Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Vic Norris | -- | 3316 | 2023-09-25 12:03:43 | | | |
| 2 | Rita Xu | Meta information modification | 3316 | 2023-09-26 03:30:29 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kohiyama, M.; Herrick, J.; Norris, V. DnaA and Hyperstructure Dynamics in the Cell Cycle. Encyclopedia. Available online: https://encyclopedia.pub/entry/49596 (accessed on 07 February 2026).
Kohiyama M, Herrick J, Norris V. DnaA and Hyperstructure Dynamics in the Cell Cycle. Encyclopedia. Available at: https://encyclopedia.pub/entry/49596. Accessed February 07, 2026.
Kohiyama, Masamichi, John Herrick, Vic Norris. "DnaA and Hyperstructure Dynamics in the Cell Cycle" Encyclopedia, https://encyclopedia.pub/entry/49596 (accessed February 07, 2026).
Kohiyama, M., Herrick, J., & Norris, V. (2023, September 25). DnaA and Hyperstructure Dynamics in the Cell Cycle. In Encyclopedia. https://encyclopedia.pub/entry/49596
Kohiyama, Masamichi, et al. "DnaA and Hyperstructure Dynamics in the Cell Cycle." Encyclopedia. Web. 25 September, 2023.
Copy Citation
The DnaA protein has long been considered to play the key role in the initiation of chromosome replication in modern bacteria.
Charles E. Helmstetter Prize
E. coli
ribonucleotide reductase
1. Introduction
The coordination of cell growth and chromosome replication is achieved by mechanisms that are still being uncovered. One approach to investigating this coordination is genetics and, over the last half century, this has led to the isolation of conditional lethal mutants of cell division or DNA synthesis. As part of these investigations, in early 1960, Kohiyama started to isolate mutants of Escherichia coli K12 that fail to grow at a high temperature. With his collaborators, he found that nearly 1% of colonies obtained at 30 °C from a mutagenized culture failed to grow at 42 °C but, on examining each clone for DNA or protein syntheses and morphological changes after transfer to 42 °C, he found only a few mutants affected in DNA synthesis, with the majority being those defective in protein synthesis, such as valyl-sRNA synthetase [1], or in cell division, without identification of the mutated genes [2]. Isolation of temperature-sensitive (ts) mutants continued at the Pasteur Institute and the resulting strain collection has been beneficial to studies on the cell cycle such as the discovery of the FtsZ ring, which is essential for division [3], and to studies on metabolism such as those on the ribonucleotide reductase [4].
The first priority was the elucidation of the regulatory mechanism of chromosome replication as hypothesized in the Replicon Theory [5], according to which DNA replication starts from the genetically defined point (oriC) by the action of an initiator. Kohiyama, therefore, sought mutants that failed to initiate replication at high temperatures and found two [6]. These mutations were mapped to the same locus and the gene was called dnaA [7].
Further characterization of these dnaA mutants demonstrated a close connection between initiation of replication and cell cycle control: at a non-permissive temperature, a dnaA mutant temporarily stops dividing and forms filamentous cells [2]; division later resumes towards one end of the filament to produce normal-sized cells that lack DNA [8]. The fact that the size of these anucleate cells is relatively constant (but see [9]) is consistent with the idea that DnaA is involved, directly or indirectly, in the positioning of the division site.
In fact, the possibility that DnaA protein acts as a regulator of gene expression was raised by Hansen a few years after the isolation of the first mutant [10], and DnaA was subsequently shown to regulate many operons [11]. These observations, therefore, help make DnaA a candidate for the role of coordinator of the cell cycle.
To explore this proposal, it is essential to characterise the biochemical properties of the DnaA protein. A large part of this was done by Kornberg and his collaborators using genetic engineering only 20 years after the first isolation of a dnaA mutant [12]. They found that the DnaA protein is an ATPase possessing a high affinity for the replication origin (oriC) via DnaA boxes constituted of nine bases. The consequence of this interaction is the opening of oriC, which allows the insertion of DNA helicase into oriC in order to start DNA synthesis after the loading of DNA polymerase III. This interaction between DnaA and DnaA boxes seems to be important in most of the processes in which DnaA is involved [13][14]. DnaA has four domains [15][16] (Figure 1). Domain I binds both the helicase, DnaB, and the DiaA protein that may help link DnaA dimers and monomers; it also has a site for a low-affinity domain I–domain I interaction to generate dimers. Domain II is a linker. Domain III has an AAA+ motif that binds ATP or ADP; this binding of ATP leads to head-to-tail homo-oligomers in a helix; it also binds ssDNA and has a region that interacts with membrane. Domain IV is the dsDNA-binding domain that recognizes DnaA boxes (for references, see [11]).
