
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Kostas A. Triantaphyllopoulos | -- | 5664 | 2023-09-08 20:21:34 | | | |
| 2 | Lindsay Dong | -36 word(s) | 5628 | 2023-09-11 03:20:34 | | |
Video Upload Options
Non-coding RNAs (ncRNA) have paved the way to new perspectives on the regulation of gene expression, not only in biology and medicine, but also in associated fields and technologies, ensuring advances in diagnostic means and therapeutic modalities. Critical in this multistep approach are the associations of long non-coding RNA (lncRNA) with diseases and their causal genes in their networks of interactions, gene enrichment and expression analysis, associated pathways, the monitoring of the involved genes and their functional roles during disease progression from one stage to another. Studies have shown that Johne’s Disease (JD), caused by Mycobacterium avium subspecies partuberculosis (MAP), shares common lncRNAs, clinical findings, and other molecular entities with Crohn’s Disease (CD). This has been a subject of vigorous investigation owing to the zoonotic nature of this condition, although results are still inconclusive.
1. Introduction
2. Morphology and Biological Characteristics of Mycobacterium avium
3. MAP, Crohn’s Disease, and Relative Pathologies
4. Long Non-Coding RNAs (LncRNAs) and Their Footprint in Gene Regulation
4.1. LncRNAs in the Disease State
4.2. Principles of Classification for Long Non-Coding RNAs
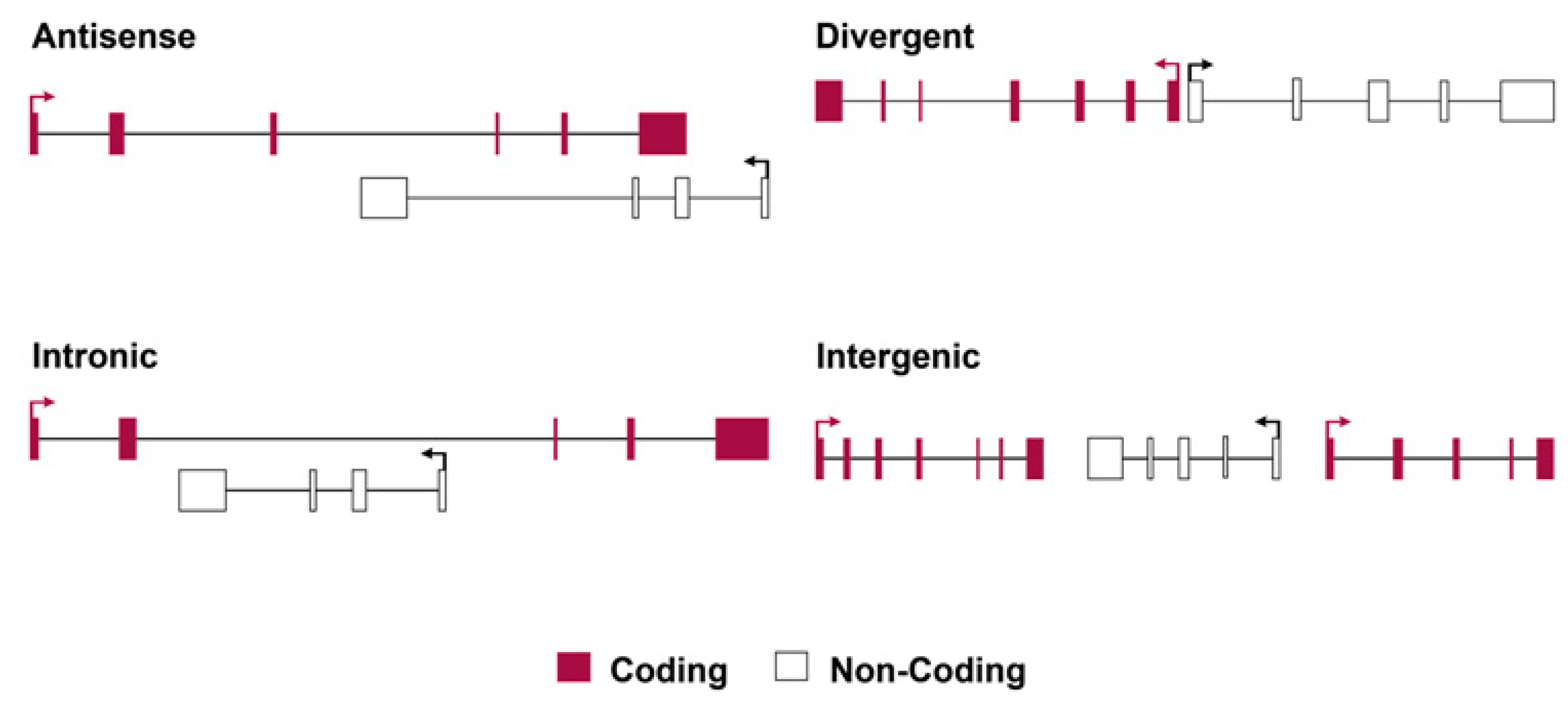
5. LncRNAs’ Involvement in Immune Dysregulation in IBD and JD
5.1. LncRNA Evidence in IBD-Related Pathologies
5.2. LncRNA Evidence from JD-Related Pathologies
5.3. Differences in LncRNA Profiles of CD and JD
