Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Agustín Valenzuela-fernández | -- | 3090 | 2023-09-01 19:19:35 | | | |
| 2 | Peter Tang | Meta information modification | 3090 | 2023-09-04 03:24:22 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Valenzuela-Fernández, A.; Cabrera-Rodríguez, R.; Casado, C.; Pérez-Yanes, S.; Pernas, M.; García-Luis, J.; Marfil, S.; Olivares, I.; Estévez-Herrera, J.; Trujillo-González, R.; et al. Contribution of the HIV-1 Envelope Glycoprotein to AIDS. Encyclopedia. Available online: https://encyclopedia.pub/entry/48750 (accessed on 08 February 2026).
Valenzuela-Fernández A, Cabrera-Rodríguez R, Casado C, Pérez-Yanes S, Pernas M, García-Luis J, et al. Contribution of the HIV-1 Envelope Glycoprotein to AIDS. Encyclopedia. Available at: https://encyclopedia.pub/entry/48750. Accessed February 08, 2026.
Valenzuela-Fernández, Agustín, Romina Cabrera-Rodríguez, Concha Casado, Silvia Pérez-Yanes, María Pernas, Jonay García-Luis, Silvia Marfil, Isabel Olivares, Judith Estévez-Herrera, Rodrigo Trujillo-González, et al. "Contribution of the HIV-1 Envelope Glycoprotein to AIDS" Encyclopedia, https://encyclopedia.pub/entry/48750 (accessed February 08, 2026).
Valenzuela-Fernández, A., Cabrera-Rodríguez, R., Casado, C., Pérez-Yanes, S., Pernas, M., García-Luis, J., Marfil, S., Olivares, I., Estévez-Herrera, J., Trujillo-González, R., Blanco, J., & Lopez-Galindez, C. (2023, September 01). Contribution of the HIV-1 Envelope Glycoprotein to AIDS. In Encyclopedia. https://encyclopedia.pub/entry/48750
Valenzuela-Fernández, Agustín, et al. "Contribution of the HIV-1 Envelope Glycoprotein to AIDS." Encyclopedia. Web. 01 September, 2023.
Copy Citation
In the absence of antiviral therapy, HIV-1 infection progresses to a wide spectrum of clinical manifestations that are the result of an entangled contribution of host, immune and viral factors. The contribution of these factors is not completely established. Several investigations have described the involvement of the immune system in the viral control. In addition, distinct HLA-B alleles, HLA-B27, -B57-58, were associated with infection control. The combination of these elements and antiviral host restriction factors results in different clinical outcomes.
HIV-1 Env function
natural control of the infection
elite controllers
1. Introduction
In the absence of antiretroviral therapy (ART), the hallmark of human immunodeficiency virus type 1 (HIV-1) infection is the gradual destruction of the naive and memory CD4+ T-lymphocytes and the associated immunological abnormalities leading to the acquired immunodeficiency syndrome (AIDS) [1][2][3][4][5][6][7][8][9][10][11] (Figure 1, progressors immune system damage, top box). The severity of the symptoms and viral transmission strongly correlate with the peak of viral load (VL) during primary infection and the subsequent viral set-point [12][13][14][15][16][17][18][19][20][21][22][23][24]. HIV-1 infection is characterized by a wide spectrum of disease outcomes according to the progression time of patients. Different nomenclatures have been used to name the distinct groups of HIV-1 individuals (reviewed in [25]). The typical HIV-1 infected patient, in the absence of ART, progresses to AIDS and death over a period of about 8–10 years after seroconversion [5][26][27]. Some patients, designated rapid progressor (RP), progress to AIDS within three years of