Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Lindsey Ray | -- | 4957 | 2023-09-12 22:34:14 | | | |
| 2 | Camila Xu | Meta information modification | 4957 | 2023-09-13 03:30:03 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Ray, L.; Medeiros, D. Duplicated Genes within Neural Crest Gene Regulatory Network. Encyclopedia. Available online: https://encyclopedia.pub/entry/49090 (accessed on 07 February 2026).
Ray L, Medeiros D. Duplicated Genes within Neural Crest Gene Regulatory Network. Encyclopedia. Available at: https://encyclopedia.pub/entry/49090. Accessed February 07, 2026.
Ray, Lindsey, Daniel Medeiros. "Duplicated Genes within Neural Crest Gene Regulatory Network" Encyclopedia, https://encyclopedia.pub/entry/49090 (accessed February 07, 2026).
Ray, L., & Medeiros, D. (2023, September 12). Duplicated Genes within Neural Crest Gene Regulatory Network. In Encyclopedia. https://encyclopedia.pub/entry/49090
Ray, Lindsey and Daniel Medeiros. "Duplicated Genes within Neural Crest Gene Regulatory Network." Encyclopedia. Web. 12 September, 2023.
Copy Citation
Neural crest cells (NCCs) are an embryonic cell type that are unique to vertebrates, which emerge from the neural plate border. During neurulation, NCCs migrate throughout the body to give rise to a diverse array of neural and non-neural cell types including cartilage, bone, smooth muscle, peripheral neurons, and melanocytes.
gene regulatory network
neural crest cells
genome duplication
vertebrates
1. Introduction
Vertebrates are the most abundant lineage of deuterostomes, comprising about 83% of the species described in the clade [1]. Compared to their invertebrate relatives, vertebrates have elaborated upon the chordate body plan with a range of new cell types tissues, organs, and structures, contributing to more complex morphologies [2][3][4]. Neural crest cells (NCCs) are an embryonic cell type that are unique to vertebrates, which emerge from the neural plate border. During neurulation, NCCs migrate throughout the body to give rise to a diverse array of neural and non-neural cell types including cartilage, bone, smooth muscle, peripheral neurons, and melanocytes [2][3][4][5]. Research on invertebrate chordates has highlighted cells that share some traits with NCCs, including the ability to migrate and give rise to neural or mesenchymal tissues, suggesting that the last common ancestor of chordates and vertebrates had NCC-like cells [2][4][6][7]. However, these invertebrate cell types lack the pluripotency, long-range migratory ability, and spatial awareness of true NCCs [2][7]. The evolution of NCCs was thus a taxon-defining change in development and is thought to have potentiated the species diversity seen in vertebrates.
The development of NCCs and their derivatives have been studied for over a century, and new technologies are allowing scientists to test the theories about their evolution. The dominant model posits that NCCs evolved by co-opting pre-existing genes and genetic subcircuits from other germ layers, building the neural crest gene regulatory network (NC GRN) in a stepwise fashion [2][3][5][7]. Developmental gene regulatory networks consist of transcription factors that bind to cis-regulatory elements to activate or suppress downstream genes, signaling molecules that mediate intercellular communication, and effector genes that determine cellular phenotype. The NC GRN is novel to vertebrates, and understanding how these gene interactions were established will shed light on how macroevolutionary novelties, like new cell types, arise.
The vertebrate head is comprised largely of NCC derivatives and is an evolutionary innovation thought to have facilitated the species richness of this taxon relative to invertebrate chordates [2][4][8]. Gans and Northcutt originally hypothesized the “new head”, as an elaboration of the pre-existing pharyngeal skeleton of chordates, was made possible by NCCs and their migratory, multipotent nature [8]. They speculated that with a muscular and fortified head skeleton, vertebrates were able to evolve forms of active predation and fill many ecological niches, permitting their immense speciation [8]. The new head hypothesis has inspired investigations into the genetic basis of NCC evolution using a variety of developmental and genetic techniques to elucidate conserved and divergent aspects of the vertebrate NC GRN [5][9][10][11].
Another interesting trait separating vertebrates from other chordates is their genome structure and content. The “2R” hypothesis posits that vertebrates quadrupled their genetic material, reducing genetic pleiotropy and permitting mutations to persist without disturbing crucial gene functions [12]. Comparative genomic studies highlight the expansion of the vertebrate genome. Ancestral chordate linkage groups correspond to multiple homologous regions in the vertebrate genomes, indicating a four-fold increase in gene content in vertebrates [13]. Additionally, a third whole genome duplication (WGD) event occurred at the base of teleosts (Figure 1), which are the most species-rich lineage of vertebrates [14]. Taken together, the increase in species diversity following 2R and 3R gene duplication events in vertebrates indicates a positive correlation between genetic material and species diversity, though it is still debated whether WGDs were necessary for the evolutionary expansion of vertebrates.
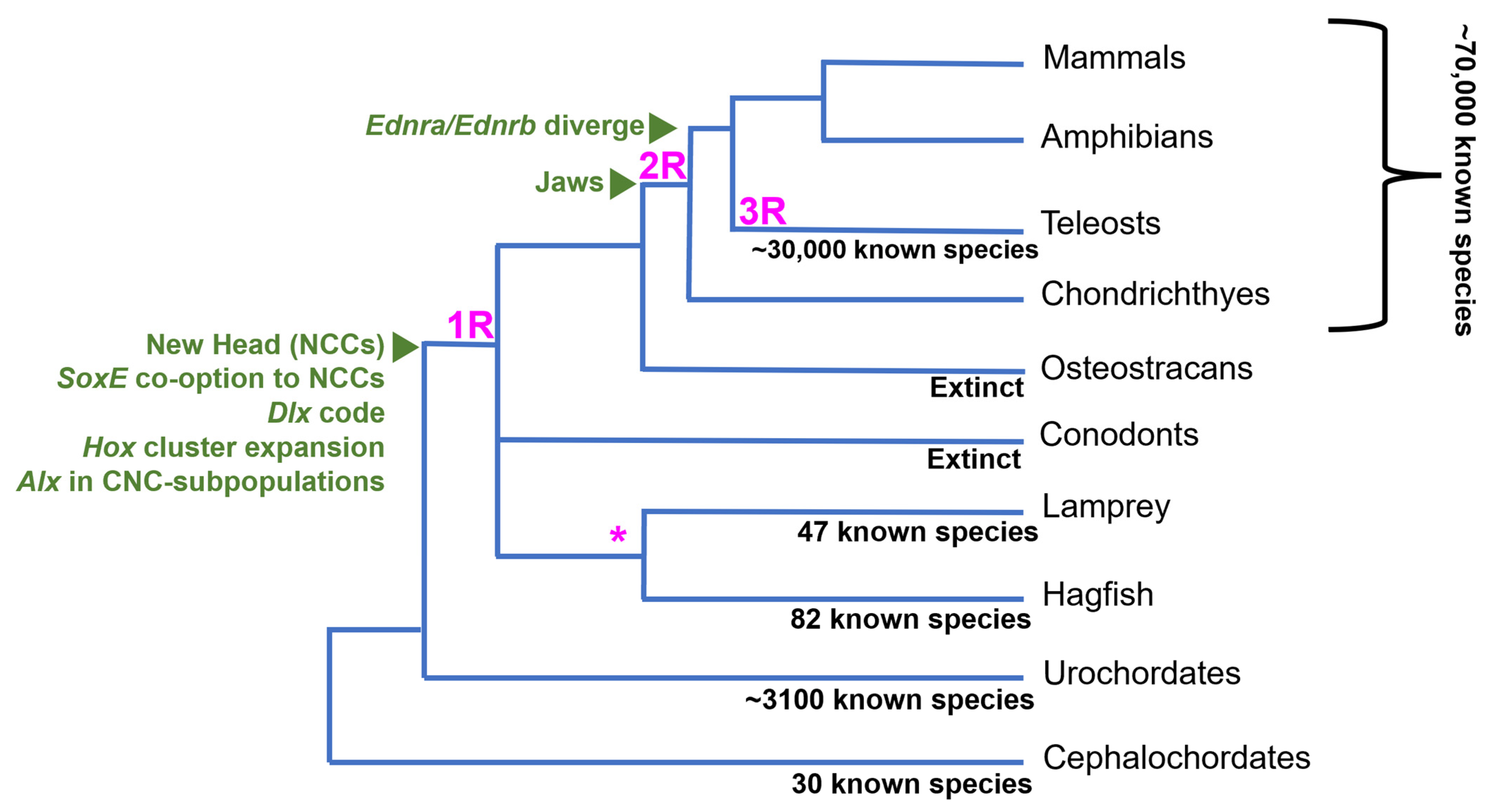
The fossil record provides some support for the new head hypothesis. While most living species of vertebrates have jaws, there was a large radiation of jawless vertebrates during the Silurian through the end of the Devonian period (~300–400 Ma) [15][17]. Muscularization of the pharyngeal basket and extension of the upper lip allowed jawless stem gnathostomes, like osteostracans, to have powerful suction power and bony cranial structures that allowed them to efficiently capture and crush prey [15]. Prior to this innovation, the chordate pharyngeal skeleton facilitated passive respiration and filter feeding with ciliated structures [18]. Further diversification of vertebrates occurred after the evolution of the jaw, as gnathostomes largely replaced agnathans in the late Devonian [15][17][19]. One study shows there was an increase in active predation after the appearance of jawed fish, indicated by bite marks on fossilized prey or finding fish within the stomachs of fossilized predatory fish [17]. This evidence supports the idea that after the evolution of the new head, jaw evolution further facilitated vertebrate species richness by increasing the predatory capability of jawed fish and allowing them to fill new ecological niches.