A notable example is a variation in the genetic locus of protein tyrosine phosphatase 2 (PTPN2) in UC and IBD, which regulates cytokine signaling by acting on multiple phosphorylated proteins [70]. A study of patients with CD demonstrated a link between the SNP rs2542151 and lower levels of the PTPN2 protein in colonic fibroblasts, as well as the formation of aberrant autophagosomes in intestinal epithelial cells (IECs) [71]. The PTPN2 locus SNP rs2542151 is related to the variation in lncRNA LINC01882, which is primarily expressed in T cells and is also involved in IL-2 expression, affecting important events such as differentiation, immune responses, and the homeostasis of lymphocytes, including Tregs mechanisms. Specifically, the transcript LINC01882 has been reported to play significant roles in autoimmune diseases, including IBD, and in peripheral blood mononuclear cells (PBMCs) of UC patients [72]. Additionally, the lncRNA ROCKI negatively regulates its cognate encoding gene, myristoylated alanine-rich protein kinase C (MARCKS), which promotes inflammatory cytokine and chemokine production. Thus, the above consecutive events show that MARCKS’ gene expression, mediated by ROCKI, contributes to the IBD pathology [73].
Consistent with the above and the contribution of lncRNAs in IBD, which has been also shown in several studies, is their expression profile which can successfully distinguish IBD patients from healthy controls [74]. Furthermore, the transcription characteristics and clinically relevant parameters of lncRNAs indicate that they have strong potential to be used as prognostic biomarkers in IBD [75].
5.4. Expression Profiling in Crohn’s Disease vs. Healthy Controls
6. The Epigenetic Role of the LncRNAs Involved in Human IBD-Related Pathologies and Mycobacterial Infections of the Host
7. The Triarchy of Infection, lncRNA Intervention, and Regulation (IIR)
7.1. The Infection Stage
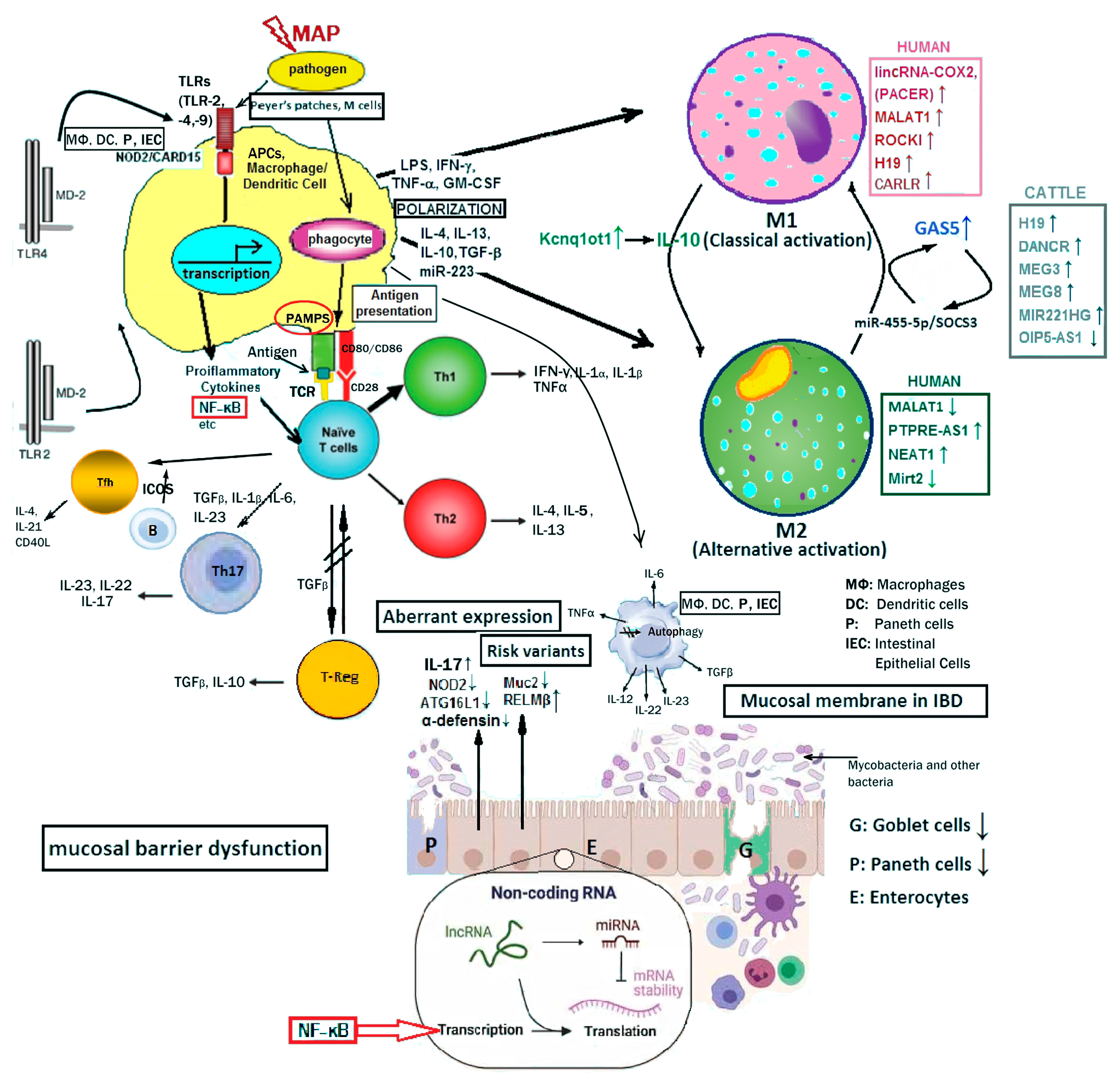
7.2. Signaling Event Pathways and lncRNA Intervention
7.3. Coding Genes and LncRNAs Regulate the Pathological Phenotype Variables