primary infection [26][28][29]. On the other side, there is a small subset of HIV-1 individuals that are able to permanently control viral replication and clinical progression and might never progress or progress very slowly [5][30] (Figure 1, non-progressors immune system damage, bottom box). In general, these subjects have been infected with HIV-1 for more than ten years, maintaining high CD4+ lymphocyte numbers, undetectable VL, without clinical symptoms, and remaining therapy naïve [31]. These individuals have been defined as long-term non-progressors (LTNPs) [32][33][34][35][36], elite controllers (ECs) [30][37], slow progressors [38][39][40], HIV controllers (HICs) [41] and elite suppressors [42]. Within this set of individuals, some subgroups can be distinguished in terms of VL: viremic LTNPs or viremic controllers (vLTNPs or LTNP-VCs) with VLs between 50 and 2000 copies/mL [32], LTNPs viremic non-controllers (LTNP-NCs) with VLs above 2000 copies/mL [43], and LTNP-Elite controllers (LTNP-ECs) with undetectable viral loads (<50 copies/mL) [26]. In the LTNP-ECs subgroup, there is a natural control of the infection without any ART, maintaining undetectable HIV-1 viral loads for long periods of time (even for more than 20–30 years) and lack of clinical progression [44]. This clinical phenotype is the consequence of the necessary cooperative interaction of host, immune, and viral factors [26][30][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66] (Figure 1, non-progressors bottom box). Several investigations described the contribution of the immune system, both at the cellular and serological level, in the primary and the subsequent control of the viral infection. This control is the result of many elements and the activity of different cell types, such as CD4+ and CD8+ T cells, natural killers (NKs), dendritic cells (DCs), different types of antibodies (Abs), cell restrictions factors, human leucocyte antigens (HLAs) genotypes and/or host factors like CCR5 protective mutations [26][44][45][48][49][50][51][52][53][54][56][61][66][67][68][69][70][71][72][73], as summarized in Figure 1. In addition, HLA-B genotypes HLA-B57/B58 or -B27 [63], HLA-B*35:01 [74][75] and HLA-C [26][76][77], such as the HLA-C*03:02 1 in an African Pediatric Population [78], are linked with the control of HIV-1 infection (Figure 1, non-progressors bottom box). In some LTNP individuals [79] that harbor viruses with low replication capacity [80][81][82][83], the HIV-1 LTNP phenotype has been associated with the presence of potent and broad cytotoxic T lymphocyte (CTL) responses [66][84] (Figure 1, non-progressors bottom box) and active NK cells.
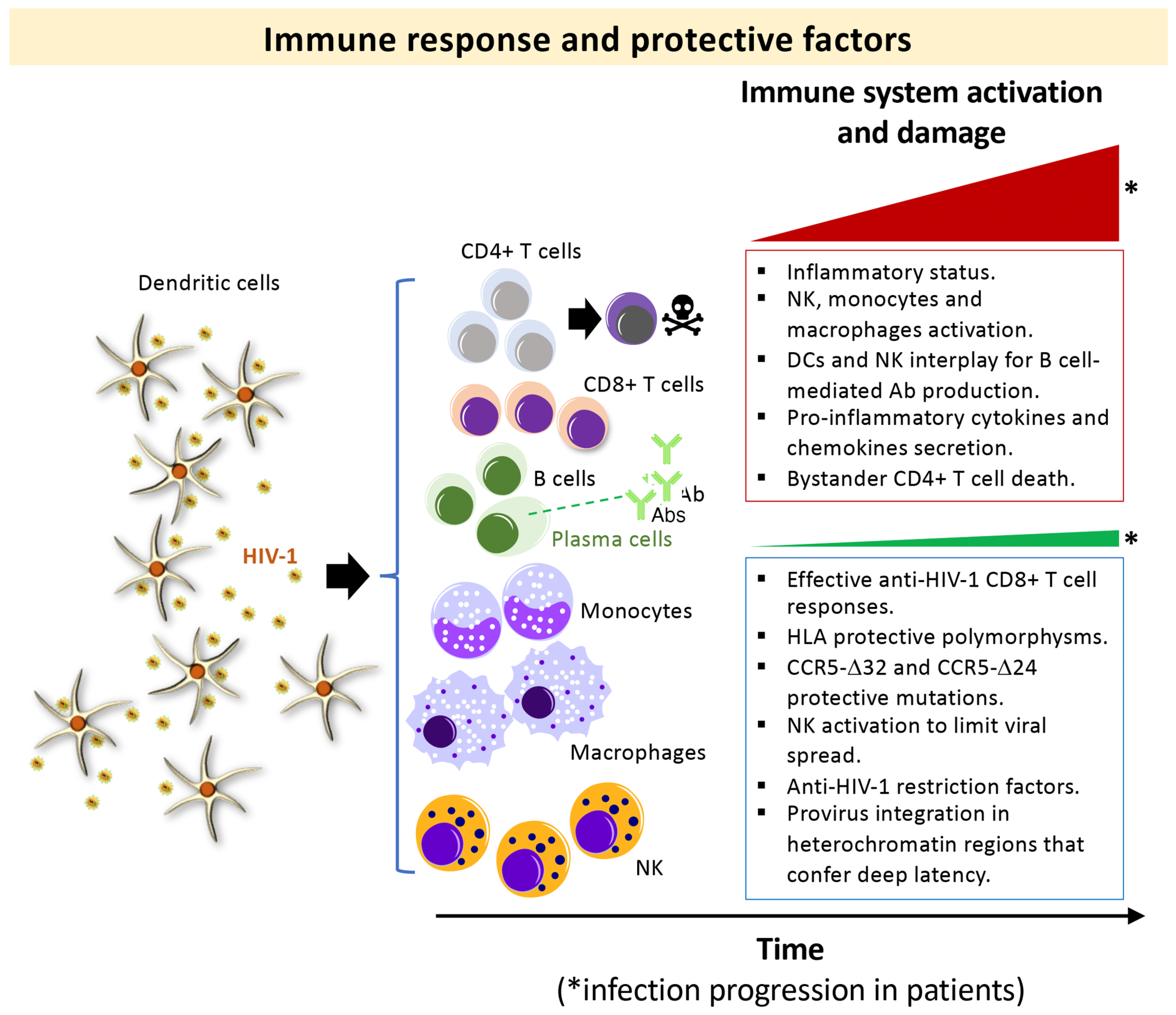
Figure 1. Scheme summarizing the events in primary HIV-1 infection associated with the immune control and damage, in HIV-1 progressor and non-progressor phenotypes: Main immune responses and damage associated with progressors HIV-1 infected individuals (top box) and non-progressors (bottom box) clinical phenotypes. Top box, in an acute phase of the HIV-1 infection, antigen-elicited rapid responses of innate immune cells lead to the activation of natural killer (NK) cell receptors together with monocytes/macrophages and the release of inflammatory cytokines/chemokines. This HIV-1 elicited-NK cells activation leads to secretion of IFN-γ and MIP-1β to limit viral spread [85], modulate the adaptive response