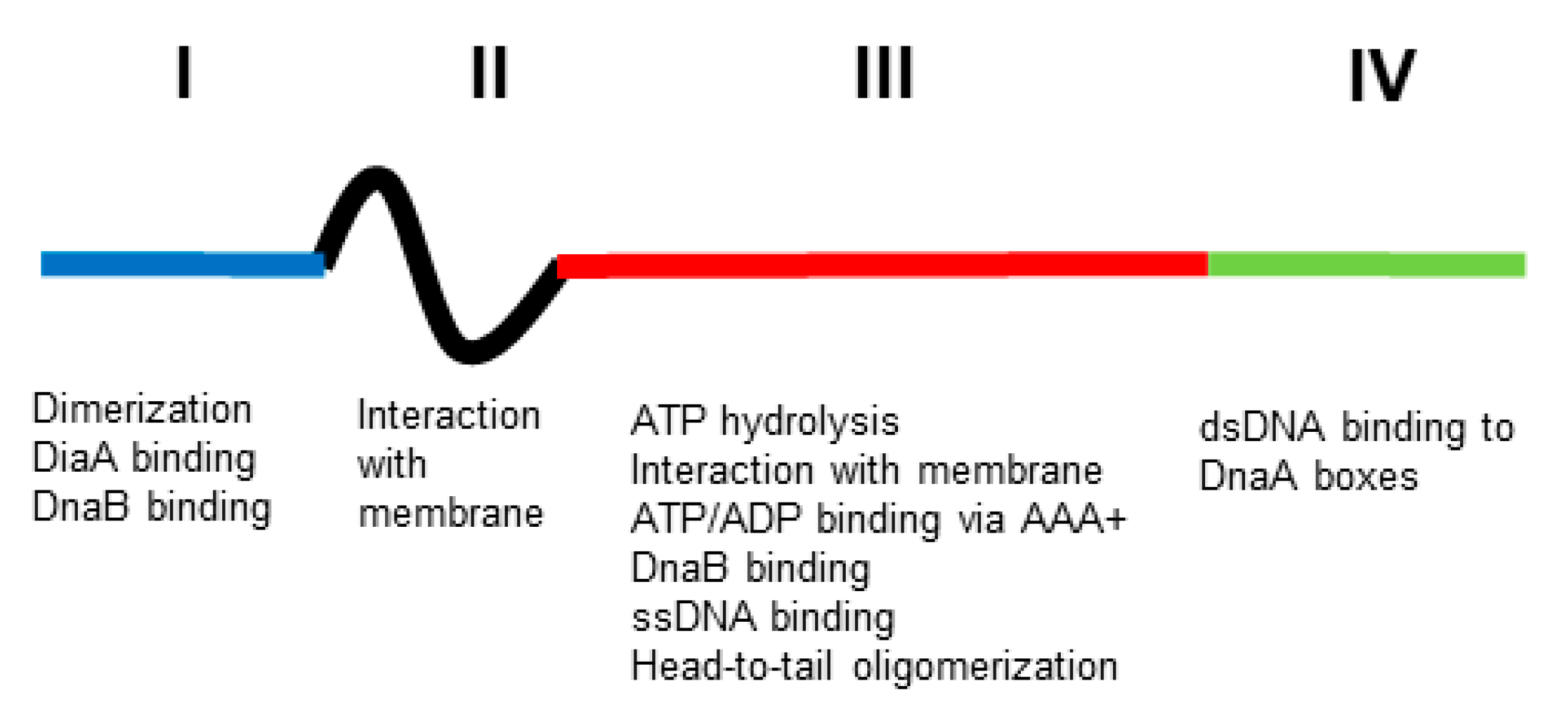
Figure 1. Functional structure of DnaA. The four domains (I, II, III and IV, blue, black, red and green, respectively) have different functions (see text for explanation). Not to scale.
Hyperstructures are large assemblies of molecules and macromolecules that have particular functions; they constitute a level of organization intermediate between the macromolecule and the cell.
2. Is There a Chromosomal DnaA Hyperstructure?
The chromosomal datA site is a 1 kb region that contains binding sites for DnaA and that helps in its inactivation [17]. An excess of datA sites results in a delay to the initiation of replication, whereas the lack of datA results in extra initiations, and it was originally suggested that the binding of DnaA to newly duplicated datA during oriC sequestration could help prevent premature reinitiation when oriC is desequestered [17]. When datA is negatively supercoiled, DnaA-ATP oligomers are stabilised, and datA-IHF interactions and DnaA-ATP hydrolysis are promoted [18]. These results are consistent with a datA-based chromosomal hyperstructure helping regulate initiation. The two chromosomal intergenic regions, DnaA-reactivating sequence 1 (DARS1) and DnaA-reactivating sequence 2 (DARS2), each contain a cluster of DnaA binding sites; these sites promote regeneration of DnaA-ATP from DnaA-ADP by nucleotide exchange, and thereby help to promote the initiation of replication [19]. (Note that DnaA-ADP is mainly monomeric and unable to go from high-affinity binding sites to nucleate polymerisation at the low-affinity binding sites in the origin of replication.) The reactivation of DnaA by DARS2 is coordinated by the site-specific binding to DARS2 of IHF and Fis; this binding of IHF is temporally regulated during the cell cycle [20], as is the binding of Fis, which occurs specifically prior to initiation [21]. It is thought that DARS1 is mainly involved in maintaining the origin concentration, whereas DARS2 is also involved in maintaining single cell synchrony [22].