The absence of genetic data from extinct, stem vertebrate species makes it difficult to directly link WGDs to vertebrate innovations, like the new head or paired fins. The current estimate of the timing of the WGDs is based on comparative studies between the genomes of invertebrate deuterostomes, cyclostomes, and crown gnathostomes [20][21][22][23]. Recent genomic analyses support at least one WGD at the base of vertebrates, with a second phase of genome-scale duplication occurring in jawed vertebrates [24]. The fossil record also shows evidence for a teleost-specific third WGD (3R) around 50–100 million years ago, as stem teleosts have cell sizes similar to modern species [14]. This suggests a link between WGDs and the evolution of key teleost morphological innovations. However, the large delay between the 3R and teleost radiation suggests it was not a direct driver of teleost diversification [14].
It has been proposed that the 2R WGDs may have facilitated the formation of the NC GRN by permitting new gene interactions while maintaining those crucial for development [3][5][22][25]. Many homologs active within the neural plate border (NBP) and underlying mesoderm of invertebrate chordates are active in the vertebrate NBP and neural crest cells that originate from it [2][4][7]. This suggests that ancient genes were co-opted into the NC-GRN from the NPB and other non-ectodermal tissues. It has been proposed that WGDs affect the regulatory landscape of genes more than the functionality of proteins, potentially facilitating gene co-option and GRN evolution [7][20]. Alternatively, elaboration of a pre-existing proto-NC GRN, active in the NPB, or blastula-stage cells, may have occurred before the WGD [25].
2. The NC GRN
2.1. Neural Crest Establishment and Migration
During the development of the central nervous system, all chordates deploy a suite of genes to separate neural and non-neural ectoderm, including homologs of Tfap2, Zic, Msx1/2, Pax3/7, and Dlx3/5 [2][5][26][27]. In vertebrates, these genes activate a suite of neural crest marker genes, including Snail, Id, Tfap2, Twist, FoxD3, and SoxE within neural plate border (NPB) cells [2][5]. These NCC specifier genes consist of transcription factors that are deployed at different stages of NCC development. In invertebrate chordates, homologs of most NCC specifiers, with the exception of snails, are absent from the neural border and expressed in other germ layers or ectodermal domains, suggesting they were co-opted to neural border cells in vertebrates [28][29]. As neurulation proceeds, the vertebrate NPB cells expressing a combination of NCC specifier genes become true neural crests.
There are many subnetworks (also known as subcircuits) within the NC GRN that regulate different aspects of the NCC phenotype during different phases of their development including pluripotency, migratory capabilities, and differentiation capacity. These subcircuits use many of the same transcription factors and signaling molecules as the core NC GRN at different times to attenuate the activation and suppression of downstream target genes. NCC marker genes regulate downstream genes that initiate their delamination from the ectoderm during the epithelial-to-mesenchymal transition (EMT), which is marked by the expression of Snail, FoxD3, Twist, Lmo4, and Zeb2 [27]. Regulation of the EMT subcircuit leads to the dynamic expression of cadherin proteins regulated by transcription factors, such as Snail and Zeb2 [30]. Some vertebrates have slight differences in cadherin protein activity during the EMT, but many express type I cadherins prior to delamination and switch to type II cadherins prior to and during migration, orchestrating changes in the cytoskeleton that allow the proper NC movement [27][30][31]. These genes, along with Ets1, c-Myc, Tfap2, Id, and SoxE, are expressed after NCCs separate from NPB and remain active during their migration throughout the body [5][27]. It is important to note there are differences between the gnathostome and cyclostome NC GRN, as pre-and post-migratory 22NCCs express various NC-specific components at different times and with different expression boundaries, but gene swap experiments show that these homologous proteins are functionally similar between lineages [2][9][27][28]. The evolutionary significance of the heterochronic expression of these components of the NC GRN between vertebrates remains unknown.
The initial positioning of NCCs along the anterior–posterior axis affects their ultimate post-migratory destination and fate. In gnathostomes, NCCs form four major subpopulations: cranial, vagal, cardiac, and trunk/sacral, all of which give rise to different structures throughout the body [28][32][33]. In the other major lineage of extant vertebrates, the jawless cyclostomes, the precise boundaries, and derivatives of non-cranial NCC subpopulations are less clear. Cranial NCCs migrate into the pharyngeal arches (PAs) in a conserved pattern across vertebrates, where they receive various signals dependent on their anterior–posterior position that determine their skeletal fate after migration [16][28][34]. At the NPB, amniote cranial NCCs (CNCCs) are marked by the co-expression of Bm3, Lhx5, and Dmbx1 that activate SoxE, Tfap2, and Est1, which is maintained throughout the migration to the PAs [10]. Recent work in mice revealed that CNCCs reactivate pluripotency marker Oct4 after delamination and reset their positional information and become transcriptionally equivalent prior to migration into the PAs [35]. Whether this is a conserved feature of the NC GRN, or unique to amniotes, is unclear.
Skates, a representative of jawed cartilaginous fish, lack the early CNCC markers but deploy SoxE, Tfap2, and Ets1 prior to and during migration, while zebrafish share a majority of their CNCC specification and migration GRN with amniotes, with the exception of bm3 [10]. This evidence highlights a stepwise acquisition of the CNCC GRN in gnathostomes as well as a high conservation of their expression patterns. Lamprey lack the vagal stream of NCCs, with trunk NC-derived Schwan precursor cells giving rise to their enteric nervous system, which stems from vagal NCCs in gnathostomes [28][36][37]. Generally, lamprey CNCCs exhibit a GRN more comparable to a trunk neural crest than those in gnathostomes, with migratory CNCCs being marked with SoxE and Tfap2 [9][10][38]. They also express some genes orthologous to amniote early cranial specifiers, Lhx5 and Dmbx1, later in the PAs [10]. Additionally, the migration patterns of CNCCs in lamprey are less restricted, with the cells destined for the posterior pharyngeal arches migrating initially as a sheet rather than distinct streams [34]. This may be a result of heterochronies between the NCC migratory GRN of jawed and jawless vertebrates [34].
2.2. Neural Crest Derivatives in the New Head
The vertebrate head skeleton can be divided into the neurocranium, which encases the brain and can have a mesodermal component, and the viscerocranium, which develops from NCCs in the pharyngeal arches [39]. NCCs populate the PAs during head development and give rise to unique structures in each arch. While there are many cranial NCC derivatives that are shared by all vertebrates, hagfish and lamprey lack some NCC derivatives, such as jaws and cranial sympathetic ganglia [34][37]. Due to the inaccessibility of hagfish embryos, the vast majority of what is known about NCC development in agnathans is from studies of lamprey embryos.
