. Targeting proteins to membranes: Structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005, 15, 213–220.
- Aviram, N.; Schuldiner, M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017, 130, 4079–4085.
- Gemmer, M.; Förster, F. A clearer picture of the ER translocon complex. J. Cell Sci. 2020, 133, jcs231340.
- Lang, S.; Nguyen, D.; Bhadra, P.; Jung, M.; Helms, V.; Zimmermann, R. Signal peptide features determining the substrate specificities of targeting and translocation components in human ER protein import. Front. Physiol. 2022, 13, 833540.
- Sicking, M.; Lang, S.; Bochen, F.; Drenth, J.P.H.; Zacharia, M.; Zimmermann, R.; Roos, A.; Linxweiler, M. Complexity and specificity of Sec61 channelopathies: Human diseases affecting gating of the Sec61 complex. Cells 2021, 10, 1036.
- Jansen, R.L.M.; van der Klei, I.J. The peroxisome biogenesis factors Pex3 and Pex19: Multitasking proteins with disputed functions. FEBS Lett. 2019, 593, 457–474.
- Dhimann, R.; Caesar, S.; Thiam, A.R.; Schrul, B. Mechanisms of protein targeting to lipid droplets: A unified cell biological and biophysical perspective. Sem. Cell Dev. Biol. 2020, 108, 4–13.
- Hansen, K.G.; Aviram, N.; Laborenz, J.; Bibi, C.; Meyer, M.; Spang, A.; Schuldiner, M.; Herrmann, J.M. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 2018, 361, 1118–1122.
- Koch, C.; Schuldiner, M.; Herrmann, J.M. ER-SURF: Riding the endoplasmic reticulum SURFace to mitochondria. Int. J. Mol. Sci. 2021, 22, 9655.
- Palade, G. Intracellular aspects of protein synthesis. Science 1975, 189, 347–358.
- Palade, G.; Porter, K.R. Studies on the endoplasmic reticulum. J. Exp. Med. 1954, 100, 641–656.
- Blobel, G.; Dobberstein, B. Transfer of proteins across membranes: I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 1975, 67, 835–851.
- Blobel, G.; Dobberstein, B. Transfer of proteins across membranes: II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 1975, 67, 852–862.
- Von Heijne, G. Signal sequences. J. Mol. Biol. 1985, 184, 99–105.
- Von Heijne, G.; Gavel, Y. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 1988, 174, 671–678.
- Hegde, R.S.; Bernstein, H. The surprising complexity of signal peptides. Trends Biochem. Sci. 2006, 31, 563–571.
- Siegel, V.; Walter, P. Functional dissection of the signal recognition particle. Trends Biochem. Sci. 1988, 13, 314–316.
- Ng, D.T.; Brown, J.D.; Walter, P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 1996, 134, 269–278.
- Halic, M.; Beckmann, R. The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Biol. 2005, 15, 116–125.
- Halic, M.; Blau, M.; Becker, T.; Mielke, T.; Pool, M.R.; Wild, K.; Sinning, I.; Beckmann, R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 2006, 444, 507–511.
- Gamerdinger, M.; Hanebuth, M.A.; Frickey, T.; Deuerling, E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 2015, 348, 201–207.
- Hsieh, H.-H.; Lee, J.H.; Chandrasekar, S.; Shan, S.-o. A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun. 2020, 11, 5840.
- Jomaa, A.; Eitzinger, S.; Zhu, Z.; Chandrasekar, S.; Kobajashi, K.; Shan, S.-o.; Ban, N. Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle. Cell Rep. 2021, 36, 109350.
- Jomaa, A.; Gamerdinger, M.; Hsieh, H.-H.; Wallisch, A.; Chandrasekaran, V.; Ulusoy, Z.; Scaiola, A.; Hegde, R.S.; Shan, S.-o.; Ban, N.; et al. Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting. Science 2022, 375, 839–844.
- Meyer, D.I.; Dobberstein, B. A membrane component essential for vectorial translocation of nascent proteins across the endoplasmic reticulum: Requirements for its extraction and reassociation with the membrane. J. Cell Biol. 1980, 87, 498–502.
- Gilmore, R.; Blobel, G.; Walter, P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982, 95, 463–469.
- Tajima, S.; Lauffer, L.; Rath, V.L.; Walter, P. The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J. Cell Biol. 1986, 103, 1167–1178.