After initiation, the ATPase activity of DnaA is stimulated by the regulatory inactivation of the DnaA (RIDA) complex composed of the Hda protein interacting with the DNA-loaded β-clamp [23]. Recently, it has been found that only the disruption of RIDA has a major effect on initiation (since DARS and datA can compensate for one another) [24]. All of this raises the question of whether the inactivation of DnaA takes place within a chromosomal DnaA hyperstructure.
DnaA is also a sequence-specific transcriptional regulator. Such regulators bind to sites that are distributed on the chromosome with a periodicity consistent with a solenoidal-type organization [25]; this organization would bring together a regulator and its sites into a hyperstructure. If this hyperstructure does indeed exist, what is its relationship with the datA and the DARS sites? Do the above constitute separate hyperstructures and, if so, how do they interact? Or are they all part of an initiation hyperstructure?
3. Is There a Membrane DnaA Hyperstructure?
The involvement of the membrane in the initiation of replication has long been known [26][27][28], and it is tempting to speculate that the initiation hyperstructure may also contain acidic phospholipids such as cardiolipin, and even that the hyperstructure is physically associated with the bilayer itself, possibly in a fluid state. Indeed, a significant proportion of the DnaA (10% or more) in the cell is associated with the membrane [29][30]. In this association, both domain II and the hydrophobic domain III of DnaA are important [31]. Mutations and deletions affecting the membrane interaction sequence in domain III of DnaA restored growth to cells with a lowered content of acidic phospholipids [32]. DnaA association with the membrane may dissociate ADP from DnaA depending on the degree of protein crowding on the membrane [33][34]. Insertion into or association with the membrane is fundamental to transertion (the coupled transcription, translation, and insertion of proteins into a membrane) and to the existence of transertion hyperstructures [35][36][37]; this raises the question of whether a DnaA hyperstructure based on transertion might exist for part of the cell cycle and, moreover, whether such transertion might be important for the existence and/or operation of a chromosomal DnaA or other hyperstructures.
4. Does the Initiation Hyperstructure Contain Glycolytic Enzymes?
Metabolism is coupled to DNA synthesis through nutrient richness and growth rate in a variety of ways. One way this occurs is via (p)ppGpp [38], with the transcription of dnaA in E. coli [39] and the level of DnaA protein in C. crescentus [40] being lowered by (p)ppGpp. Another way is via a central carbon metabolism that, in E. coli, can: (1) promote DnaA to its active DnaA-ATP form and its binding to oriC by cAMP (a regulator of this part of metabolism) [41]; (2) suppress the defects of the dnaA46 mutant by changes in pyruvate and acetate metabolism [42]; (3) inhibit DnaA conversion to DnaA-ATP and its binding to oriC (via acetylation of DnaA, with acetyl-CoA and acetyl-phosphate as donors) (for references see [43]). Evidence for the involvement of the central carbon metabolism in DNA replication in Bacillus subtilis includes: (1) subunits of pyruvate dehydrogenase (PdhC) and related enzymes bind the origin of replication region, DnaC and DnaG inhibit the initiation of replication; (2) mutations in the genes of central carbon metabolism suppress initiation and elongation defects in dnaC, dnaG, and dnaE mutants; (3) mutations in gapA (which encodes glyceraldehyde 3-phosphate dehydrogenase) perturb the metabolic control of replication; (4) pyruvate kinase (PykA) can both inhibit initiation and stimulate elongation via proposed interactions with DnaC, DnaG, and DnaE as modulated, perhaps, by phosphorylation [43][44].
Given that metabolic enzymes can exist as hyperstructures in their own right, a question that arises here is whether metabolic enzymes are also part of an initiation hyperstructure. And a related question is whether the metabolites themselves are directly part of initiation and/or replication hyperstructures, not just by binding to the constituent proteins (as in the case of DnaA and cAMP [41]) or by being used to modify the protein (as in the case of DnaA and acetyl-CoA), but also by binding directly to RNA or DNA. If metabolites were indeed to bind the origin region, this would make the connection with the Ring World, an origins-of-life scenario in which small, double-stranded DNA rings were selected firstly because they catalysed the reactions of central carbon metabolism [45].