Intercellular signals that are received by CNCCs as they migrate into the head are generally conserved between jawed and jawless vertebrates [16]. Within the PAs, both cyclostomes and gnathostomes develop NC-derived skeletal structures. However, the cartilage composition differs in the pharyngeal skeleton, and lampreys totally lack bone [40]. Fibroblast growth factor (Fgf) expression is activated by retinoic acid (RA) signaling within the pharyngeal pouch endoderm and is crucial for cartilage development in all vertebrates. FGF receptors on CNCCs receive that signal and activate a downstream cartilage regulatory module containing Dlx, SoxE, Twist, and Ets [16][40]. A similar GRN is deployed in the cartilage of amphioxus oral cirri, implying an ancient cartilage regulatory subcircuit may have been co-opted into CNCCs during vertebrate evolution [4]. Chondrocytes differentiate and give rise to cellular and “soft” cartilages that permit the stability and flexibility of the viscerocranium. In addition to the cartilage differentiation genes mentioned above, gnathostomes require Barx and Runx for proper facial cartilage/bone development, while lamprey only deploy those genes in the branchial vasculature of the PAs [41].
All vertebrates deploy a conserved subset of transcription factors within CNCCs, filling the PAs that give rise to different structures and skeletal fates [16]. Within the head, CNCCs give rise to both dermal and endochondral bone [42] Dermal bones, such as mandibular bones forming from the first PA, ossify directly from mesenchymal cell matrices. Endochondral bone, such as the CNC-derived parietal bone of the skull, requires a cartilage intermediate prior to ossification [42]. Just before FGF signaling activates chondrogenesis, other signaling pathways including BMP, endothelin (Edn), and Notch initiate the patterning of the CNC and resulting head [16][23]. Twist, Ets, Id, Alx, and SoxE orthologs are expressed in all vertebrate skeletogenic CNCCs during migration and the population of the PAs [41]. Skeletal elements differentiate through the polarized and combinatorial expression of Alx, Hand, Msx, and Prrx around a core of nested Dlx expression along the dorsal–ventral (DV) axis that corresponds to unique structures in the head [16][41]. These genes cooperate with Hox genes expressed along the anterior–posterior (AP) axis of the PAs to give rise to transcriptionally distinct CNCC populations in each arch that give rise to skeletal elements of various shapes and properties [16][43]. Dlx and Hox genes are thought to pattern the DV and AP axes of the vertebrate head in a highly conserved, code-like pattern [16][44].
3. Duplicated Genes within the NC GRN
The term “ohnolog” was coined to distinguish gene duplicates that were products of vertebrate-specific WGDs from paralogs that are exclusive to a single lineage and orthologs shared by multiple lineages [12]. All genes in the NC GRN appear to have been duplicated during the vertebrate WGDs, with the resulting ohnologs being differentially retained across lineages (Figure 2). However, virtually all NC GRN ohnologs function at some point during neural crest development. It would be highly unlikely for each duplicated member of these gene families to be independently co-opted into the same GRN. Thus, the fact that virtually all NC GRN ohnologs are active at some point in the NC GRN, or its various subcircuits, serves as robust evidence that the co-option of these genes to the neural border, and the assembly of the NC GRN, occurred before the WGDs.
In this context, the differential expression of NC GRN ohnologs likely reflects the temporal and spatial subfunctionalization of NC GRN components after the WGDs (depicted in Figure 3). The functional consequences of this extensive subfunctionalization of NC GRN ohnologs remain unclear. However, in general terms, these duplications appear to have substantially increased the overall complexity of the modern NC GRN by creating temporally and spatially restricted subcircuits [2][3][7][10][11]. The dedication of individual NC GRN ohnologs to particular phases of the NC GRN, or particular NC subpopulations, may have increased the modularity of the GRN, allowing different portions to evolve without interfering with its other functions. Consider an NC GRN gene that was ancestrally involved in the initial activation of the NC GRN (i.e., NCC specification) and later during the differentiation of two NCC derivations, derivative 1 and derivative 2. Suppose this gene was duplicated during the WGDs, and its three retained ohnologs, A, B, and C, became temporally and spatially subfunctionalized: A being expressed early and dedicated to NCC induction, and B and C expressed later and dedicated to derivative 1 and 2, respectively. If a mutation affected the expression or function of the B ohnolog, causing an adaptive change in derivative 1, it would have minimal effect on NCC specification or the formation of derivative 2. Conceivably, this subfunctionalization of ohnologs could then lead to biochemical or regulatory neofunctionalization, as they evolved new biochemical properties or expression domains.
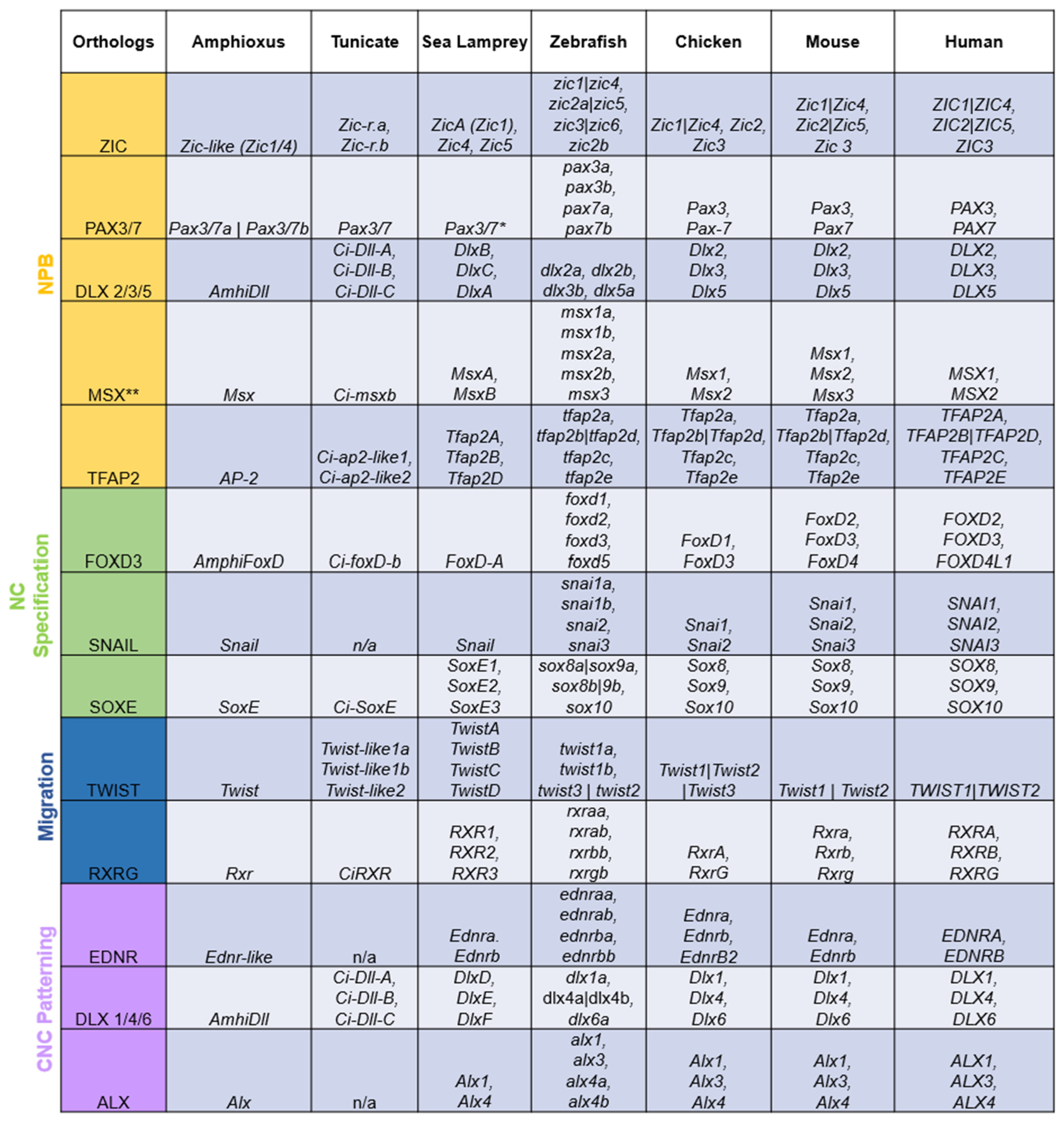
Figure 2. Duplicated orthologs of the NC GRN—amphioxus, tunicate, and sea lamprey orthologs were found in the literature and the NCBI and Stowers Institute databases. Amphioxus [29][45][46][47][48]; tunicate [29][49][50][51][52][53][54][55][56][57][58]; sea lamprey [9][16][23][29][41][59][60][61]. The jawed vertebrate orthologs were collected from the Ohnologs Data Repository [62] and the NCBI gene database. The (|) between gene names indicates that genes are not ohnologs and were duplicated before/after vertebrate-specific WGDs. The (**) on Msx orthologs indicates that these genes were marked as pre-2R duplicates according to [63]. The (*) on lamprey Pax3/7 means that there were discrepancies between the annotations of the lamprey genome [64], NCBI, and past work [9].