As shown in Figure 2, LncRNAs in humans, such as PACER, MALAT1, HOTAIR and CARLR, directly or indirectly, interact with NF-κB pathway components to regulate target gene transcription. Likewise bovine lncRNAs such as, H19, DANCR, MEG3, MEG8, MIR221HG and OIP5-AS1 are differentially expressed upon MAP challenge to regulate biological functions and disease state.
8. The Prospect of LncRNAs in Contemporary Therapies of GI
9. Conclusions
References
- Cullen, C.M.; Aneja, K.K.; Beyhan, S.; Cho, C.E.; Woloszynek, S.; Convertino, M.; McCoy, S.J.; Zhang, Y.; Anderson, M.Z.; Alvarez-Ponce, D.; et al. Emerging Priorities for Microbiome Research. Front. Microbiol. 2020, 11, 136.
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197.
- Huang, Z.; Liu, J.; Li, L.; Guo, Y.; Luo, Q.; Li, J. Long non-coding RNA expression profiling of macrophage line RAW264.7 infected by Mycobacterium tuberculosis. Biotech. Histochem. 2020, 95, 403–410.
- Wei, L.; Liu, K.; Jia, Q.; Zhang, H.; Bie, Q.; Zhang, B. The Roles of Host Noncoding RNAs in Mycobacterium tuberculosis Infection. Front. Immunol. 2021, 12, 664787.
- Yang, X.; Yang, J.; Wang, J.; Wen, Q.; Wang, H.; He, J.; Hu, S.; He, W.; Du, X.; Liu, S.; et al. Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Sci. Rep. 2016, 6, 38963.
- Pekarek, L.; Torres-Carranza, D.; Fraile-Martinez, O.; Garcia-Montero, C.; Pekarek, T.; Saez, M.A.; Rueda-Correa, F.; Pimentel-Martinez, C.; Guijarro, L.G.; Diaz-Pedrero, R.; et al. An Overview of the Role of MicroRNAs on Carcinogenesis: A Focus on Cell Cycle, Angiogenesis and Metastasis. Int. J. Mol. Sci. 2023, 24, 7268.
- Chen, Y.; Li, Z.; Chen, X.; Zhang, S. Long non-coding RNAs: From disease code to drug role. Acta Pharm. Sin. B 2021, 11, 340–354.
- Zhang, Z.; Shi, S.; Li, J.; Costa, M. Long Non-Coding RNA MEG3 in Metal Carcinogenesis. Toxics 2023, 11, 157.
- Wang, B.; Wang, M.; Jia, S.; Li, T.; Yang, M.; Ge, F. Systematic Survey of the Regulatory Networks of the Long Noncoding RNA BANCR in Cervical Cancer Cells. J. Proteome Res. 2022, 21, 1137–1152.