5. Does the DnaA-Initiation Hyperstructure Contain SeqA?
E. coli avoids multiple reinitiations of chromosome replication by a sequestration mechanism that depends on the SeqA protein binding preferentially to newly replicated, hemi-methylated GATC sites, many of which are clustered in oriC. This sequestration, which involves the membrane, occurs only when oriC is hemi-methylated [46] and when SeqA, which has an affinity for the membrane, is present [47][48]; the result is an inhibition of initiation [49]. SeqA forms multimers and SeqA-DNA complexes can cover 100 kb of DNA and are close or integral to the replication hyperstructure(s), and have a bidirectional movement that differs from that of the origins (which goes to the poles) [50][51]. GATC sites are clustered not only in the oriC region but also in many genes involved in the replication and repair of DNA (such as dnaA, dnaC, dnaE, gyrA, topA, hepA, lhr, parE, mukB, recB, recD, and uvrA), as well as genes involved in the synthesis of the precursors of DNA (such as nrdA, purA, purF, purL, pyrD and pyrI), consistent, perhaps, with the presence of these genes and their products in a replication hyperstructure [52]. One related question is whether the DnaA-initiation hyperstructure in its earliest form contains SeqA and, reciprocally, a second question is whether the replication hyperstructure contains DnaA?
6. What Is the Relationship between Strand Opening and DnaA Binding?
Kornberg’s group first showed that DnaA opens oriC in vitro depending on the presence of ATP [53]. The opening of oriC was detected by P1 nuclease sensitivity in this case, and repeatedly shown by other techniques such as KMnO4 modification [54] and DMS footprinting [55]. oriC contains (1) the DnaA-ATP-Oligomerization Region (DOR), with twelve DnaA boxes, and (2) the neighbouring Duplex Unwinding Element (DUE), which contains three AT-rich 13-mer repeats along with DnaA binding motifs. DnaA-ATP assembles into a pentamer via its binding to DnaA boxes in one half of the DOR and, progressively, via its binding to the single-stranded motifs in the DUE, which stabilizes the unwound DUE. This unwinding is promoted by the nucleoid-associated protein (NAP), IHF, or indeed by HU [14]. Katayama’s group analysed, also by P1 nuclease sensitivity, the opening of M13 oriC DNA by DnaA in vitro at the DUE using various types of mutated oriC and IHF; the results obtained were consistent with those obtained in vivo [56]. That said, it is difficult to follow the kinetics of oriC opening with these techniques.
Strick’s group performed a single molecule analysis on DnaA–oriC(2kb) interaction using an optical magnetic tweezer to follow the rapid kinetics of double-stranded DNA opening. They observed formation of stable complexes between supposedly DnaA-ATP oligomers and oriC with different degrees of positive supercoiling. The formation of these complexes occurred using an oriC that lacked the DUE, raising the question of whether they were studying a non-canonical reaction. Other questions include why the kinetics of the complex formation was not studied, why the formation of the complex did not occur constantly [57], and whether DnaA was actually present in the complex. It should be pointed out that the use of optical magnetic tweezers is technically demanding: it requires the attachment of oriC DNA to a magnetic bead followed by the selection of intact oriC-containing beads (which are easily damaged and consequently in a minority).
Techniques based on minicircles of DNA facilitate the detection of fine-scale modifications to the DNA structure. Using an oriC minicircle of 641 bp with three negative supercoils, Landoulsi and Kohiyama found that around 80% of this substrate was positively twisted three times during incubation with DnaA and that the efficiency of unwinding was affected by the degree of negative superhelicity of the minicircle (three negative turns proved more effective in causing unwinding than two or four negative turns). Unwinding of this oriC minicircle by DnaA was verified by Bal31 sensitivity (rather than by P1 nuclease sensitivity), whilst the presence of DnaA on the unwound minicircle was confirmed by an anti-DnaA antiserum. The problem raised by this work is that the unwinding did not require ATP [58]. It should also be noted that the above work on oriC minicircles depends on a sophisticated technique that requires the formation of circles from a linear 641 bp oriC fragment that can only be achieved in a glass capillary after overnight incubation in the presence of DNA ligase and ethidium bromide, which introduces superhelicity; modification of the superhelicity of minicircles resulting from DnaA action is scored after Topo I treatment and is not directly measured.
This work raised the question of whether or not the ATP-dependent opening of oriC by DnaA (as demonstrated by P1 nuclease sensitivity) is the unique pathway for the initiation of replication. The fact that the mutant isolated first, dnaA46, which has lost the ATP binding site, can grow normally at a low temperature indicates the existence of an alternative pathway whereby oriC can fire without ATP. Consistent with this, the growth of dnaA46 is more sensitive than the wild type to gyrase inhibitors [59], whilst the opening of oriC minicircles by DnaA is sensitive to negative supercoiling densities [58] (see above). Another explanation, offered by Kaguni’s group, is that the DnaA46 protein, with the aid of DnaK, can form a structure similar to that of DnaA-ATP [60]. Although no data are presently available from X-ray crystallography of the whole molecule of DnaA or from cryoEM analysis of DnaA-oriC, significant advances have been made by the Berger group using DnaA from Aquifex aeolicus along with the nonhydrolyzable ATP analog AMP-PCP [15], and by Katayama and collaborators using a combination of biochemistry and computer simulations to model the central part of the oriC-DnaA-IHF complex [16].