- Jan, C.H.; Williams, C.C.; Weissman, J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 2014, 346, 1257521.
- Chartron, J.W.; Hunt, K.C.L.; Frydman, J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 2016, 536, 224–228.
- Görlich, D.; Prehn, S.; Hartmann, E.; Kalies, K.-U.; Rapoport, T.A. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 1992, 71, 489–503.
- Görlich, D.; Rapoport, T.A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 1993, 75, 615–630.
- Simon, S.M.; Blobel, G. A protein-conducting channel in the endoplasmic reticulum. Cell 1991, 65, 371–380.
- Wirth, A.; Jung, M.; Bies, C.; Frien, M.; Tyedmers, J.; Zimmermann, R.; Wagner, R. The Sec61p complex is a dynamic precursor activated channel. Mol. Cell 2003, 12, 261–268.
- Beckmann, R.; Spahn, C.M.; Eswar, N.; Helmers, J.; Penczek, P.A.; Sali, A.; Frank, J.; Blobel, G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 2001, 107, 361–372.
- Van den Berg, B.; Clemons, W.M.; Collinson, I.; Modis, Y.; Hartmann, E.; Harrison, S.C.; Rapoport, T.A. X-ray structure of a protein-conducting channel. Nature 2004, 427, 36–44.
- Pfeffer, S.; Brandt, F.; Hrabe, T.; Lang, S.; Eibauer, M.; Zimmermann, R.; Förster, F. Structure and 3D arrangement of ER-membrane associated ribosomes. Structure 2012, 20, 1508–1518.
- Pfeffer, S.; Dudek, J.; Gogala, M.; Schorr, S.; Linxweiler, J.; Lang, S.; Becker, T.; Beckmann, R.; Zimmermann, R.; Förster, F. Structure of the mammalian oligosaccharyltransferase in the native ER protein translocon. Nat. Commun. 2014, 5, 3072.
- Voorhees, R.M.; Fernández, I.S.; Scheres, S.H.W.; Hegde, R.S. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 2014, 157, 1632–1643.
- Voorhees, R.M.; Hegde, R.S. Structure of the Sec61 channel opened by a signal peptide. Science 2016, 351, 88–91.
- Pfeffer, S.; Burbaum, L.; Unverdorben, P.; Pech, M.; Chen, Y.; Zimmermann, R.; Beckmann, R.; Förster, F. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 2015, 6, 8403.
- Wiedmann, M.; Kurzchalia, T.V.; Hartmann, E.; Rapoport, T.A. A signal sequence receptor in the endoplasmic reticulum membrane. Nature 1987, 328, 830–833.
- Fons, R.D.; Bogert, B.A.; Hegde, R.S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003, 160, 529–539.
- Menetret, J.F.; Hegde, R.S.; Aguiar, M.; Gygi, S.P.; Park, E.; Rapoport, T.A.; Akey, C.W. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure 2008, 16, 1126–1137.
- Sommer, N.; Junne, T.; Kalies, K.-U.; Spiess, M.; Hartmann, E. TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim. Biophys. Acta 2013, 1833, 3104–3111.
- Pfeffer, S.; Dudek, J.; Ng, B.; Schaffa, M.; Albert, S.; Plitzko, J.; Baumeister, W.; Zimmermann, R.; Freeze, H.F.; Engel, B.D.; et al. Dissecting the molecular organization of the translocon-associatecd protein complex. Nat. Commun. 2017, 8, 14516.
- Jaskolowski, M.; Jomaa, A.; Gamerdinger, M.; Shresta, S.; Leibundgut, M.; Deuerling, E.; Ban, N. Molecular basis of the TRAP complex function in ER protein biogenesis. Nat. Struct. Mol. Biol. 2023, 375, 839–844.
- Pauwels, E.; Shewakramani, N.R.; De Wijngaert, B.; Camps, A.; Provinciael, B.; Stroobants, J.; Kalies, K.-U.; Hartmann, E.; Maes, P.; Vermeire, K.; et al. Structural insights into TRAP association with ribosome-Sec61 complex and translocon inhibition by a CADA derivative. Sci. Adv. 2023, 9, eadf0797.