- Song, X.; Gao, F.; Li, H.; Qin, W.; Chai, C.; Shi, G.; Yang, H. Long noncoding RNA THRIL promotes foam cell formation and inflammation in macrophages. Cell Biol. Int. 2023, 47, 156–166.
- Bates, A.; O’Brien, R.; Liggett, S.; Griffin, F. The effect of sub-clinical infection with Mycobacterium avium subsp. paratuberculosis on milk production in a New Zealand dairy herd. BMC Vet. Res. 2018, 14, 93.
- Zarei-Kordshouli, F.; Geramizadeh, B.; Khodakaram-Tafti, A. Prevalence of Mycobacterium avium subspecies paratuberculosis IS 900 DNA in biopsy tissues from patients with Crohn’s disease: Histopathological and molecular comparison with Johne’s disease in Fars province of Iran. BMC Infect. Dis. 2019, 19, 23.
- King, H.C.; Khera-Butler, T.; James, P.; Oakley, B.B.; Erenso, G.; Aseffa, A.; Knight, R.; Wellington, E.M.; Courtenay, O. Environmental reservoirs of pathogenic mycobacteria across the Ethiopian biogeographical landscape. PLoS ONE 2017, 12, e0173811.
- Riojas, M.A.; McGough, K.J.; Rider-Riojas, C.J.; Rastogi, N.; Hazbon, M.H. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic synonyms of Mycobacterium tuberculosis. Int. J. Syst. Evol. Microbiol. 2018, 68, 324–332.
- Rossini, N.O.; Dias, M.V.B. Mutations and insights into the molecular mechanisms of resistance of Mycobacterium tuberculosis to first-line. Genet. Mol. Biol. 2023, 46, e20220261.
- Mataragka, A.; Leousi, E.; Liandris, E.; Ntafis, V.; Leontides, L.; Aggelidou, E.; Bossis, I.; Triantaphyllopoulos, K.A.; Theodoropoulou, I.; Ikonomopoulos, J. Faecal shedding of Mycobacterium avium subspecies paratuberculosis reduces before parturition in sheep? Small Rumin. Res. 2017, 147, 32–36.
- Mataragka, A.; Sotirakoglou, K.; Gazouli, M.; Triantaphyllopoulos, K.A.; Ikonomopoulos, J. Parturition affects test-positivity in sheep with subclinical paratuberculosis; investigation following a preliminary analysis. J. King Saud Univ. Sci. 2019, 31, 1399–1403.
- Garcia, A.B.; Shalloo, L. Invited review: The economic impact and control of paratuberculosis in cattle. J. Dairy Sci. 2015, 98, 5019–5039.
- Thoen, C.; Jarnagin, J.; Richards, W. Isolation and identification of mycobacteria from porcine tissues: A three-year summary. Am. J. Vet. Res. 1975, 36, 1383–1386.
- Greig, A.; Stevenson, K.; Perez, V.; Pirie, A.; Grant, J.; Sharp, J. Paratuberculosis in wild rabbits (Oryctolagus cuniculus). Vet. Rec. 1997, 140, 141–143.
- Beard, P.; Daniels, M.; Henderson, D.; Pirie, A.; Rudge, K.; Buxton, D.; Rhind, S.; Greig, A.; Hutchings, M.; McKendrick, I.; et al. Paratuberculosis infection of nonruminant wildlife in Scotland. J. Clin. Microbiol. 2001, 39, 1517–1521.
- Fechner, K.; Matz-Rensing, K.; Lampe, K.; Kaup, F.J.; Czerny, C.P.; Schafer, J. Detection of Mycobacterium avium subsp. paratuberculosis in non-human primates. J. Med. Primatol. 2017, 46, 211–217.
- Byrne, A.; Ollier, S.; Tahlan, K.; Biet, F.; Bissonnette, N. Genomic epidemiology of Mycobacterium avium subsp. paratuberculosis isolates from Canadian dairy herds provides evidence for multiple infection events. Front. Genet. 2023, 14, 1043598.
- Davis, W.C. On deaf ears, Mycobacterium avium paratuberculosis in pathogenesis Crohn’s and other diseases. World J. Gastroenterol. 2015, 21, 13411–13417.
- McNees, A.L.; Markesich, D.; Zayyani, N.R.; Graham, D.Y. Mycobacterium paratuberculosis as a cause of Crohn’s disease. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 1523–1534.
- Sechi, L.A.; Dow, C.T. Mycobacterium avium ss. paratuberculosis Zoonosis—The Hundred Year War—Beyond Crohn’s Disease. Front. Immunol. 2015, 6, 96.