7. Does DnaA Participate in Differentiation?
In the strand segregation hypothesis, a coherent phenotypic diversity is generated by the segregation of certain hyperstructures with only one of the parental DNA strands [61]; candidate hyperstructures for such asymmetric segregation include those containing the NAPs and the topoisomerases. An asymmetric segregation of a chromosomal DnaA hyperstructure is another seductive possibility: could DnaA play a particular role in generating phenotypic diversity (e.g., in preparing a population to confront stresses via its role in modulating gene expression) or in connecting different phenotypes with different patterns of the cell cycle—or indeed both?
8. What Modifications Does DnaA Undergo and What Are Their Roles?
It has been proposed that a hyperstructure might be assembled if enzymes (such as protein kinases and acetyltransferases) and their NAP substrates were to associate with one another in a positive feedback loop in which, for example, the modification of an NAP by its cognate enzyme increases the probability of colocation of both the NAPs and the enzyme [61]. In line with this, the acetylation of a lysine residue (K178) prevents DnaA from binding to ATP and inhibits initiation, whilst the acetylation of another lysine residue (K243) also inhibits initiation but does not affect the ATP/ADP binding affinity of DnaA or the ability of DnaA to bind to the dnaA promoter region and to DARS1 [62].
DnaA binds cAMP with a Kd of a similar order to that with which it binds to ATP; indeed, the affinity of DnaA for cAMP is such that most of the cell’s DnaA should be bound to cAMP when the latter is present at the physiological concentration of 1 µM [41]. cAMP bound to DnaA is chased by ATP but not by ADP (note that there is only one cAMP binding site on the protein [41]). In vitro, cAMP stimulates DnaA binding to oriC and to DnaA sites elsewhere in the chromosome [41]; in vivo, the addition of cAMP to a cya mutant (which encodes the adenylate cyclase that catalyses the production of cAMP) increased the level of DnaA [63]. Significantly, despite DnaA’s stability in vivo, it has recently been shown to be degraded in vivo in ATP depletion conditions [64], and one possibility is that cAMP helps to both protect DnaA from degradation and regenerate DnaA-ATP from DnaA-ADP (by causing the release of the bound ADP). This raises the question of whether the state of the environment as reflected in a cAMP signal is transduced by the level of DnaA and by the DnaA-ATP: DnaA-ADP ratio into the expression of DnaA-regulated genes and cell cycle timing.
In the case of Caulobacter crescentus, the phosphorylation status of CtrA is central to cell cycle progress [65]. The many possible post-translational modifications to DnaA and to other proteins in the initiation and replication hyperstructures, therefore, include phosphorylation, and several other modifications, such as succinylation, methylation, proprionylation, malonylation, deamidation of asparagines, and glycosylation (for references see [61]). Another post-translational modification—and one that is largely ignored—is the covalent addition of poly-(R)-3-hydroxybutyrate (PHB) to proteins [66]; one proposed function of such addition to NAPs would be to regulate their interaction with nucleic acids [67]. An important question is, therefore, whether DnaA undergoes modifications like the addition of PHB and, if so, does such modification help the type of hyperstructure into which DnaA assembles?
9. Is DnaA a Controller of Chromosomal Copy Numbers Rather Than a Timer?
Fralick found that the timing of initiation and the number of replicating chromosomes per cell (and the DNA/mass ratio) could be varied independently of one another in a temperature-sensitive dnaA(ts) mutant grown at different temperatures. These results were interpreted as DnaA being an essential component of the “replication apparatus” but not itself being the signal that triggers initiation [68][69]. This interpretation would be consistent with the finding that the time of initiation is not advanced by a 50% increase in the concentration of DnaA-ATP, with the authors concluding that although DnaA protein is required for initiation of synchronous and well-timed replication cycles, the accumulation of DnaA-ATP does not control the time of initiation [70]. It should be noted that stopping the transcription of dnaA only led to a small increase in cell size, as DnaA was diluted by growth, whilst only disrupting RIDA had a major effect on initiation [24]. Finally, a mathematical model has recently been proposed that combines the titration- and activation-of-DnaA strategies to explain how initiation might be timed at fast and slow growth rates and to give both a precise volume per origin and a constant volume between initiations [71]. Finally, Liu and his group have shown how easily arbitrary choices of parameter settings and insufficient controls can affect evaluation of the constant-initiation-mass hypothesis [72].
References
- Yaniv, M.; Kohiyama, M.; Jacob, F.; Gros, F. On the properties of Valyl-sRNA synthetase in various thermosensitive Escherichia coli mutants. C. R. Acad. Hebd. Seances Acad. Sci. D 1965, 260, 6734–6737.
- Kohiyama, M.; Cousin, D.; Ryter, A.; Jacob, F. Thermosensitive mutants of Escherichia coli K12. I. Isolation and rapid characterization. Ann. L’institut Pasteur 1966, 110, 465–486.
- Bi, E.F.; Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 1991, 354, 161–164.