- Dierks, T.; Volkmer, J.; Schlenstedt, G.; Jung, C.; Sandholzer, U.; Zachmann, K.; Schlotterhose, P.; Neifer, K.; Schmidt, B.; Zimmermann, R. A microsomal ATP-binding protein involved in efficient protein transport into the mammalian endoplasmic reticulum. EMBO J. 1996, 15, 6931–6942.
- Tyedmers, J.; Lerner, M.; Bies, C.; Dudek, J.; Skowronek, M.H.; Haas, I.G.; Heim, N.; Nastainczyk, W.; Volkmer, J.; Zimmermann, R. Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc. Natl. Acad. Sci. USA 2000, 97, 7214–7219.
- Mayer, H.-A.; Grau, H.; Kraft, R.; Prehn, S.; Kalies, K.-U.; Hartmann, E. Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 2000, 275, 14550–14557.
- Tyedmers, J.; Lerner, M.; Wiedmann, M.; Volkmer, J.; Zimmermann, R. Polypeptide chain binding proteins mediate completion of cotranslational protein translocation into the mammalian endoplasmic reticulum. EMBO Rep. 2005, 4, 505–510.
- Lakkaraju, A.K.K.; Thankappan, R.; Mary, C.; Garrison, J.L.; Taunton, J.; Strub, K. Efficient secretion of small proteins in mammalian cells relies on Sec62-dependent posttranslational translocation. Mol. Biol. Cell 2012, 23, 2712–2722.
- Lang, S.; Benedix, J.; Fedeles, S.V.; Schorr, S.; Schirra, C.; Schäuble, N.; Jalal, C.; Greiner, M.; Haßdenteufel, S.; Tatzelt, J.; et al. Different effects of Sec61α-, Sec62 and Sec63-depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J. Cell Sci. 2012, 125, 1958–1969.
- Jadhav, B.; McKenna, M.; Johnson, N.; High, S.; Sinning, I.; Pool, M.R. Mammalian SRP receptor switches the Sec61 translocase from Sec62 to SRP-dependent translocation. Nat. Commun. 2015, 6, 10133.
- Conti, B.J.; Devaraneni, P.K.; Yang, Z.; David, L.L.; Skach, W.R. Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol. Cell 2015, 58, 269–283.
- Haßdenteufel, S.; Johnson, N.; Paton, A.W.; Paton, J.C.; High, S.; Zimmermann, R. Chaperone-mediated Sec61 channel gating during ER import of small precursor proteins overcomes Sec61 inhibitor-reinforced energy barrier. Cell Rep. 2018, 23, 1373–1386.
- Haßdenteufel, S.; Nguyen, D.; Helms, V.; Lang, S.; Zimmermann, R. Components and mechanisms for ER import of small human presecretory proteins. FEBS Lett. 2019, 593, 2506–2524.
- Itskanov, S.; Park, E. Structure of the posttranslational Sec protein-translocation channel complex from yeast. Science 2019, 363, 84–87.
- Wu, X.; Cabanos, C.; Rapoport, T.A. Structure of the post-translational protein translocation machinery of the ER membrane. Nature 2019, 566, 136–139.
- Itskanov, S.; Kuo, K.M.; Gumbart, J.C.; Park, E. Stepwise gating of the Sec61 protein-conducting channel by Sec62 and Sec63. Nat. Struct. Mol. Biol. 2021, 28, 162–172.
- Weng, T.-H.; Steinchen, W.; Beatrix, B.; Berninghausen, O.; Becker, T.; Bange, G.; Cheng, J.; Beckmann, R. Architecture of the active post-translational SEC translocon. EMBO J. 2021, 40, e105643.
- Görlich, D.; Hartmann, E.; Prehn, S.; Rapoport, T.A. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature 1992, 357, 47–52.
- High, S.; Martoglio, B.; Görlich, D.; Andersen, S.S.L.; Ashford, A.A.; Giner, A.; Hartmann, E.; Prehn, S.; Rapoport, T.A.; Dobberstein, B.; et al. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J. Biol. Chem. 1993, 268, 26745–26751.
- Hegde, R.S.; Voigt, S.; Rapoport, T.A.; Lingappa, V.R. TRAM regulates the exposure of nascent secretory proteins to the cytosol during translocation into the endoplasmic reticulum. Cell 1998, 92, 621–631.
- Voigt, S.; Jungnickel, B.; Hartmann, E.; Rapoport, T.A. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 1996, 134, 25–35.