- Elmagzoub, W.A.; Idris, S.M.; Isameldin, M.; Arabi, N.; Abdo, A.; Ibrahim, M.; Khan, M.A.A.; Tanneberger, F.; Bakhiet, S.M.; Okuni, J.B.; et al. Mycobacterium avium subsp. paratuberculosis and microbiome profile of patients in a referral gastrointestinal diseases centre in the Sudan. PLoS ONE 2022, 17, e0266533.
- Dow, C.T. Hermon-Taylor: M. paratuberculosis and Crohn’s Disease-The Book of Revelation According to John. Pathogens 2021, 10, 1469.
- Agrawal, G.; Clancy, A.; Huynh, R.; Borody, T. Profound remission in Crohn’s disease requiring no further treatment for 3-23 years: A case series. Gut Pathog. 2020, 12, 16.
- Agrawal, G.; Hamblin, H.; Clancy, A.; Borody, T. Anti-Mycobacterial Antibiotic Therapy Induces Remission in Active Paediatric Crohn’s Disease. Microorganisms 2020, 8, 1112.
- Ekundayo, T.C.; Okoh, A.I. Systematic Assessment of Mycobacterium avium Subspecies Paratuberculosis Infections from 1911-2019: A Growth Analysis of Association with Human Autoimmune Diseases. Microorganisms 2020, 8, 1212.
- Gupta, S.; Chaubey, K.K.; Agarwal, P.; Kuenstner, J.T.; Parashar, D.; Singh, S.V. Therapeutic management of Mycobacterium avium subspecies paratuberculosis infection with complete resolution of symptoms and disease in a patient with advanced inflammatory bowel syndrome. Mol. Biol. Rep. 2021, 48, 7013–7020.
- Savarino, E.; Bertani, L.; Ceccarelli, L.; Bodini, G.; Zingone, F.; Buda, A.; Facchin, S.; Lorenzon, G.; Marchi, S.; Marabotto, E.; et al. Antimicrobial treatment with the fixed-dose antibiotic combination RHB-104 for Mycobacterium avium subspecies paratuberculosis in Crohn’s disease: Pharmacological and clinical implications. Expert Opin. Biol. Ther. 2019, 19, 79–88.
- Eslami, M.; Shafiei, M.; Ghasemian, A.; Valizadeh, S.; Al-Marzoqi, A.H.; Shokouhi Mostafavi, S.K.; Nojoomi, F.; Mirforughi, S.A. Mycobacterium avium paratuberculosis and Mycobacterium avium complex and related subspecies as causative agents of zoonotic and occupational diseases. J. Cell. Physiol. 2019, 234, 12415–12421.
- Timms, V.J.; Daskalopoulos, G.; Mitchell, H.M.; Neilan, B.A. The Association of Mycobacterium avium subsp. paratuberculosis with Inflammatory Bowel Disease. PLoS ONE 2016, 11, e0148731.
- Pierce, E.S. Could Mycobacterium avium subspecies paratuberculosis cause Crohn’s disease, ulcerative colitis… and colorectal cancer? Infect. Agents Cancer 2018, 13, 1.
- Okuni, J.B.; Hansen, S.; Eltom, K.H.; Eltayeb, E.; Amanzada, A.; Omega, J.A.; Czerny, C.P.; Abd El Wahed, A.; Ojok, L. Paratuberculosis: A Potential Zoonosis and a Neglected Disease in Africa. Microorganisms 2020, 8, 1007.
- Zhang, H.; Xue, C.; Wang, Y.; Shi, J.; Zhang, X.; Li, W.; Nunez, S.; Foulkes, A.S.; Lin, J.; Hinkle, C.C.; et al. Deep RNA Sequencing Uncovers a Repertoire of Human Macrophage Long Intergenic Noncoding RNAs Modulated by Macrophage Activation and Associated with Cardiometabolic Diseases. J. Am. Heart Assoc. 2017, 6, e007431.
- Palazzo, A.F.; Koonin, E.V. Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell 2020, 183, 1151–1161.
- Villa, T.; Porrua, O. Pervasive transcription: A controlled risk. FEBS J. 2022, 290, 3723–3736.
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79.
- Li, J.; Xu, Q.; Huang, Z.J.; Mao, N.; Lin, Z.T.; Cheng, L.; Sun, B.; Wang, G. CircRNAs: A new target for the diagnosis and treatment of digestive system neoplasms. Cell Death Dis. 2021, 12, 205.
- Wang, F.; Nazarali, A.J.; Ji, S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 2016, 6, 1167–1176.
- Zhou, Z.; Wang, X.; Hu, Q.; Yang, Z. CircZfp609 contributes to cerebral infarction via sponging miR-145a-5p to regulate BACH1. Metab. Brain Dis. 2023, 38, 1971–1981.
- Das, K.; Garnica, O.; Dhandayuthapani, S. Modulation of Host miRNAs by Intracellular Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 79.