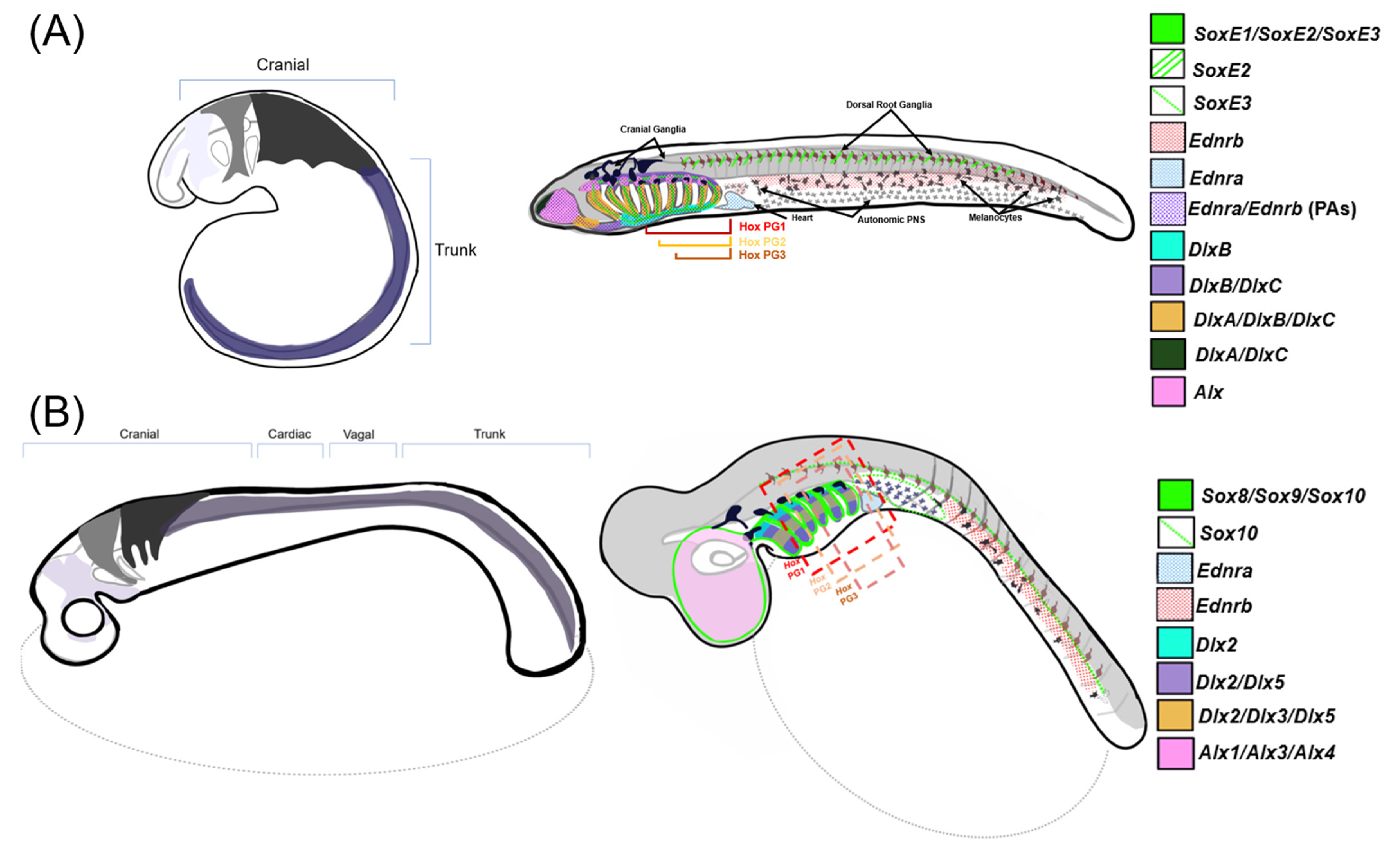
Figure 3. Comparisons of NCC migration and patterning between gnathostomes and lamprey—a stylized diagram of neural crest cell migration and post-migratory gene expression in NCCs within (A) sea lamprey and (B) a representative gnathostome (catshark). Left: three homologous cranial neural crest streams are displayed migrating into prospective PAs and one generalized trunk stream that migrates posterior of the head. Right: gene expression map of the described NC-GRN orthologs within respective model organisms at the later, pharyngula stage. Note that Alx1, Alx3, and Alx4 are expressed differentially along the mediolateral axis in gnathostomes but are depicted as a single expression pattern within the nasopharyngeal region in this diagram [65][66][67].
3.1. SoxE
Transcription factors of the SoxE family participate in multiple steps of NCC development. While there is one SoxE in invertebrate chordates, SoxE is duplicated in vertebrates with three each in non-teleost gnathostomes (Sox8, Sox9, and Sox10) and lamprey (SoxE1, SoxE2, and SoxE3) and five in teleosts [4][68][69]. SoxE3 is in lamprey and gnathostome is in Sox9, and both have a conserved role in regulating cartilage morphogenesis. SoxE2 is comparable to Sox10, as both have similar melanogenic and glial-inducing roles [68][69]. All vertebrate SoxE paralogs are expressed during induction, migration, and differentiation of NCCs but with heterochronic differences across lineages [22][68][69]. The most parsimonious explanation for this is that SoxE was co-opted into the NC GRN before the first WGD at the base of the vertebrates. 1R ohnologs were then duplicated in gnathostomes during the 2R WGD, while one or both 1R ohnologs were independently duplicated in the cyclostome lineage. Based on the similar expression of SoxE3/Sox9 in CNCCs and SoxE2/Sox10 in the trunk, the subfunctionalization of R1 SoxE duplicates likely occurred before the gnathostome/agnathan split.
Interestingly, AmphiSoxE is expressed in the fibrillar cartilage of the oral cirri in amphioxus, pointing to an ancestral role for SoxE in cartilage development [4]. AmphiSoxE is capable of activating downstream genes in NC GRN modules, such as neurogenic Phox2 and melanogenic Mitf, within transgenic zebrafish, expressing AmphiSoxE and other NC specifiers in transgenic chicks [22][68]. This indicates an ancestral function in differentiation and NCC induction in the ancestral SoxE. However, AmphiSoxE regulatory elements are not capable of driving reporter expression within NCCs in transgenic zebrafish, indicating that a cis change permitted SoxE function in neural crest GRN [4]. Additionally, transgenic mice expressing the Drosophila ortholog of SoxE (Sox100B) in place of Sox10 developed many NC derivatives, including the peripheral nervous system (PNS) and melanocytes, showing that Sox10 has retained a highly conserved role of inductive capabilities [70]. However, when Sox8 is expressed in place of Sox10, transgenic mice fail to develop melanocytes, indicating that gnathostome Sox8/9/10 proteins have diverged in functionality [3][71]. Taken together, these data suggest that after an ancestral SoxE was co-opted into the NC GRN by a cis-regulatory mutation, it underwent duplication and both temporal and spatial subfunctionalization of its ancestral expression pattern. This was followed by some divergence of the SoxE protein function, reflecting either subfunctionalization or neofunctionalization of SoxE ohnologs.