- Holmgren, A.; Reichard, P.; Thelander, L. Enzymatic synthesis of deoxyribonucleotides, 8. The effects of ATP and dATP in the CDP reductase system from E. coli. Proc. Natl. Acad. Sci. USA 1965, 54, 830–836.
- Jacob, F.; Brenner, S. On the regulation of DNA synthesis in bacteria: The hypothesis of the replicon. Comptes. Rendus Hebd. Seances L’academie Sci. 1963, 256, 298–300.
- Kohiyama, M.; Lanfrom, H.; Brenner, S.; Jacob, F. Modifications of indispensable functions in thermosensitive Escherichia coli mutants. On a mutation preventing replication of the bacterial chromosome. Comptes. Rendus Hebd. Seances L’academie Sci. 1963, 257, 1979–1981.
- Hirota, Y.; Ryter, A.; Jacob, F. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harb. Symp. Quant. Biol. 1968, 33, 677–693.
- Hirota, Y.; Jacob, F. Production of bacteria without DNA. C. R. Acad. Hebd. Seances Acad. Sci. D 1966, 263, 1619–1621.
- Mulder, E.; Woldringh, C.L. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J. Bacteriol. 1989, 171, 4303–4314.
- Hansen, F.G.; Rasmussen, K.V. Regulation of the DnaA product in Escherichia coli. Mol. Gen. Genet. 1977, 155, 219–225.
- Hansen, F.G.; Atlung, T. The DnaA tale. Front. Microbiol. 2018, 9, 319.
- Fuller, R.S.; Kornberg, A. Purified DnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc. Natl. Acad. Sci. USA 1983, 80, 5817–5821.
- Grimwade, J.E.; Leonard, A.C. Blocking, bending, and binding: Regulation of initiation of chromosome replication during the Escherichia coli cell cycle by transcriptional modulators that interact with origin DNA. Front. Microbiol. 2021, 12, 732270.
- Yoshida, R.; Ozaki, S.; Kawakami, H.; Katayama, T. Single-stranded DNA recruitment mechanism in replication origin unwinding by DnaA initiator protein and HU, an evolutionary ubiquitous nucleoid protein. Nucleic Acids Res. 2023, 51, 6286–6306.
- Erzberger, J.P.; Mott, M.L.; Berger, J.M. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006, 13, 676–683.
- Shimizu, M.; Noguchi, Y.; Sakiyama, Y.; Kawakami, H.; Katayama, T.; Takada, S. Near-atomic structural model for bacterial DNA replication initiation complex and its functional insights. Proc. Natl. Acad. Sci. USA 2016, 113, E8021–E8030.
- Kitagawa, R.; Ozaki, T.; Moriya, S.; Ogawa, T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998, 12, 3032–3043.
- Kasho, K.; Tanaka, H.; Sakai, R.; Katayama, T. Cooperative DnaA binding to the negatively supercoiled datA locus stimulates DnaA -ATP hydrolysis. J. Biol. Chem. 2017, 292, 1251–1266.
- Fujimitsu, K.; Senriuchi, T.; Katayama, T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP- DnaA. Genes Dev. 2009, 23, 1221–1233.
- Kasho, K.; Fujimitsu, K.; Matoba, T.; Oshima, T.; Katayama, T. Timely binding of IHF and Fis to dars2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 2014, 42, 13134–13149.
- Miyoshi, K.; Tatsumoto, Y.; Ozaki, S.; Katayama, T. Negative feedback for dars2-Fis complex by ATP-DnaA supports the cell cycle-coordinated regulation for chromosome replication. Nucleic Acids Res. 2021, 49, 12820–12835.
- Frimodt-Moller, J.; Charbon, G.; Krogfelt, K.A.; Lobner-Olesen, A. DNA replication control is linked to genomic positioning of control regions in Escherichia coli. PLoS Genet. 2016, 12, e1006286.
- Kato, J.; Katayama, T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001, 20, 4253–4262.
- Knoppel, A.; Brostrom, O.; Gras, K.; Elf, J.; Fange, D. Regulatory elements coordinating initiation of chromosome replication to the Escherichia coli cell cycle. Proc. Natl. Acad. Sci. USA 2023, 120, e2213795120.
- Kepes, F. Periodic transcriptional organization of the E. coli genome. J. Mol. Biol. 2004, 340, 957–964.
- Yung, B.Y.; Kornberg, A. Membrane attachment activates DnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 85, 7202–7205.
- Castuma, C.E.; Crooke, E.; Kornberg, A. Fluid membranes with acidic domains activate DnaA, the initiator protein of replication in Escherichia coli. J. Biol. Chem. 1993, 268, 24665–24668.
- Xia, W.; Dowhan, W. In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 783–787.
- Newman, G.; Crooke, E. DnaA, the initiator of Escherichia coli chromosomal replication, is located at the cell membrane. J. Bacteriol. 2000, 182, 2604–2610.
- Regev, T.; Myers, N.; Zarivach, R.; Fishov, I. Association of the chromosome replication initiator DnaA with the Escherichia coli inner membrane in vivo: Quantity and mode of binding. PLoS ONE 2012, 7, e36441.