- Aguilar, C.; Mano, M.; Eulalio, A. Multifaceted Roles of microRNAs in Host-Bacterial Pathogen Interaction. Microbiol. Spectr. 2019, 7, 10–1128.
- Kim, J.K.; Kim, T.S.; Basu, J.; Jo, E.K. MicroRNA in innate immunity and autophagy during mycobacterial infection. Cell. Microbiol. 2017, 19, e12687.
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118.
- Chedin, F. Nascent Connections: R-Loops and Chromatin Patterning. Trends Genet. TIG 2016, 32, 828–838.
- Lan, Y.; Xiao, X.; He, Z.; Luo, Y.; Wu, C.; Li, L.; Song, X. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018, 46, 5809–5821.
- Qin, W.; Li, X.; Xie, L.; Li, S.; Liu, J.; Jia, L.; Dong, X.; Ren, X.; Xiao, J.; Yang, C.; et al. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Res. 2016, 44, 6423–6433.
- Sridhar, B.; Rivas-Astroza, M.; Nguyen, T.C.; Chen, W.; Yan, Z.; Cao, X.; Hebert, L.; Zhong, S. Systematic Mapping of RNA-Chromatin Interactions In Vivo. Curr. Biol. 2017, 27, 610–612.
- Asim, M.N.; Ibrahim, M.A.; Imran Malik, M.; Dengel, A.; Ahmed, S. Advances in Computational Methodologies for Classification and Sub-Cellular Locality Prediction of Non-Coding RNAs. Int. J. Mol. Sci. 2021, 22, 8719.
- Laham-Karam, N.; Laitinen, P.; Turunen, T.A.; Yla-Herttuala, S. Activating the Chromatin by Noncoding RNAs. Antioxid. Redox Signal. 2018, 29, 813–831.
- Yang, F.; Yang, Y.; Chen, L.; Zhang, Z.; Liu, L.; Zhang, C.; Mai, Q.; Chen, Y.; Chen, Z.; Lin, T.; et al. The gut microbiota mediates protective immunity against tuberculosis via modulation of lncRNA. Gut Microbes 2022, 14, 2029997.
- Weikard, R.; Demasius, W.; Kuehn, C. Mining long noncoding RNA in livestock. Anim. Genet. 2017, 48, 3–18.
- Koufariotis, L.T.; Chen, Y.P.; Chamberlain, A.; Vander Jagt, C.; Hayes, B.J. A catalogue of novel bovine long noncoding RNA across 18 tissues. PLoS ONE 2015, 10, e0141225.
- Zhou, Z.Y.; Li, A.M.; Adeola, A.C.; Liu, Y.H.; Irwin, D.M.; Xie, H.B.; Zhang, Y.P. Genome-wide identification of long intergenic noncoding RNA genes and their potential association with domestication in pigs. Genome Biol. Evol. 2014, 6, 1387–1392.
- Gruber, A.J.; Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 2019, 20, 599–614.
- Kern, C.; Wang, Y.; Chitwood, J.; Korf, I.; Delany, M.; Cheng, H.; Medrano, J.F.; Van Eenennaam, A.L.; Ernst, C.; Ross, P.; et al. Genome-wide identification of tissue-specific long non-coding RNA in three farm animal species. BMC Genom. 2018, 19, 684.
- Kern, C.; Wang, Y.; Xu, X.; Pan, Z.; Halstead, M.; Chanthavixay, G.; Saelao, P.; Waters, S.; Xiang, R.; Chamberlain, A.; et al. Functional annotations of three domestic animal genomes provide vital resources for comparative and agricultural research. Nat. Commun. 2021, 12, 1821.
- Mele, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37.
- Washietl, S.; Kellis, M.; Garber, M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014, 24, 616–628.
- Ma, S.; Ming, Z.; Gong, A.Y.; Wang, Y.; Chen, X.; Hu, G.; Zhou, R.; Shibata, A.; Swanson, P.C.; Chen, X.M. A long noncoding RNA, lincRNA-Tnfaip3, acts as a coregulator of NF-kappaB to modulate inflammatory gene transcription in mouse macrophages. FASEB J. 2017, 31, 1215–1225.
- Zhao, K.; Tan, J.Y.; Mao, Q.D.; Ren, K.Y.; He, B.G.; Zhang, C.P.; Wei, L.Z. Overexpression of long non-coding RNA TUG1 alleviates TNF-alpha-induced inflammatory injury in interstitial cells of Cajal. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 312–320.
- Yu, X.; Zheng, H.; Chan, M.T.; Wu, W.K. HULC: An oncogenic long non-coding RNA in human cancer. J. Cell. Mol. Med. 2017, 21, 410–417.