in an interplay with DCs [86], and shape the induction of antibodies through the elimination of follicular T cells (Tfh) [87]. Macrophages and microglia that survive after acute HIV-1 infection could become viral reservoirs [88]. High VL is associated with predominant destruction of bystander non-infected CD4+ T mediated by HIV-1 Env (reviewed in [2][89][90]), a process the researchers have previously reported to be dependent on the HIV-1 gp41-Env subunit by promoting autophagy and apoptosis [91][92][93][94][95][96]. Bottom box, some host factors, such as viral antigen-elicited CD8+ T cell response and Th1-type cytokine production [66][97][98][99], NK cell receptors [67], HLA polymorphisms [61] and CCR5 protective mutations (i.e., homo and heterozygous CCR5-Δ32 and heterozygous CCR5-Δ24 deletions/genotypes) [45][48][49][50][51][52][53][54][56] together with a limited pro-viral reservoir [68] and some restriction factors [69] have been related to a protective phenotype against HIV-1 infection. *Red and *green triangles represent how the viral infection progresses in HIV-1 infected patients of extreme different clinical phenotype: progressors (red triangle) and non-progressors (green triangle). The boxes summarized the main associated immune responses observed for the related clinical phenotype.
2. HIV-1 Envs from LTNP-EC Individuals Present Inefficient Viral Functions, Associated with the Natural Control of the Infection and the Non-Progressor Clinical Phenotype
The investigation of the HIV-1 env/Env functions was undertaken analyzing viruses from HIV-1 individuals with different clinical phenotypes: LTNP-EC, viremic non-progressor, progressor and rapid progressor (RPs) [26][44][70][71][72][73][83][100][101][102][103][104][105][106]. In the initial studies, the researchers focused on LTNP individuals infected for long periods of time (i.e., more than 10 years and with more than 25–30 years of clinical follow-up) [26][44][70][72][100][106]. The isolated viral env sequences (full-length viral env) from infected individuals were cloned into expression plasmids (Figure 2a). The viral clones were completely sequenced at the nucleotide level and submitted to phylogenetic analysis (Figure 2a). These viral Envs were then characterized by multiple phenotypic test/assays to disclose the principal properties of their viruses (Figure 2b–e). In order to do this, the researchers developed several techniques to study the functions of viral Env during the first steps of the viral cycle. This phase of the viral cycle is a complex multistage process with highly regulated steps involving many cellular molecules mobilized by the Env viral binding to CD4, CXCR4 or CCR5 receptors [107][108][109][110][111][112][113][114][115][116][117][118]. As the researchers and others described, these interactions end in the formation of a fusion pore through which the viral capsid enters the cell [102][104][119][120][121]. The efficient pore fusion formation relies on key signals triggered by Env-CD4 interaction promoting cytoskeleton modifications [102][104][119][121], such as microtubules (MTs) acetylation in the α-tubulin subunit [121], F-actin severing, and capping reorganization [102][104]. A deficiency in these HIV-1-Env-mediated signals leads to a defect in the early steps of viral infection and replication [102][104][119][120][122][123][124], which ends in a limited viral replication.