- Hou, Y.; Kumar, P.; Aggarwal, M.; Sarkari, F.; Wolcott, K.M.; Chattoraj, D.K.; Crooke, E.; Saxena, R. The linker domain of the initiator DnaA contributes to its ATP binding and membrane association in E. coli chromosomal replication. Sci. Adv. 2022, 8, eabq6657.
- Zheng, W.; Li, Z.; Skarstad, K.; Crooke, E. Mutations in DnaA protein suppress the growth arrest of acidic phospholipid-deficient Escherichia coli cells. EMBO J. 2001, 20, 1164–1172.
- Aranovich, A.; Gdalevsky, G.Y.; Cohen-Luria, R.; Fishov, I.; Parola, A.H. Membrane-catalyzed nucleotide exchange on DnaA. Effect of surface molecular crowding. J. Biol. Chem. 2006, 281, 12526–12534.
- Aranovich, A.; Braier-Marcovitz, S.; Ansbacher, E.; Granek, R.; Parola, A.H.; Fishov, I. N-terminal-mediated oligomerization of DnaA drives the occupancy-dependent rejuvenation of the protein on the membrane. Biosci. Rep. 2015, 35, e00250.
- Norris, V.; Madsen, M.S. Autocatalytic gene expression occurs via transertion and membrane domain formation and underlies differentiation in bacteria: A model. J. Mol. Biol. 1995, 253, 739–748.
- Woldringh, C.L. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 2002, 45, 17–29.
- Fishov, I.; Namboodiri, S. A nonstop thrill ride from genes to the assembly of the T3SS injectisome. Nat. Commun. 2023, 14, 1973.
- Fernandez-Coll, L.; Maciag-Dorszynska, M.; Tailor, K.; Vadia, S.; Levin, P.A.; Szalewska-Palasz, A.; Cashel, M. The absence of (p)ppGpp renders initiation of Escherichia coli chromosomal DNA synthesis independent of growth rates. mBio 2020, 11, e03223-19.
- Chiaramello, A.E.; Zyskind, J.W. Coupling of DNA replication to growth rate in Escherichia coli: A possible role for guanosine tetraphosphate. J. Bacteriol. 1990, 172, 2013–2019.
- Gonzalez, D.; Collier, J. Effects of (p)ppGpp on the progression of the cell cycle of Caulobacter crescentus. J. Bacteriol. 2014, 196, 2514–2525.
- Hughes, P.; Landoulsi, A.; Kohiyama, M. A novel role for cAMP in the control of the activity of the E. coli chromosome replication initiator protein, DnaA. Cell 1988, 55, 343–350.
- Tymecka-Mulik, J.; Boss, L.; Maciag-Dorszynska, M.; Matias Rodrigues, J.F.; Gaffke, L.; Wosinski, A.; Cech, G.M.; Szalewska-Palasz, A.; Wegrzyn, G.; Glinkowska, M. Suppression of the Escherichia coli dnaA46 mutation by changes in the activities of the pyruvate-acetate node links DNA replication regulation to central carbon metabolism. PLoS ONE 2017, 12, e0176050.
- Horemans, S.; Pitoulias, M.; Holland, A.; Pateau, E.; Lechaplais, C.; Ekaterina, D.; Perret, A.; Soultanas, P.; Janniere, L. Pyruvate kinase, a metabolic sensor powering glycolysis, drives the metabolic control of DNA replication. BMC Biol. 2022, 20, 87.
- Holland, A.; Pitoulias, M.; Soultanas, P.; Janniere, L. The replicative DnaE polymerase of Bacillus subtilis recruits the glycolytic pyruvate kinase (PykA) when bound to primed DNA templates. Life 2023, 13, 965.
- Norris, V.; Demongeot, J. The ring world: Eversion of small double-stranded polynucleotide circlets at the origin of DNA double helix, RNA polymerization, triplet code, twenty amino acids, and strand asymmetry. Int. J. Mol. Sci. 2022, 23, 12915.
- Ogden, G.B.; Pratt, M.J.; Schaechter, M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell 1988, 54, 127–135.
- Shakibai, N.; Ishidate, K.; Reshetnyak, E.; Gunji, S.; Kohiyama, M.; Rothfield, L. High-affinity binding of hemimethylated oriC by Escherichia coli membranes is mediated by a multiprotein system that includes SeqA and a newly identified factor, SeqB. Proc. Natl. Acad. Sci. USA 1998, 95, 11117–11121.
- d’Alencon, E.; Taghbalout, A.; Kern, R.; Kohiyama, M. Replication cycle dependent association of SeqA to the outer membrane fraction of E. coli. Biochimie 1999, 81, 841–846.
- Landoulsi, A.; Malki, A.; Kern, R.; Kohiyama, M.; Hughes, P. The E. coli cell surface specifically prevents the initiation of DNA replication at oriC on hemimethylated DNA templates. Cell 1990, 63, 1053–1060.