3.2. Dlx
The Dlx transcription factor family, which underwent an ancient tandem duplication in the last common ancestor of vertebrates and urochordates, is homologous to the distal-less gene (Dll) in insects [72]. This tandem gene pair was duplicated in gnathostomes into pairs of multiple copies across different chromosomes in a pattern that agrees with 2R duplication events [72][73]. As with other gene families, Dlx orthology is not perfect between cyclostomes and gnathostomes, and teleosts have more Dlx paralogs than other gnathostomes. Each tandem duplicate has its own orthology group, annotated Dlx1/4/6 and Dlx2/3/5, in gnathostomes. While in lamprey, Dlx orthologs are annotated as DlxD/E/F and DlxA/B/C, respectively [16][73]. The last common vertebrate ancestor is hypothesized to have duplicated its tandem Dlx clusters during the 1R event with lineage-specific duplications followed by differential retention, resulting in all vertebrates having six Dlx genes [16][72][73].
The Dlx family has a highly conserved role in patterning, especially within the vertebrate head where it exhibits a nested expression pattern along the DV axis of the Pas; however, the precise borders of these expression patterns vary between jawed and jawless vertebrates [16][74]. The sea lamprey exhibits nested DV expression of DlxA-D paralogs and regionalized Hand1 and Msx transcription, reminiscent of the Dlx-Hand-Msx code in gnathostomes [75]. This nesting is less obvious in the Japanese lamprey with the exception of DlxF, which is differentially expressed in the nasal region [43]. As in gnathostomes, the combinatorial differences in Dlx, Hand, and Msx expression domains along the DV axis also correlate with different cartilage types within the lamprey head [75].
As with other NC GRN genes, all Dlx paralogs are expressed in NCC at some point during NCC development, strongly supporting the idea that the ancestral Dlx gene pair was co-opted into the NC GRN before the WGDs [16]. The distinct expression patterns of Dlx ohnologs further suggest the subfunctionalization of an ancestral, pan-NCC expression pattern. The fact that different combinations of Dlx genes mark different portions of the head skeleton is consistent with the participation of different Dlx ohnologs in separate regulatory subcircuits in the NC GRN. The degree to which the biochemical function of the transcription factors encoded Dlx genes was affected by WGDs is unclear, as there is limited evidence that Dlx proteins have qualitatively different DNA binding properties. However, the divergent expression of Dlx ohnologs supports the idea that the WGDs allowed the regulatory landscape of the Dlx family to diverge, resulting in the nested expression of the “Dlx Code” [40]. The complex expression of Dlx ohnologs within the vertebrate head may be consistent with the idea that the patterning of NCC derivative became more elaborate following the vertebrate-specific expansion of this gene family.
3.3. Hox Clusters
The Hox gene family has been presented as evidence for the 2R hypothesis, one reason being that amphioxus possesses a single, syntenic cluster of tandem Hox homologs, while gnathostomes have four homologous clusters across separate chromosomes [76][77][78]. Hox clusters in teleosts are also evidence for teleost-specific genome duplication (3R) and are distributed across seven chromosomes [72]. Alternatively, some have used data from phylogenetic analyses of human Hox genes as evidence against WGDs, and propose that this gene family was expanded by small-scale events that occurred in vertebrates [79][80][81]. There are ancient paralogical groups of Hox genes that are annotated numerically within clusters (i.e., Hox1, Hox2, Hox3…). These groups are cis-tandem duplicates that occur throughout metazoans, and Hox ohnologs are specified by letters (i.e., Hoxa1, Hoxb1, Hoxc1…) and occupy separate chromosomes in vertebrates. These transcription factor genes are expressed in a conserved, colinear manner across bilaterians along the anterior–posterior axis and have been shown to induce homeotic transformations if mutated [82][83][84][85].
Within the pharyngeal arches of all vertebrates, the first PA (PA1) is Hox-negative, PA2 is marked by Hox2 paralogs, and PA3, along with the posterior arches, are marked by Hox3 paralogs [43][84][86][87]. The ectopic expression of Hox cis-paralogs within the PAs can lead to homeotic transformations, for instance when Hoxa2 is expressed in the first PA of mice, which is Hox-negative, embryos lose the mandibular structures or take on a second arch identity [88]. Additionally, certain levels of ectopic Hoxa2 expression can alter skeletal element identity in mice, highlighting the possibility that the regulation of Hox gene expression could have more of an influence on skeletal identity than protein function [88]. Knock-downs of hox2 ohnologs in zebrafish have shown that these genes are partially redundant, as removing the function of one ohnolog often results in a less dramatic phenotypic effect than targeting the whole group [85][89]. This highlights the functional differences between Hox tandem duplicates, yet the degree to which Hox ohnologs differ regarding DNA binding specificity is still unclear.
In cyclostomes, the Hox gene orthology is disparate from gnathostomes, providing another line of evidence for cyclostome-specific duplications subsequent to their divergence from jawed vertebrates and the possibility of segmental gene duplications in this family [44][64]. Differences in the Hox cluster number and organization between different species of lamprey and hagfish add difficulty to solving the phylogeny of Hox clusters [44][84]. Although Hox orthologues are hard to assign between jawed and jawless vertebrates, there is evidence that cis-regulatory elements (CREs) upstream of Hoxa1/Hoxb1 in gnathostomes and hoxα1 in lamprey are capable of driving similar expression in NC [44]. Similar CRE activity provides minor support for orthology assignment as well as the nested colinear expression pattern of tandem duplicates in the pharyngeal arches [44][84]. Hox ohnologs have been shown to have both redundant and diverged traits in NC patterning, making them a complex gene family for understanding their evolution after duplication events. Recent work in zebrafish has shown that within the PAs, certain levels of ectopic hoxa2 expression can alter skeletal element identity, highlighting the possibility that the regulation of Hox gene expression could have more of an influence on skeletal identity than protein function [88].
3.4. EdnR
Cranial neural crest cells have endothelin receptors that interpret signals from the surrounding tissues to activate downstream genes, including NCC specifier genes and Dlx [16][23][41]. The endothelin receptor genes (Ednra/Ednrb) are both critical components of the NC GRN, suggesting they are likely “1R” duplicates of a single ancestral Ednr present in the last common vertebrate ancestor [23][90]. The 2R WGD in gnathostomes was presumably followed by the loss of two Ednrs, leaving only one Ednra and one Ednrb, while the teleost 3R resulted in four Ednr genes in this group. Following the gnathostome–cyclostome split, Ednrs evolved somewhat divergent roles in CNCC differentiation [23][91]. Within gnathostomes, Ednra has a key role in skeletogenesis and vascular development, while Ednrb contributes to melanocyte and peripheral nervous system development, and Ednrb mutants have no craniofacial defects [92]. In contrast, lamprey Ednra and Ednrb both are required for proper PA formation, and those lacking either Ednra or Ednrb fail to form branchial arch cartilages properly [23][92]. Recently, it was discovered that skate embryos also express Ednrb in the ventral and intermediate NCCs in all pharyngeal arches in a way that resembles lamprey expression [93]. Together, these data imply that after the R2 WGD, Ednr ohnolog expression diverged in chondrichthyans and bony fish, with the developmental roles of Ednra and Ednrb becoming more specialized in the latter [23][94].
Endothelin receptors on the surface of NCCs activate genes that contribute to the skeletal phenotype within each arch. Within gnathostomes, Ednra activates Hand and Dlx5/6 genes within the ventral portion of the PAs in zebrafish, mice, and frogs, where they contribute to the lower jaw and joint formation [23][92][95]. Lamprey exhibit similar Dlx expression within the ventral Pas; however, hand expression is not regulated by Ednra [23]. Duplicated genes can subfunctionalize or neofunctionalize following duplications; Ednra and Ednrb appear to have diverged in terms of expression pattern and downstream gene targets [23][41]. It is possible that a stem or independent duplication provided the flexibility necessary for agnathans and gnathostomes to alter endothelin signaling, supporting the morphological specialization of their head skeletons. Gene swap experiments between cyclostome and gnathostome Ednra/Ednrb orthologues will highlight the divergent aspects of these paralogous signaling pathways. Ednra/Ednrb divergence following duplication is a potential link between head specialization and “2nd R”, but further research will have to be conducted to deem it as a necessary event for that divergence.