- Sauri, A.; McCormick, P.J.; Johnson, A.E.; Mingarro, I. Sec61alpha and TRAM are sequentially adjacent to a nascent viral membrane protein during its ER integration. J. Mol. Biol. 2007, 366, 366–374.
- Cohen, N.; Aviram, N.; Schuldiner, M. A systematic proximity ligation approach to studying protein-substrate specificity identifies the substrate spectrum of the Ssh1 translocon. EMBO J. 2023, 43, e113385.
- O’Keefe, S.; Pool, M.R.; High, S. Membrane protein biogenesis at the ER: The highways and byways. FEBS J. 2022, 289, 6835–6862.
- Hegde, R.S.; Keenan, R.J. The mechanism of integral membrane protein biogenesis. Nat. Rev. Mol. Cell Biol. 2022, 23, 107–124.
- Schrul, B.; Kopito, R.R. Peroxin-dependent targeting of a lipid-droplet-destined membrane protein to ER subdomains. Nat. Cell Biol. 2016, 18, 740–751.
- Schrul, B.; Schliebs, W. Intracellular communication between lipid droplets and peroxisomes: The Janus face of PEX19. Biol. Chem. 2018, 399, 741–749.
- Yamamoto, Y.; Sakisaka, T. The peroxisome biogenesis factors posttranslationally target reticulon homology-domain containing proteins to the endoplasmic reticulum membrane. Sci. Rep. 2018, 8, 2322.
- Aviram, N.; Ast, T.; Costa, E.A.; Arakel, E.; Chuartzman, S.G.; Jan, C.H.; Haßdenteufel, S.; Dudek, J.; Jung, M.; Schorr, S.; et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 2016, 540, 134–138.
- Hirata, T.; Yang, J.; Tomida, S.; Tokoro, Y.; Kinoshita, T.; Fujita, M.; Kizuka, Y. ER entry pathway and glycosylation of GPI-anchored proteins are determined by N-terminal signal sequence and C-terminal GPI-attachment sequence. J. Biol. Chem. 2022, 298, 102444.
- Kalies, K.-U.; Rapoport, T.A.; Hartmann, E. The beta-subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J. Cell Biol. 1998, 141, 887–894.
- Chen, X.; Van Valkenburgh, C.; Liang, H.; Fang, H.; Green, N. Signal peptidase and oligosaccharyltransferase interact in a sequential and dependent manner within the endoplasmic reticulum. J. Biol. Chem. 2001, 276, 2411–2416.
- Liaci, A.M.; Steigenberger, B.; Tamara, S.; de Souza, P.T.; Gröllers-Mulderij, M.; Ogrissek, P.; Marrink, S.-J.; Scheltema, R.A.; Förster, F. Structure of the human signal peptidase complex reveals the determinants for signal petide cleavage. Mol. Cell 2021, 81, 3934–3948.
- Braunger, K.; Pfeffer, S.; Shrimal, S.; Gilmore, R.; Berninhausen, O.; Mandon, E.C.; Becker, T.; Förster, F.; Beckmann, R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 2018, 360, 215–219.
- Wild, R.; Kowal, J.; Eyring, J.; Ngwa, E.M.; Aebi, M.; Locher, K.P. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science 2018, 359, 545–550.
- Gemmer, M.; Chaillet, M.; van Loenhout, J.; Arenas, R.C.; Vismpas, D.; Gröllers-Mulderji, M.; Kohl, F.A.; Albanese, P.; Scheltema, R.A.; Howes, S.C.; et al. Visualization of translation and protein biogenesis at the ER membrane. Nature 2023, 614, 160–167.
- Müller, G.; Zimmermann, R. Import of honeybee prepromelittin into the endoplasmic reticulum: Structural basis for independence of SRP and docking protein. EMBO J. 1987, 6, 2099–2107.
- Müller, G.; Zimmermann, R. Import of honeybee prepromelittin into the endoplasmic reticulum: Energy requirements for membrane insertion. EMBO J. 1988, 7, 639–648.
- Schlenstedt, G.; Zimmermann, R. Import of frog prepropeptide GLa into microsomes requires ATP but does not involve docking protein or ribosomes. EMBO J. 1987, 6, 699–703.