- Hiraga, S.; Ichinose, C.; Niki, H.; Yamazoe, M. Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA-protein complexes in E. coli. Mol. Cell 1998, 1, 381–387.
- Helgesen, E.; Saetre, F.; Skarstad, K. Topoisomerase IV tracks behind the replication fork and the SeqA complex during DNA replication in Escherichia coli. Sci. Rep. 2021, 11, 474.
- Norris, V.; Fralick, J.; Danchin, A. A SeqA hyperstructure and its interactions direct the replication and sequestration of DNA. Mol. Microbiol. 2000, 37, 696–702.
- Sekimizu, K.; Bramhill, D.; Kornberg, A. ATP activates DnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 1987, 50, 259–265.
- Messer, W.; Egan, B.; Gille, H.; Holz, A.; Schaefer, C.; Woelker, B. The complex of oriC DNA with the DnaA initiator protein. Res. Microbiol. 1991, 142, 119–125.
- Nievera, C.; Torgue, J.J.; Grimwade, J.E.; Leonard, A.C. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-rc. Mol. Cell 2006, 24, 581–592.
- Sakiyama, Y.; Kasho, K.; Noguchi, Y.; Kawakami, H.; Katayama, T. Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli. Nucleic Acids Res. 2017, 45, 12354–12373.
- Zorman, S.; Seitz, H.; Sclavi, B.; Strick, T.R. Topological characterization of the DnaA-oriC complex using single-molecule nanomanipuation. Nucleic Acids Res. 2012, 40, 7375–7383.
- Landoulsi, A.; Kohiyama, M. DnaA protein dependent denaturation of negative supercoiled oriC DNA minicircles. Biochimie 2001, 83, 33–39.
- Filutowicz, M. Requirement of DNA gyrase for the initiation of chromosome replication in Escherichia coli k-12. Mol. Gen. Genet. 1980, 177, 301–309.
- Hupp, T.R.; Kaguni, J.M. Activation of mutant forms of DnaA protein of Escherichia coli by DnaK and GrpeE proteins occurs prior to DNA replication. J. Biol. Chem. 1993, 268, 13143–13150.
- Norris, V.; Kayser, C.; Muskhelishvili, G.; Konto-Ghiorghi, Y. The roles of nucleoid-associated proteins and topoisomerases in chromosome structure, strand segregation and the generation of phenotypic heterogeneity in bacteria. FEMS Microbiol. Rev. 2022.
- Li, S.; Zhang, Q.; Xu, Z.; Yao, Y.F. Acetylation of lysine 243 inhibits the oric binding ability of DnaA in Escherichia coli. Front. Microbiol. 2017, 8, 699.
- Landoulsi, A.; Kohiyama, M. Initiation of DNA replication in delta cya mutants of Escherichia coli K12. Biochimie 1999, 81, 827–834.
- Charbon, G.; Mendoza-Chamizo, B.; Campion, C.; Li, X.; Jensen, P.R.; Frimodt-Moller, J.; Lobner-Olesen, A. Energy starvation induces a cell cycle arrest in Escherichia coli by triggering degradation of the DnaA initiator protein. Front. Mol. Biosci. 2021, 8, 629953.
- Coppine, J.; Kaczmarczyk, A.; Petit, K.; Brochier, T.; Jenal, U.; Hallez, R. Regulation of bacterial cell cycle progression by redundant phosphatases. J. Bacteriol. 2020, 202, e00345-20.
- Seebach, D. No life on this planet without PHB. Helv. Chim. Acta 2023, 106, e202200205.
- Reusch, R.N.; Shabalin, O.; Crumbaugh, A.; Wagner, R.; Schroder, O.; Wurm, R. Posttranslational modification of E. coli histone-like protein H-NS and bovine histones by short-chain poly-(r)-3-hydroxybutyrate (cPHB). FEBS Lett. 2002, 527, 319–322.
- Fralick, J.A. Studies on the regulation of initiation of chromosome replication in Escherichia coli. J. Mol. Biol. 1978, 122, 271–286.
- Fralick, J.A. Is DnaA the ‘pace-maker’ of chromosome replication? An old paper revisited. Mol. Microbiol. 1999, 31, 1011–1012.
- Flatten, I.; Fossum-Raunehaug, S.; Taipale, R.; Martinsen, S.; Skarstad, K. The DnaA protein is not the limiting factor for initiation of replication in Escherichia coli. PLoS Genet. 2015, 11, e1005276.
- Berger, M.; Wolde, P.R.T. Robust replication initiation from coupled homeostatic mechanisms. Nat. Commun. 2022, 13, 6556.
- Qian’andong Cao, Wenqi Huang, Zheng Zhang, Pan Chu, Ting Wei, Hai Zheng and Chenli Liu. The Quantification of Bacterial Cell Size: Discrepancies Arise from Varied Quantification Methods.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
599
Revisions:
2 times
(View History)
Update Date:
26 Sep 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