- Gupta, P.; Peter, S.; Jung, M.; Lewin, A.; Hemmrich-Stanisak, G.; Franke, A.; von Kleist, M.; Schutte, C.; Einspanier, R.; Sharbati, S.; et al. Analysis of long non-coding RNA and mRNA expression in bovine macrophages brings up novel aspects of Mycobacterium avium subspecies paratuberculosis infections. Sci. Rep. 2019, 9, 1571.
- Marete, A.; Ariel, O.; Ibeagha-Awemu, E.; Bissonnette, N. Identification of Long Non-coding RNA Isolated From Naturally Infected Macrophages and Associated with Bovine Johne’s Disease in Canadian Holstein Using a Combination of Neural Networks and Logistic Regression. Front. Vet. Sci. 2021, 8, 639053.
- Talyan, S.; Andrade-Navarro, M.A.; Muro, E.M. Identification of transcribed protein coding sequence remnants within lincRNAs. Nucleic Acids Res. 2018, 46, 8720–8729.
- Bussieres-Marmen, S.; Vinette, V.; Gungabeesoon, J.; Aubry, I.; Perez-Quintero, L.A.; Tremblay, M.L. Loss of T-cell protein tyrosine phosphatase in the intestinal epithelium promotes local inflammation by increasing colonic stem cell proliferation. Cell. Mol. Immunol. 2018, 15, 367–376.
- Lin, L.; Zhou, G.; Chen, P.; Wang, Y.; Han, J.; Chen, M.; He, Y.; Zhang, S. Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease? Cell Death Dis. 2020, 11, 456.
- Houtman, M.; Shchetynsky, K.; Chemin, K.; Hensvold, A.H.; Ramskold, D.; Tandre, K.; Eloranta, M.L.; Ronnblom, L.; Uebe, S.; Catrina, A.I.; et al. T cells are influenced by a long non-coding RNA in the autoimmune associated PTPN2 locus. J. Autoimmun. 2018, 90, 28–38.
- Zhang, Q.; Chao, T.C.; Patil, V.S.; Qin, Y.; Tiwari, S.K.; Chiou, J.; Dobin, A.; Tsai, C.M.; Li, Z.; Dang, J.; et al. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019, 38, e100041.
- Xu, J.; Xu, H.M.; Yang, M.F.; Liang, Y.J.; Peng, Q.Z.; Zhang, Y.; Tian, C.M.; Wang, L.S.; Yao, J.; Nie, Y.Q.; et al. New Insights Into the Epigenetic Regulation of Inflammatory Bowel Disease. Front. Pharmacol. 2022, 13, 813659.
- Yarani, R.; Mirza, A.H.; Kaur, S.; Pociot, F. The emerging role of lncRNAs in inflammatory bowel disease. Exp. Mol. Med. 2018, 50, 1–14.
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47.
- Qiao, C.; Yang, L.; Wan, J.; Liu, X.; Pang, C.; You, W.; Zhao, G. Long noncoding RNA ANRIL contributes to the development of ulcerative colitis by miR-323b-5p/TLR4/MyD88/NF-kappaB pathway. Biochem. Biophys. Res. Commun. 2019, 508, 217–224.
- Wang, S.; Hou, Y.; Chen, W.; Wang, J.; Xie, W.; Zhang, X.; Zeng, L. KIF9-AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease. Mol. Med. Rep. 2018, 17, 2195–2202.
- Gomes, C.P.; Nobrega-Pereira, S.; Domingues-Silva, B.; Rebelo, K.; Alves-Vale, C.; Marinho, S.P.; Carvalho, T.; Dias, S.; Bernardes de Jesus, B. An antisense transcript mediates MALAT1 response in human breast cancer. BMC Cancer 2019, 19, 771.
- Li, Y.; Zhu, L.; Chen, P.; Wang, Y.; Yang, G.; Zhou, G.; Li, L.; Feng, R.; Qiu, Y.; Han, J.; et al. MALAT1 Maintains the Intestinal Mucosal Homeostasis in Crohn’s Disease via the miR-146b-5p-CLDN11/NUMB Pathway. J. Crohn’s Colitis 2021, 15, 1542–1557.
- Bure, I.V.; Nemtsova, M.V.; Kuznetsova, E.B. Histone Modifications and Non-Coding RNAs: Mutual Epigenetic Regulation and Role in Pathogenesis. Int. J. Mol. Sci. 2022, 23, 5801.
- Triantaphyllopoulos, K.A.; Ikonomopoulos, I.; Bannister, A.J. Epigenetics and inheritance of phenotype variation in livestock. Epigenetics Chromatin 2016, 9, 31.
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9.