3.5. Alx
Alx genes are part of NC GRN subcircuits that confer cellular identity in NCC-derived skeletal elements, contributing to their unique shapes and different components [16][41][65]. This gene family has a conserved skeletal role across deuterostomes and is deployed in the skeletal GRNs of echinoderms and vertebrates [96]. Amphioxus possesses two copies of Alx (Bf-alx1, Bf-alx2)m whose loci are near each other and share highly similar intron/exon structures, indicating a lineage-specific, tandem duplication [96]. Non-teleost gnathostomes have three Alx homologs, Alx1/Cart1, Alx3, and Alx4, and teleosts possess additional paralogs of Alx4 (alx4a, alx4b) that are generated during the vertebrate and teleost WGDs, demonstrating that these paralogs have been differentially retained [65][96]. Lamprey possesses two Alx homologs that were likely generated during the first vertebrate WGD and orthologous to gnathostomes Alx1 and Alx4 [41]. Only the expression of lamprey Alx4 has been reported, and like gnathostome Alx ohnologs, it is expressed in skeletogenic CNCCs [41].
The expression of different Alx paralogs corresponds to different cellular cartilage shapes within the vertebrate head [16][65][66][75][96]. In lamprey, Alx4 expression is largely coincident with stiffer, more rigid cartilage phenotypes, indicating an ancestral role in the differentiation of skeletal tissue subtypes [41]. In zebrafish, combinations of Alx paralogs label NCC-derived chondrocytes with distinct cellular phenotypes. Detailed functional analyses suggest that this “Alx code” contributes to the intricate shapes of the gnathostome head skeleton [65]. In mice, Alx1 is activated earlier and is concentrated in the midline regions of the frontal nasal prominence (FNP), while Alx3 and Alx4 are largely limited to the lateral tissues of the FNP [66][67]. Experiments in mice have also shown that Alx paralogs have diverged in function, with homozygous deletions of Alx1 or Alx3 resulting in different craniofacial malformations, while Alx4 loss-of-function results in minimal defects [66]. Deciphering the evolution of the gnathostome “Alx code” would be aided by gene swap experiments within and between species or the overexpression experiments of various Alx1/3/4 paralogs. Furthermore, analyzing the open chromatin regions surrounding paralogs within various lineages will help determine whether regulatory changes are at the base of their divergence or if there are protein sequences that have diverged biochemically.
References
- Bánki, O.; Roskov, Y.; Döring, M.; Ower, G.; Hernández Robles, D.R.; Plata Corredor, C.A.; Stjernegaard Jeppesen, T.; Örn, A.; Vandepitte, L.; Hobern, D.; et al. Catalogue of Life Checklist. Cat. Life 2023.
- Medeiros, D.M. The evolution of the neural crest: New perspectives from lamprey and invertebrate neural crest-like cells. WIREs Dev. Biol. 2013, 2, 1–15.
- Van Otterloo, E.; Cornell, R.A.; Medeiros, D.M.; Garnett, A.T. Gene regulatory evolution and the origin of macroevolutionary novelties: Insights from the neural crest. Genesis 2013, 51, 457–470.
- Jandzik, D.; Garnett, A.T.; Square, T.A.; Cattell, M.V.; Yu, J.-K.; Medeiros, D.M. Evolution of the new vertebrate head by co-option of an ancient chordate skeletal tissue. Nature 2015, 518, 534–537.
- Martik, M.L.; Bronner, M.E. Riding the crest to get a head: Neural crest evolution in vertebrates. Nat. Rev. Neurosci. 2021, 22, 616–626.
- Cheung, M.; Tai, A.; Lu, P.J.; Cheah, K.S. Acquisition of multipotent and migratory neural crest cells in vertebrate evolution. Curr. Opin. Genet. Dev. 2019, 57, 84–90.
- York, J.R.; McCauley, D.W. The origin and evolution of vertebrate neural crest cells. Open Biol. 2020, 10, 190285.
- Gans, C.; Northcutt, R.G. Neural Crest and the Origin of Vertebrates: A New Head. Science 1983, 220, 268–273.
- Sauka-Spengler, T.; Meulemans, D.; Jones, M.; Bronner-Fraser, M. Ancient Evolutionary Origin of the Neural Crest Gene Regulatory Network. Dev. Cell 2007, 13, 405–420.
- Martik, M.L.; Gandhi, S.; Uy, B.R.; Gillis, J.A.; Green, S.A.; Simoes-Costa, M.; Bronner, M.E. Evolution of the new head by gradual acquisition of neural crest regulatory circuits. Nature 2019, 574, 675–678.
- Betancur, P.; Bronner-Fraser, M.; Sauka-Spengler, T. Assembling Neural Crest Regulatory Circuits into a Gene Regulatory Network. Annu. Rev. Cell Dev. Biol. 2010, 26, 581–603.
- Ohno, S. Evolution by Gene Duplication; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013.
- Putnam, N.H.; Butts, T.; Ferrier, D.E.K.; Furlong, R.F.; Hellsten, U.; Kawashima, T.; Robinson-Rechavi, M.; Shoguchi, E.; Terry, A.; Yu, J.-K.; et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 2008, 453, 1064–1071.
- Davesne, D.; Friedman, M.; Schmitt, A.D.; Fernandez, V.; Carnevale, G.; Ahlberg, P.E.; Sanchez, S.; Benson, R.B.J. Fossilized cell structures identify an ancient origin for the teleost whole-genome duplication. Proc. Natl. Acad. Sci. USA 2021, 118, e2101780118.
- Donoghue, P.C.J.; Keating, J.N. Early vertebrate evolution. Palaeontology 2014, 57, 879–893.
- Square, T.; Jandzik, D.; Romášek, M.; Cerny, R.; Medeiros, D.M. The origin and diversification of the developmental mechanisms that pattern the vertebrate head skeleton. Dev. Biol. 2017, 427, 219–229.
- Randle, E.; Sansom, R.S. Bite marks and predation of fossil jawless fish during the rise of jawed vertebrates. Proc. R. Soc. B Biol. Sci. 2019, 286, 20191596.
- Glenn Northcutt, R. The new head hypothesis revisited. J. Exp. Zool. B Mol. Dev. Evol. 2005, 304, 274–297.
- Purnell, M.A. Feeding in extinct jawless heterostracan fishes and testing scenarios of early vertebrate evolution. Proc. R. Soc. Lond. B Biol. Sci. 2002, 269, 83–88.
- Marlétaz, F.; Firbas, P.N.; Maeso, I.; Tena, J.J.; Bogdanovic, O.; Perry, M.; Wyatt, C.D.R.; de la Calle-Mustienes, E.; Bertrand, S.; Burguera, D.; et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 2018, 564, 64–70.
- Holland, L.Z.; Ocampo Daza, D. A new look at an old question: When did the second whole genome duplication occur in vertebrate evolution? Genome Biol. 2018, 19, 209.
- Tai, A.; Cheung, M.; Huang, Y.-H.; Jauch, R.; Bronner, M.E.; Cheah, K.S.E. SOXE neofunctionalization and elaboration of the neural crest during chordate evolution. Sci. Rep. 2016, 6, 34964.
- Square, T.A.; Jandzik, D.; Massey, J.L.; Romášek, M.; Stein, H.P.; Hansen, A.W.; Purkayastha, A.; Cattell, M.V.; Medeiros, D.M. Evolution of the endothelin pathway drove neural crest cell diversification. Nature 2020, 585, 563–568.
- Simakov, O.; Marlétaz, F.; Yue, J.-X.; O’Connell, B.; Jenkins, J.; Brandt, A.; Calef, R.; Tung, C.-H.; Huang, T.-K.; Schmutz, J.; et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 2020, 4, 820–830.
- Schock, E.N.; York, J.R.; LaBonne, C. The developmental and evolutionary origins of cellular pluripotency in the vertebrate neural crest. Semin. Cell Dev. Biol. 2023, 138, 36–44.
- Thawani, A.; Groves, A.K. Building the Border: Development of the Chordate Neural Plate Border Region and Its Derivatives. Front. Physiol. 2020, 11, 608880.
- Simões-Costa, M.; Bronner, M.E. Establishing neural crest identity: A gene regulatory recipe. Development 2015, 142, 242–257.
- Rothstein, M.; Simoes-Costa, M. On the evolutionary origins and regionalization of the neural crest. Semin. Cell Dev. Biol. 2023, 138, 28–35.