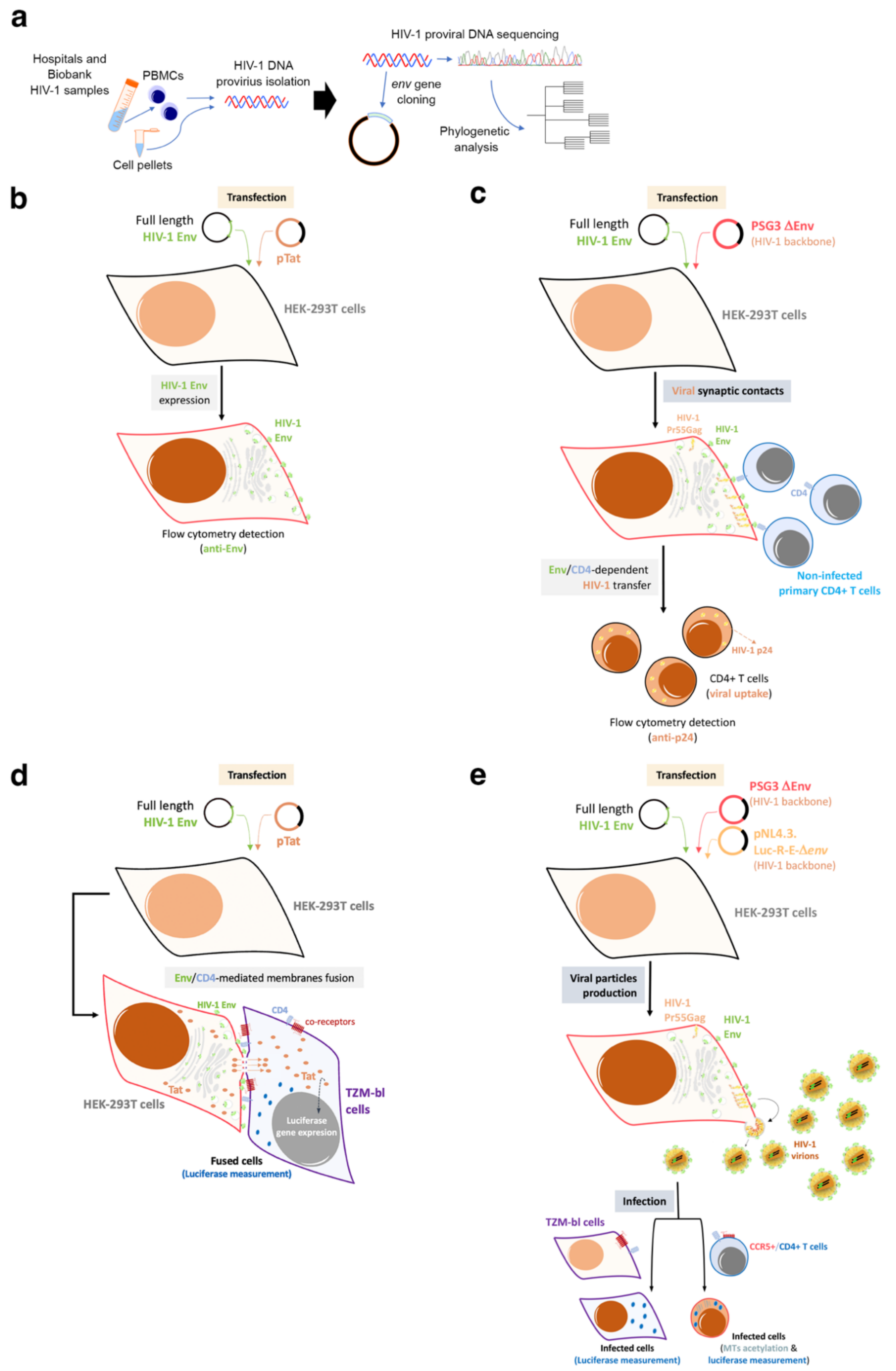
Figure 2. Schemes of the experimental procedures followed to characterize primary HIV-1 envs/Envs from HIV-1 infected patients: The researchers studied viral Envs of viruses from individuals with different clinical outcomes, such as LTNP-ECs, vLTNPs, VNPs, Progressors and RPs to investigate the role of the viral Env protein in HIV-1 pathogenesis. (a) The researchers cloned full-length viral envs in expression plasmids from the viruses of these individuals. The viral clones were completely sequenced at the nucleotide level and submitted to phylogenetic analysis. These viral envs/Envs were analyzed by multiple phenotypic characterizations to see the principal properties of their viruses as presented in the following panels; (b) Env expression: HEK-293T cells were co-transfected with a ptat Δenv HIV-1 expression plasmid together with reference or primary full-length viral env HIV-1 expression plasmid. By using the specific anti-Env antibody, cell-surface Env expression was analyzed by flow cytometry; (c) Env-mediated viral transfer: HEK-293T cells producing HIV-1 virions bearing reference or primary Envs were co-cultured with primary CD4+ T cells. Flow cytometry was used together with specific anti-p24 antibody to measure HIV-1 transfer to CD4+ T cells; (d) Env-mediated fusion activity: HEK-293T cells transfected with the env defective pSG3-HIV-1 backbone and primary envs plasmids (i.e., producing HIV-1 virions) or cells over-expressing the viral Env together with Tat viral protein (pTat construct) were co-cultured with target TZM-bl cells. Then, Env fusion capacity was measured by the magnitude of Tat-induced Luciferase activity in fused cells; (e) Env-mediated viral infection: TZM-bl cells were infected with serial dilutions of HIV-1 virions isolated from HEK-293T cells cotransfected with Δenv pSG3-HIV-1 and with primary or reference HIV-1 Envs. Infectivity capacity was determined in TZM-bl cells by measuring the luciferase activity in HIV-1 infected TZM-bl cells or in CD4+ T CEM.NKR-CCR5 cells by quantifying luciferase activity associated with the enter infectious Δenv pNL4-3.Luc.R-E- pseudovirus carrying primary Env.