- Bujold, D.; Morais, D.A.L.; Gauthier, C.; Cote, C.; Caron, M.; Kwan, T.; Chen, K.C.; Laperle, J.; Markovits, A.N.; Pastinen, T.; et al. The International Human Epigenome Consortium Data Portal. Cell Syst. 2016, 3, 496–499 e492.
- Consortium, E.P.; Bernstein, B.E.; Birney, E.; Dunham, I.; Green, E.D.; Gunter, C.; Snyder, M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74.
- Gonzalez-Serna, D.; Villanueva-Martin, G.; Acosta-Herrera, M.; Marquez, A.; Martin, J. Approaching Shared Pathophysiology in Immune-Mediated Diseases through Functional Genomics. Genes 2020, 11, 1482.
- Deng, W.; Zhang, Y.; Cai, J.; Zhang, J.; Liu, X.; Yin, J.; Bai, Z.; Yao, H.; Zhang, Z. LncRNA-ANRIL promotes gastric cancer progression by enhancing NF-kB signaling. Exp. Biol. Med. 2019, 244, 953–959.
- Rajeev, R.; Dwivedi, A.P.; Sinha, A.; Agarwaal, V.; Dev, R.R.; Kar, A.; Khosla, S. Epigenetic interaction of microbes with their mammalian hosts. J. Biosci. 2021, 46, 94.
- Ramos, G.P.; Papadakis, K.A. Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin. Proc. 2019, 94, 155–165.
- Schoultz, I.; Keita, A.V. Cellular and Molecular Therapeutic Targets in Inflammatory Bowel Disease-Focusing on Intestinal Barrier Function. Cells 2019, 8, 193.
- Nakamura, Y.; Kimura, S.; Hase, K. M cell-dependent antigen uptake on follicle-associated epithelium for mucosal immune surveillance. Inflamm. Regen. 2018, 38, 15.
- Nishino, K.; Nishida, A.; Inoue, R.; Kawada, Y.; Ohno, M.; Sakai, S.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Kawahara, M.; et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 2018, 53, 95–106.
- Gao, P.; Liu, H.; Huang, H.; Sun, Y.; Jia, B.; Hou, B.; Zhou, X.; Strober, W.; Zhang, F. The Crohn Disease-associated ATG16L1(T300A) polymorphism regulates inflammatory responses by modulating TLR- and NLR-mediated signaling. Autophagy 2022, 18, 2561–2575.
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51.
- Qu, X.; Zheng, C.; Wang, B.; Wang, F.; Sun, X.; Gao, Y.; Xia, Q.; Kong, X. Comprehensive analysis of circular RNAs from steatotic livers after ischemia and reperfusion injury by next-generation RNA sequencing. FEBS Lett. 2021, 595, 99–109.
- Wehkamp, J.; Stange, E.F. An Update Review on the Paneth Cell as Key to Ileal Crohn’s Disease. Front. Immunol. 2020, 11, 646.
- Ntunzwenimana, J.C.; Boucher, G.; Paquette, J.; Gosselin, H.; Alikashani, A.; Morin, N.; Beauchamp, C.; Thauvette, L.; Rivard, M.E.; Dupuis, F.; et al. Functional screen of inflammatory bowel disease genes reveals key epithelial functions. Genome Med. 2021, 13, 181.
- Seva, J.; Sanes, J.M.; Mas, A.; Ramis, G.; Sanchez, J.; Parraga-Ros, E. Prevalence of Mycobacterium avium Subsp. paratuberculosis in Feral Pigeons (Columba livia) Associated with Difficulties Controlling Paratuberculosis in a Bovine Herd (Fighting Bull Breed). Animals 2022, 12, 3314.
- Johnson, A.M.F.; DePaolo, R.W. Infectious Scarring: Setting the Trigger for Intestinal Inflammation. Cell Host Microbe 2018, 23, 154–156.
- Yang, W.H.; Heithoff, D.M.; Aziz, P.V.; Sperandio, M.; Nizet, V.; Mahan, M.J.; Marth, J.D. Recurrent infection progressively disables host protection against intestinal inflammation. Science 2017, 358, eaao5610.
- Ge, Q.; Dong, Y.; Lin, G.; Cao, Y. Long Noncoding RNA Antisense Noncoding RNA in the INK4 Locus Correlates With Risk, Severity, Inflammation and Infliximab Efficacy in Crohn’s Disease. Am. J. Med. Sci. 2019, 357, 134–142.
- Aslam, N.; Lo, S.W.; Sikafi, R.; Barnes, T.; Segal, J.; Smith, P.J.; Limdi, J.K. A review of the therapeutic management of ulcerative colitis. Ther. Adv. Gastroenterol. 2022, 15, 17562848221138160.