- Yu, J.-K.; Meulemans, D.; McKeown, S.J.; Bronner-Fraser, M. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 2008, 18, 1127–1132.
- Leathers, T.A.; Rogers, C.D. Time to go: Neural crest cell epithelial-to-mesenchymal transition. Dev. Camb. Engl. 2022, 149, dev200712.
- Taneyhill, L.A.; Schiffmacher, A.T. Should I stay or should I go? Cadherin function and regulation in the neural crest. Genesis 2017, 55, e23028.
- Rothstein, M.; Bhattacharya, D.; Simoes-Costa, M. The molecular basis of neural crest axial identity. Dev. Biol. 2018, 444, S170–S180.
- Srinivasan, A.; Toh, Y.-C. Human Pluripotent Stem Cell-Derived Neural Crest Cells for Tissue Regeneration and Disease Modeling. Front. Mol. Neurosci. 2019, 12, 39.
- McCauley, D.W.; Bronner-Fraser, M. Neural crest contributions to the lamprey head. Development 2003, 130, 2317–2327.
- Zalc, A.; Sinha, R.; Gulati, G.S.; Wesche, D.J.; Daszczuk, P.; Swigut, T.; Weissman, I.L.; Wysocka, J. Reactivation of the pluripotency program precedes formation of the cranial neural crest. Science 2021, 371, eabb4776.
- York, J.R.; Yuan, T.; McCauley, D.W. Evolutionary and Developmental Associations of Neural Crest and Placodes in the Vertebrate Head: Insights from Jawless Vertebrates. Front. Physiol. 2020, 11, 986.
- Green, S.A.; Uy, B.R.; Bronner, M.E. Ancient evolutionary origin of vertebrate enteric neurons from trunk-derived neural crest. Nature 2017, 544, 88–91.
- Nikitina, N.V.; Bronner-Fraser, M. Gene regulatory networks that control the specification of neural-crest cells in the lamprey. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2009, 1789, 274–278.
- Martin, W.M.; Bumm, L.A.; McCauley, D.W. Development of the viscerocranial skeleton during embryogenesis of the sea lamprey, Petromyzon Marinus. Dev. Dyn. 2009, 238, 3126–3138.
- Jandzik, D.; Hawkins, M.B.; Cattell, M.V.; Cerny, R.; Square, T.A.; Medeiros, D.M. Roles for FGF in lamprey pharyngeal pouch formation and skeletogenesis highlight ancestral functions in the vertebrate head. Development 2014, 141, 629–638.
- Cattell, M.; Lai, S.; Cerny, R.; Medeiros, D.M. A New Mechanistic Scenario for the Origin and Evolution of Vertebrate Cartilage. PLoS ONE 2011, 6, e22474.
- Matsuoka, T.; Ahlberg, P.E.; Kessaris, N.; Iannarelli, P.; Dennehy, U.; Richardson, W.D.; McMahon, A.P.; Koentges, G. Neural crest origins of the neck and shoulder. Nature 2005, 436, 347–355.
- Parker, H.J.; Pushel, I.; Krumlauf, R. Coupling the roles of Hox genes to regulatory networks patterning cranial neural crest. Dev. Biol. 2018, 444, S67–S78.
- Parker, H.J.; De Kumar, B.; Green, S.A.; Prummel, K.D.; Hess, C.; Kaufman, C.K.; Mosimann, C.; Wiedemann, L.M.; Bronner, M.E.; Krumlauf, R. A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates. Nat. Commun. 2019, 10, 1189.
- Sumiyama, K.; Irvine, S.Q.; Ruddle, F.H. The role of gene duplication in the evolution and function of the vertebrate Dlx/distal-less bigene clusters. In Genome Evolution: Gene and Genome Duplications and the Origin of Novel Gene Functions; Meyer, A., Van de Peer, Y., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 151–159.
- Yu, J.-K.; Holland, N.D.; Holland, L.Z. Tissue-specific expression of FoxD reporter constructs in amphioxus embryos. Dev. Biol. 2004, 274, 452–461.
- Tocchini-Valentini, G.D.; Rochel, N.; Escriva, H.; Germain, P.; Peluso-Iltis, C.; Paris, M.; Sanglier-Cianferani, S.; Dorsselaer, A.V.; Moras, D.; Laudet, V. Structural and Functional Insights into the Ligand-binding Domain of a Nonduplicated Retinoid X Nuclear Receptor from the Invertebrate Chordate Amphioxus. J. Biol. Chem. 2009, 284, 1938–1948.
- Meulemans, D.; Bronner-Fraser, M. Insights from Amphioxus into the Evolution of Vertebrate Cartilage. PLoS ONE 2007, 2, e787.
- Satou, Y.; Imai, K.S. Ascidian Zic Genes. In Zic Family: Evolution, Development and Disease; Aruga, J., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2018; pp. 87–106.
- Kim, K.; Orvis, J.; Stolfi, A. Pax3/7 regulates neural tube closure and patterning in a non-vertebrate chordate. Front. Cell Dev. Biol. 2022, 10, 999511.
- Caracciolo, A.; Di Gregorio, A.; Aniello, F.; Di Lauro, R.; Branno, M. Identification and developmental expression of three Distal-less homeobox containing genes in the ascidian Ciona intestinalis. Mech. Dev. 2000, 99, 173–176.
- Aniello, F.; Locascio, A.; Villani, M.G.; Di Gregorio, A.; Fucci, L.; Branno, M. Identification and developmental expression of Ci-msxb: A novel homologue of Drosophila msh gene in Ciona intestinalis. Mech. Dev. 1999, 88, 123–126.
- Jeffery, W.R.; Chiba, T.; Krajka, F.R.; Deyts, C.; Satoh, N.; Joly, J.-S. Trunk lateral cells are neural crest-like cells in the ascidian Ciona intestinalis: Insights into the ancestry and evolution of the neural crest. Dev. Biol. 2008, 324, 152–160.
- Kerner, P.; Hung, J.; Béhague, J.; Le Gouar, M.; Balavoine, G.; Vervoort, M. Insights into the evolution of the snail superfamily from metazoan wide molecular phylogenies and expression data in annelids. BMC Evol. Biol. 2009, 9, 94.
- Yamada, L.; Kobayashi, K.; Degnan, B.; Satoh, N.; Satou, Y. A genomewide survey of developmentally relevant genes in Ciona intestinalis. Dev. Genes Evol. 2003, 213, 245–253.
- Nagatomo, K.; Ishibashi, T.; Satou, Y.; Satoh, N.; Fujiwara, S. Retinoic acid affects gene expression and morphogenesis without upregulating the retinoic acid receptor in the ascidian Ciona intestinalis. Mech. Dev. 2003, 120, 363–372.
- McGonnell, I.M.; Graham, A.; Richardson, J.; Fish, J.L.; Depew, M.J.; Dee, C.T.; Holland, P.W.; Takahashi, T. Evolution of the Alx homeobox gene family: Parallel retention and independent loss of the vertebrate Alx3 gene. Evol. Dev. 2011, 13, 343–351.
- Imai, K.S.; Satoh, N.; Satou, Y. A Twist-like bHLH gene is a downstream factor of an endogenous FGF and determines mesenchymal fate in the ascidian embryos. Development 2003, 130, 4461–4472.
- Meulemans, D.; Bronner-Fraser, M. Amphioxus and lamprey AP-2 genes: Implications for neural crest evolution and migration patterns. Development 2002, 129, 4953–4962.
- York, J.R.; Zehnder, K.; Yuan, T.; Lakiza, O.; McCauley, D.W. Evolution of Snail-mediated regulation of neural crest and placodes from an ancient role in bilaterian neurogenesis. Dev. Biol. 2019, 453, 180–190.
- Manzon, L.A.; Youson, J.H.; Holzer, G.; Staiano, L.; Laudet, V.; Manzon, R.G. Thyroid hormone and retinoid X receptor function and expression during sea lamprey (Petromyzon marinus) metamorphosis. Gen. Comp. Endocrinol. 2014, 204, 211–222.
- Singh, P.P.; Isambert, H. OHNOLOGS v2: A comprehensive resource for the genes retained from whole genome duplication in vertebrates. Nucleic Acids Res. 2019, 48, gkz909.