Based on these experimental strategies, the researchers reported that Envs from viruses of a cluster of LTNP-EC individuals showed a limited ability to induce both cortical F-actin reorganization and capping, and a low signal stabilizing acetylated MTs in the α-tubulin subunit [100] (Figure 3). These HIV-1 Envs were well expressed at the cell-surface of virus producing cells (Figure 2b). These data correlated with the fact that these Envs were not able to bind to CD4 with high affinity [70][71][72][100][102][104][119][121] (Figure 2c–e and Figure 3). The inability of these ECs’ HIV-1 Envs to bind to CD4 and trigger cell signals to reorganize and modify the cytoskeleton, to generate a pseudopod where cytoskeleton and cell-surface receptors for HIV-1 infection concentrate [100][102][104] (Figure 3), accounts for the defect of the HIV-1 Envs for the promotion of pore fusion formation and transfer of viral material to primary non-infected CD4+ T cells [100] (Figure 2c,d and Figure 3). In addition, a significant correlation was observed between the HIV-1 transfer values, mediated by Env/CD4 binding capacity and fusogenicity, thus linking the fusion defect to a low CD4 affinity (Figure 2c,d). Before characterizing these mechanisms underlying inefficient HIV-1 Envs, the researchers reported non-functional viral Envs in a previous study with non-progressor individuals [26][73][83]. Taken together, these data further confirm the deficient Env fusion capacity observed in the cluster LTNP envelopes, correlating with the inefficiency to infect target cells and replicate at high levels [26][100] (Figure 2e and Figure 3).