- Schlenstedt, G.; Gudmundsson, G.H.; Boman, H.G.; Zimmermann, R. A large presecretory protein translocates both cotranslationally, using signal recognition particle and ribosome, and posttranslationally, without these ribonucleoparticles, when synthesized in the presence of mammalian microsomes. J. Biol. Chem. 1990, 265, 13960–13968.
- Kutay, U.; Hartmann, E.; Rapoport, T.A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993, 3, 72–75.
- Ast, T.; Cohen, G.; Schuldiner, M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 2013, 152, 1134–1145.
- Schuldiner, M.; Metz, J.; Schmid, V.; Denic, V.; Rakwalska, M.; Schmitt, H.D.; Schwappach, B.; Weissman, J.S. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 2008, 134, 634–645.
- Rabu, C.; Schmid, V.; Schwappach, B.; High, S. Biogenesis of tail-anchored proteins: The beginning for the end? J. Cell Sci. 2009, 122, 3605–3612.
- Mariappan, M.; Li, X.; Stefanovic, S.; Sharma, A.; Mateja, A.; Keenan, R.J.; Hegde, R.S. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 2010, 466, 1120–1124.
- Leznicki, P.; Clancy, A.; Schwappach, B.; High, S. Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 2010, 123, 2170–2178.
- Borgese, N.; Fasana, E. Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta 2011, 1808, 937–946.
- Vilardi, F.; Lorenz, H.; Dobberstein, B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J. Cell Sci. 2011, 124, 1301–1307.
- Leznicki, P.; Warwicker, J.; High, S. A biochemical analysis of the constraints of tail-anchored protein biogenesis. Biochem. J. 2011, 436, 719–727.
- Yamamoto, Y.; Sakisaka, T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol. Cell 2012, 48, 387–397.
- Vilardi, F.; Stephan, M.; Clancy, A.; Janshoff, A.; Schwappach, B. WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS ONE 2014, 9, e85033.
- Wang, F.; Chan, C.; Weir, N.R.; Denic, V. The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature 2014, 512, 441–444.
- Borgese, N.; Coy-Vergara, J.; Colombo, S.F.; Schwappach, B. The ways of tails: The GET pathway and more. Proteins 2019, 38, 289–305.
- Leznicki, P.; High, S. SGTA associates with nascent membrane protein precursors. EMBO Rep. 2020, 21, e48835.
- Leznicki, P.; Schneider, H.O.; Harvey, J.V.; Shi, W.Q.; High, S. Co-translational biogenesis of lipid droplet integral membrane proteins. J. Cell Sci. 2021, 132, jcs.259220.
- Johnson, N.; Vilardi, F.; Lang, S.; Leznicki, P.; Zimmermann, R.; High, S. TRC-40 can deliver short secretory proteins to the Sec61 translocon. J. Cell Sci. 2012, 125, 3612–3620.
- Casson, J.; McKenna, M.; Haßdenteufel, S.; Aviram, N.; Zimmermann, R.; High, S. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 2017, 130, 3851–3861.
- Haßdenteufel, S.; Sicking, M.; Schorr, S.; Aviram, N.; Fecher-Trost, C.; Schuldiner, M.; Jung, M.; Zimmermann, R.; Lang, S. hSnd2 protein represents an alternative targeting factor to the endoplasmic reticulum in human cells. FEBS Lett. 2017, 591, 3211–3224.
- Talbot, B.E.; Vandorpe, D.H.; Stotter, B.R.; Alper, S.L.; Schlondorff, J. Transmembrane insertases and N-glycosylation crtically determine synthesis, trafficking, and activity of the nonselective cation channel TRPC6. J. Biol. Chem. 2019, 294, 12655–12669.
- Yang, J.; Hirata, T.; Liu, Y.-S.; Guo, X.-Y.; Gao, X.-D.; Kinoshita, T.; Fujita, M. Human SND2 mediates ER targeting of GPI-anchored proteins with low hydrophobic GPI attachment signals. FEBS Lett. 2021, 595, 1542–1558.
- Tirincsi, A.; O’Keefe, S.; Nguyen, D.; Sicking, M.; Dudek, J.; Förster, F.; Jung, M.; Hadzibeganovic, D.; Helms, V.; High, S.; et al. Proteomics identifies substrates and a novel component in hSnd2-dependent ER protein targeting. Cells 2022, 11, 2925.