- Holland, P.W.; Garcia-Fernàndez, J.; Williams, N.A.; Sidow, A. Gene duplications and the origins of vertebrate development. Development 1994, 1994, 125–133.
- Smith, J.J.; Timoshevskaya, N.; Ye, C.; Holt, C.; Keinath, M.C.; Parker, H.J.; Cook, M.E.; Hess, J.E.; Narum, S.R.; Lamanna, F.; et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 2018, 50, 270–277.
- Mitchell, J.M.; Sucharov, J.; Pulvino, A.T.; Brooks, E.P.; Gillen, A.E.; Nichols, J.T. The alx3 gene shapes the zebrafish neurocranium by regulating frontonasal neural crest cell differentiation timing. Development 2021, 148, dev197483.
- Beverdam, A.; Brouwer, A.; Reijnen, M.; Korving, J.; Meijlink, F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development 2001, 128, 3975–3986.
- Iyyanar, P.P.R.; Wu, Z.; Lan, Y.; Hu, Y.-C.; Jiang, R. Alx1 Deficient Mice Recapitulate Craniofacial Phenotype and Reveal Developmental Basis of ALX1-Related Frontonasal Dysplasia. Front. Cell Dev. Biol. 2022, 10, 777887.
- Lakiza, O.; Miller, S.; Bunce, A.; Lee, E.M.-J.; McCauley, D.W. SoxE gene duplication and development of the lamprey branchial skeleton: Insights into development and evolution of the neural crest. Dev. Biol. 2011, 359, 149–161.
- Lee, E.M.; Yuan, T.; Ballim, R.D.; Nguyen, K.; Kelsh, R.N.; Medeiros, D.M.; McCauley, D.W. Functional constraints on SoxE proteins in neural crest development: The importance of differential expression for evolution of protein activity. Dev. Biol. 2016, 418, 166–178.
- Cossais, F.; Sock, E.; Hornig, J.; Schreiner, S.; Kellerer, S.; Bösl, M.R.; Russell, S.; Wegner, M. Replacement of mouse Sox10 by the Drosophila ortholog Sox100B provides evidence for co-option of SoxE proteins into vertebrate-specific gene-regulatory networks through altered expression. Dev. Biol. 2010, 341, 267–281.
- Kellerer, S.; Schreiner, S.; Stolt, C.C.; Scholz, S.; Bösl, M.R.; Wegner, M. Replacement of the Sox10 transcription factor by Sox8 reveals incomplete functional equivalence. Development 2006, 133, 2875–2886.
- Stock, D.W.; Ellies, D.L.; Zhao, Z.; Ekker, M.; Ruddle, F.H.; Weiss, K.M. The evolution of the vertebrate Dlx gene family. Proc. Natl. Acad. Sci. USA 1996, 93, 10858–10863.
- Takechi, M.; Adachi, N.; Hirai, T.; Kuratani, S.; Kuraku, S. The Dlx genes as clues to vertebrate genomics and craniofacial evolution. Semin. Cell Dev. Biol. 2013, 24, 110–118.
- Kuraku, S.; Takio, Y.; Sugahara, F.; Takechi, M.; Kuratani, S. Evolution of oropharyngeal patterning mechanisms involving Dlx and endothelins in vertebrates. Dev. Biol. 2010, 341, 315–323.
- Cerny, R.; Cattell, M.; Sauka-Spengler, T.; Bronner-Fraser, M.; Yu, F.; Medeiros, D.M. Evidence for the prepattern/cooption model of vertebrate jaw evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 17262–17267.
- Zhang, G.; Cohn, M.J. Genome duplication and the origin of the vertebrate skeleton. Curr. Opin. Genet. Dev. 2008, 18, 387–393.
- Wada, H.; Makabe, K. Genome duplications of early vertebrates as a possible chronicle of the evolutionary history of the neural crest. Int. J. Biol. Sci. 2006, 2, 133–141.
- Carroll, S.B. Homeotic genes and the evolution of arthropods and chordates. Nature 1995, 376, 479–485.
- Abbasi, A.A. Diversification of four human HOX gene clusters by step-wise evolution rather than ancient whole-genome duplications. Dev. Genes Evol. 2015, 225, 353–357.
- Pervaiz, N.; Shakeel, N.; Qasim, A.; Zehra, R.; Anwar, S.; Rana, N.; Xue, Y.; Zhang, Z.; Bao, Y.; Abbasi, A.A. Evolutionary history of the human multigene families reveals widespread gene duplications throughout the history of animals. BMC Evol. Biol. 2019, 19, 128.
- Abbasi, A.A. Unraveling ancient segmental duplication events in human genome by phylogenetic analysis of multigene families residing on HOX-cluster paralogons. Mol. Phylogenet. Evol. 2010, 57, 836–848.
- Kanzler, B.; Kuschert, S.J.; Liu, Y.-H.; Mallo, M. Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development 1998, 125, 2587–2597.
- Pasqualetti, M.; Ori, M.; Nardi, I.; Rijli, F.M. Ectopic Hoxa2 induction after neural crest migration results in homeosis of jaw elements in Xenopus. Development 2000, 127, 5367–5378.
- Parker, H.J.; Bronner, M.E.; Krumlauf, R. An atlas of anterior hox gene expression in the embryonic sea lamprey head: Hox-code evolution in vertebrates. Dev. Biol. 2019, 453, 19–33.
- Hunter, M.P.; Prince, V.E. Zebrafish Hox Paralogue Group 2 Genes Function Redundantly as Selector Genes to Pattern the Second Pharyngeal Arch. Dev. Biol. 2002, 247, 367–389.
- Minoux, M.; Antonarakis, G.S.; Kmita, M.; Duboule, D.; Rijli, F.M. Rostral and caudal pharyngeal arches share a common neural crest ground pattern. Development 2009, 136, 637–645.
- Couly, G.; Grapin-Botton, A.; Coltey, P.; Ruhin, B.; Douarin, N.M.L. Determination of the identity of the derivatives of the cephalic neural crest: Incompatibility between Hox gene expression and lower jaw development. Development 1998, 125, 3445–3459.
- Kitazawa, T.; Minoux, M.; Ducret, S.; Rijli, F.M. Different Ectopic Hoxa2 Expression Levels in Mouse Cranial Neural Crest Cells Result in Distinct Craniofacial Anomalies and Homeotic Phenotypes. J. Dev. Biol. 2022, 10, 9.
- Singh, N.P.; Krumlauf, R. Diversification and Functional Evolution of HOX Proteins. Front. Cell Dev. Biol. 2022, 10, 798812.
- Braasch, I.; Volff, J.-N.; Schartl, M. The Endothelin System: Evolution of Vertebrate-Specific Ligand–Receptor Interactions by Three Rounds of Genome Duplication. Mol. Biol. Evol. 2009, 26, 783–799.
- Square, T.; Jandzik, D.; Cattell, M.; Hansen, A.; Medeiros, D.M. Embryonic expression of endothelins and their receptors in lamprey and frog reveals stem vertebrate origins of complex Endothelin signaling. Sci. Rep. 2016, 6, 34282.
- Clouthier, D.E.; Garcia, E.; Schilling, T.F. Regulation of facial morphogenesis by endothelin signaling: Insights from mice and fish. Am. J. Med. Genet. A 2010, 152, 2962–2973.
- Hirschberger, C.; Sleight, V.A.; Criswell, K.E.; Clark, S.J.; Gillis, J.A. Conserved and unique transcriptional features of pharyngeal arches in the skate (Leucoraja erinacea) and evolution of the jaw. Mol. Biol. Evol. 2021, 38, 4187–4204.
- Medeiros, D.M.; Crump, J.G. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev. Biol. 2012, 371, 121–135.
- Miller, C.T.; Yelon, D.; Stainier, D.Y.R.; Kimmel, C.B. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development 2003, 130, 1353–1365.
- Khor, J.M.; Ettensohn, C.A. Transcription Factors of the Alx Family: Evolutionarily Conserved Regulators of Deuterostome Skeletogenesis. Front. Genet. 2020, 11, 569314.
More
Information
Subjects:
Developmental Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
643
Revisions:
2 times
(View History)
Update Date:
13 Sep 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