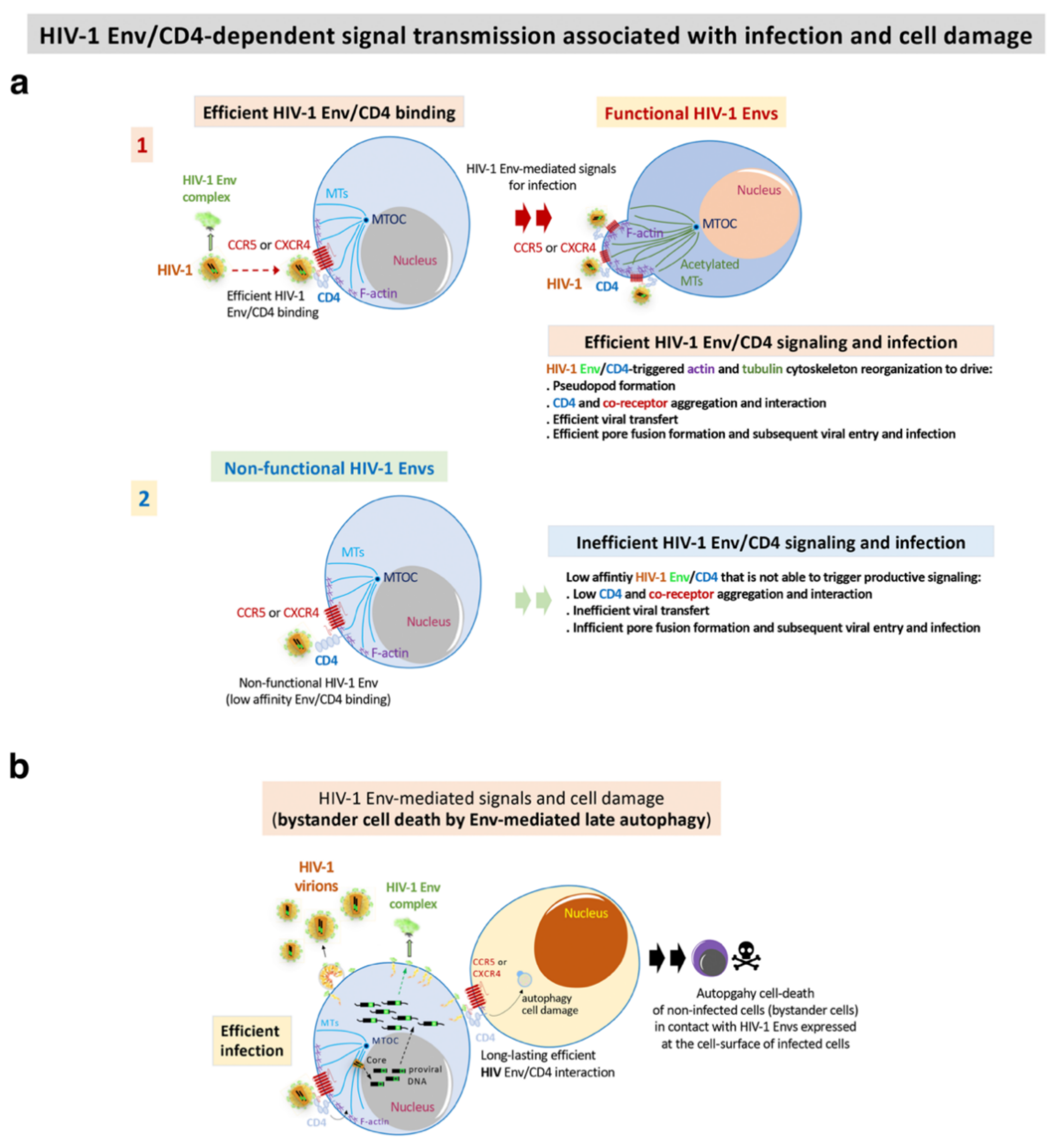
Figure 3. HIV-1 Env-mediated productive cell signals and cell damage: (a) functional HIV-1 Envs of viral isolates from progressors (progressors and RPs) and viremic (VNP) patients efficiently binds to CD4 for the promotion of the F-actin and MTs reorganizations and post-translational modifications. This signal drives pseudopod formation in CD4+ T cells where CD4 and chemokine co-receptors for HIV-1 infection reorganize, aggregate and interact (step 1) [70][72][100][102][104][105][119][121]. These events are required for efficient pore fusion formation, viral transfer, virus entry and infection. On the contrary, non-functional HIV-1 Envs of viral isolates from non-progressors’ patients (LTNP-ECs, LTNP and vLTNP)) are not able to bind to CD4 with high affinity, and thereby are unable to reorganize the cytoskeleton and favour all the events for productive viral transfer and infection (step 2) [70][72][100]; (b) functional HIV-1 Envs of viral isolates from viremic and progressors (VNP and RPs) patients efficiently bind to CD4 to trigger late autophagy with subsequent cell death of non-infected CD4+ T cells (bystander cells) by contact [72].
3. Fully Functional HIV-1 Envs Are Linked to Viremia and Progressor Clinical Phenotypes
For the investigation into the role of viral Env in the control of HIV-1 infection and pathogenesis, the researchers also analyzed viral envs/Envs from other sets of viruses from non-clustered LTNP-EC individuals, followed for more than ten years, in comparison with viruses from patients infected at the same period of time but with progressor phenotypes [70].