- Seiser, R.M.; Nicchitta, C.V. The fate of membrane-bound ribosomes following the termination of protein synthesis. J. Biol. Chem. 2000, 275, 33820–33827.
- Potter, M.D.; Seiser, R.M.; Nicchitta, C.V. Ribosome exchange revisited: A mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 2001, 11, 112–115.
- Berkovits, B.D.; Mayr, C. Alternative 3′UTRs act as scaffolds to regulate membrane protein localization. Nature 2015, 522, 363–367.
- Ma, W.; Mayr, C. A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein-protein interactions. Cell 2018, 175, 1492–1506.
- Hsu, J.C.-C.; Reid, D.W.; Hoffman, A.M.; Sarkar, D.; Nicchitta, C.V. Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA 2018, 24, 688–703.
- Bhadra, P.; Schorr, S.; Lerner, M.; Nguyen, D.; Dudek, J.; Förster, F.; Helms, V.; Lang, S.; Zimmermann, R. Quantitative proteomics and differential protein abundance analysis after depletion of putative mRNA receptors in the ER membrane of human cells identifies novel aspects of mRNA targeting to the ER. Molecules 2021, 26, 3591.
- Savitz, A.J.; Meyer, D.I. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature 1990, 346, 540–544.
- Wiedmann, B.; Saki, H.; Davis, T.A.; Wiedmann, M. A protein complex required for signal-sequence-specific sorting and translocation. Nature 1994, 370, 434–440.
- Gamerdinger, M.; Kobayashi, K.; Wallisch, A.; Kreft, S.G.; Sailer, C.; Schlömer, R.; Sachs, N.; Jomaa, A.; Stengel, F.; Ban, N.; et al. Early scanning of nascent polypeptides inside the ribosomal tunnel by NAC. Mol. Cell 2019, 75, 996–1006.
- Moeller, I.; Jung, M.; Beatrix, B.; Levy, R.; Kreibich, G.; Zimmermann, R.; Wiedmann, M.; Lauring, B. A general mechanism for regulation of access to the translocon: Competition for a membrane attachment site on ribosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 13425–13430.
- Goder, V.; Spiess, M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 2003, 22, 3645–3653.
- Goder, V.; Junne, T.; Spiess, M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol. Biol. Cell 2004, 15, 1470–1478.
- Baker, J.A.; Wong, W.-C.; Eisenhaber, B.; Warwicker, J.; Eisenhaber, F. Charged residues next to transmembrane regions revisited: “Positive-inside rule” is complemented by the “negative inside depletion/outside enrichment rule”. BMC Biol. 2017, 15, 66.
- Devaraneni, P.K.; Conti, B.; Matsumara, Y.; Yang, Z.; Johnson, A.E.; Skach, W.R. Stepwise insertion and inversion of a type II signal anchor sequence in the ribosome-Sec61 translocon complex. Cell 2011, 146, 134–147.
- Meacock, S.L.; Lecomte, F.J.L.; Crawshaw, S.G.; High, S. Different transmembrane domains associate with distinct endoplasmic reticulum components during membrane integration of a polytopic protein. Mol. Biol. Cell 2002, 13, 4114–4129.
- Ismail, N.; Crawshaw, S.G.; High, S. Active and passive displacement of transmembrane domains both occur during opsin biogenesis at the Sec61 translocon. J. Cell Sci. 2006, 119, 2826–2836.
- Anghel, S.A.; McGilvray, P.T.; Hegde, R.S.; Keenan, R.J. Identification of Oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Rep. 2017, 21, 3708–3716.
- Chitwood, P.J.; Juszkiewicz, S.; Guna, A.; Shao, S.; Hegde, R.S. EMC is required to initiate accurate membrane protein topogenesis. Cell 2018, 175, 1507–1519.
- Shurtleff, M.J.; Itzhak, D.N.; Hussmann, J.A.; Schirle Oakdale, N.T.; Costa, E.A.; Jonikas, M.; Weibezahn, J.; Popova, K.D.; Jan, C.H.; Sinitcyn, P.; et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. eLife 2018, 7, e37018.
- McGilvray, P.T.; Anghel, S.A.; Sundaram, A.; Zhong, F.; Trnka, M.J.; Fuller, J.R.; Hu, H.; Burlingame, A.L.; Keenan, R.J. An ER translocon for multi-pass mambrane protein biogenesis. eLife 2020, 9, e56889.
- O’Donnel, J.P.; Philips, B.P.; Yagita, Y.; Juszkiewicz, S.; Wagner, A.; Malinverni, D.; Keenan, R.J.; Mille, E.A.; Hegde, R.S. The architecture of EMC reveals a path for membrane protein nsertion. eLife 2020, 9, e57887.
- Pleiner, T.; Tomaleri, G.P.; Januszyk, K.; Inglis, A.J.; Hazu, M.; Voorhees, R.M. Structural basis for membrane insertion by the human ER membrane protein complex. Science 2020, 369, 433–436.
- Bai, L.; You, Q.; Feng, X.; Kovach, A.; Li, H. Structure of the ER membrane complex, a transmembrane insertase. Nature 2020, 584, 475–478.
- Wu, H.; Hegde, R.S. Mechanism of signal-anchor triage during early steps of membrane protein insertion. Cell 2023, 83, 961–973.
- Sundaram, A.; Yamsek, M.; Zhong, F.; Hooda, Y.; Hegde, R.S.; Keenan, R.J. Substrate-driven assembly of a translocon for multipass membrane proteins. Nature 2022, 611, 167–172.
- Samlinskaite, L.; Kim, M.K.; Lewis, A.J.O.; Keenan, R.J.; Hegde, R.S. Mechanism of an intramembrane chaperone for multipass membrane proteins. Nature 2022, 611, 161–166.
- Reid, D.W.; Nicchitta, C.V. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J. Biol. Chem. 2012, 287, 5518–5527.
- Hannigan, M.M.; Hoffman, A.M.; Thompson, J.W.; Zheng, T.; Nicchitta, C.V. Quantitative proteomics links the LRRC59 interactome to mRNA translation on the ER membrane. Mol. Cell. Proteom. 2020, 19, 1826–1849.
- Nguyen, D.; Stutz, R.; Schorr, S.; Lang, S.; Pfeffer, S.; Freeze, H.F.; Förster, F.; Helms, V.; Dudek, J.; Zimmermann, R. Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat. Commun. 2018, 9, 37639.
- Schorr, S.; Nguyen, D.; Haßdenteufel, S.; Nagaraj, N.; Cavalié, A.; Greiner, M.; Weissgerber, P.; Loi, M.; Paton, A.W.; Paton, J.C.; et al. Proteomics identifies signal peptide features determining the substrate specificity in human Sec62/Sec63-dependent ER protein import. FEBS J. 2020, 287, 4612–4640.
- Klein, M.-C.; Lerner, M.; Nguyen, D.; Pfeffer, S.; Dudek, J.; Förster, F.; Helms, V.; Lang, S.; Zimmermann, R. TRAM1 protein may support ER protein import by modulating the phospholipid bilayer near the lateral gate of the Sec61 channel. Channels 2020, 14, 28–44.
- Tian, S.; Wu, Q.; Zhou, B.; Choi, M.Y.; Ding, B.; Yang, W.; Dong, M. Proteomic analysis indentifies membrane proteins dependent on the ER membrane protein complex. Cell Rep. 2019, 28, 2517–2526.
- Zimmermann, R.; Lang, S.; Lerner, M.; Förster, F.; Nguyen, D.; Helms, V.; Schrul, B. Quantitative proteomics and differential protein abundance ananalysis after depletion of PEX3 from human cells identifies additional aspects of protein targeting tot he ER. Int. J. Mol. Sci. 2021, 22, 13028.
- Zimmermann, R. Rules of engagement for components of membrane protein biogenesis at the human ER. Int. J. Mol. Sci. 2025, 26, 8823.
- Gamerdinger, M.; Hanebuth, M.A.; Frickey, T.; Deuerling, E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 2015, 348, 201-207.
- Hsieh, H.-H.; Lee, J.H.; Chandrasekar, S.; Shan, S-ou. A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun. 2020, 11, 5840.
- Jomaa, A.; Gamerdinger, M.; Hsieh, H.-H.; Wallisch, A.; Chandrasekaran, V.; Ulusoy, Z.; Scaiola, A.; Hegde, R.S.; Shan, S.-ou; Ban, N.; Deuerling, E. Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting. Science 2022, 375, 839-844.