In contrast to the Envs from the LTNP EC subjects, the viruses from the progressor individuals (viremic and progressors) showed the opposite properties, with a good affinity for CD4, cell fusion and viral transfer [70][72][100][106]. Therefore, functional HIV-1 Envs are associated with infectious virus and cytopathic activity [71][72], which characterize viremic and progressor/RPs clinical outcomes [70][72][100][106] (Figure 3 and Figure 4). Functional HIV-1 Envs favor the accumulation of mutations that could result in function gains of the Envs and the evasion from immune responses (Figure 3 and Figure 4).
During viral transmission to a new host, a selection for viral variants with shorter variable regions and a reduced degree of PNGs occurs in viruses from the HIV-1 subtype B [125]. An increase in viral infectivity and replication capacity has been associated with genetic variability in the env gene [126][127][128][129][130][131]. This viral replication could favor the gain of function of the HIV-1 env by increasing viral fitness and could result in the escape from the immune response and ART [132][133][134][135][136][137][138][139][140]. In the studies, the researchers detected the loss of the N362 PNGs (HXB2 isolate; group M, subtype B (HIV-1 M:B_HXB2R:NCBI:txid11706)), which is frequently observed in the Envs of non-progressor phenotypes (EC and viremic patients) and in long-lasting progressors, but not in HIV-1 Envs from more recent progressors. This change could be related to a gain of functionality observed in these Envs [70]. However, in a study of some Australian viruses presenting the N362 glycosylation site, the viruses showed efficient fusion and transfer capacity [141]. These data reflect the significant effect that point mutations could have in the viral characteristics and HIV pathogenesis [70][100][142][143]. These results indicate that deficient viral Envs are associated with non-progressor, controller individuals and that fully functional HIV-1 Envs are mainly linked to viremic and progressor clinical phenotypes.
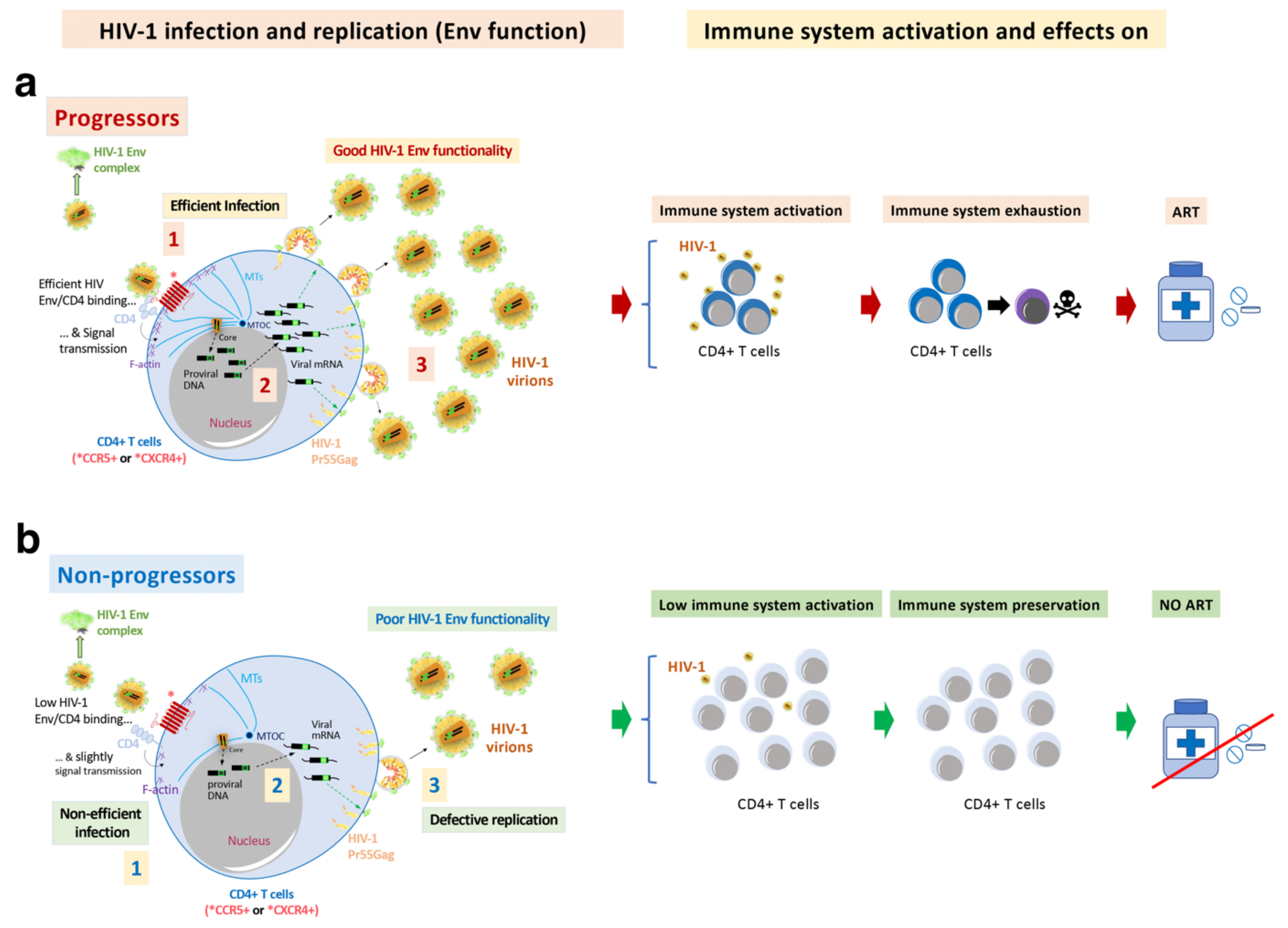
Figure 4. HIV-1 Env function correlates with clinical outcome, immune system activation, exhaustion and cell damage: (a) The researchers' works indicate that HIV-1 Envs of viruses from viremic and progressors patients (i.e., Progressors, RPs and VNPs) are associated with efficient viral infection (step 1, in Progressors) and replication (step 3, in Progressors), favouring viral diversity (step 2, in Progressors) and Env gain of functionality (step 1). This gain is linked with an increasing length of the variable loops (VLs) of the gp120 subunit of the Env viral complex and of N-linked glycosylation sites (PNGs) over the course of the epidemic. These functional Envs trigger cell signals activating target CD4+ T cells. Long-lasting activation leads to immune exhaustion. Progressors HIV-1 patients must follow ART, in order to avoid the development of AIDS; (b) On the contrary, non-functional HIV-1 Envs are associated with viruses of non-progressors HIV-1 individuals (i.e., LTNPs, LTNP-ECs and viremic LTNPs (vLTNPs)) presenting deficiencies in infection (step 1, in non-progressors), viral diversity (step 2, in non-progressors) and replication (step 3, in non-progressors). The HIV-1 envs’ sequences of LTNP-ECs, after thirty years of natural control of viral replication and viral pathogenesis, are close to the sequence of the T/F virus. This poor Env functionality could help the immune system to control the infection, preserving its functions. There are LTNP individuals and particularly LTNP-ECs or exceptional ECs that control HIV-1 infection for up to thirty years without any ART [44]. In this last study as well as in other investigating cases of HIV-1 functional cure, although there was not a direct analysis of Env functionality, all individuals showed undetectable VLs, and defects in viral replication and in the viral genome [44][68][144][145].
4. Role of the Viral Env Complex in Signal Transmission in Other Cellular Process and Cell Death
In addition to these direct viral effects on viral replication, the Env complex is also associated with other important cellular processes like fusion pore formation and autophagy/cell death dysregulation. These processes are summarized in Figure 3. In the characterization of these HIV-1 Envs properties, the researchers analyzed HIV-1 Envs from viremic non-progressors (VNPs), progressors and rapid progressors (RPs) patients [44][70][72][73][100][106]. The phenotypic characterization of the Envs from HIV-1 progressors (Figure 3 and Figure 4) indicated higher replication capacity for these HIV-1 viruses when compared with HIV-1 Envs of viruses from the LTNP-EC cluster [100]. HIV-1 Envs from progressors patients are associated with functional Env showing efficient CD4 binding and signaling that promote cytoskeleton reorganization and the formation of the pseudopod-hot region. This process allows an efficient HIV-1 infection (Figure 3a) which leads to higher fusogenic, viral transfer and infection capacities than viruses from LTNP-ECs [70][100]. It is noteworthy that the researchers reported that the HIV-1 gp41-Env subunit promotes bystander cell-death by autophagy and apoptosis [91][92][93][94][95][96]. The researchers' works indicate that functional HIV-1 Envs from VNPs and RPs promote bystander cell death in uninfected CD4+ T cells by triggering late autophagy [71][72] (Figure 3b). In line with this Env functionality, the researchers found significant env gene diversity in sequences isolated from VNPs compared with RPs, correlating with the efficient ability of these VNP HIV-1 Envs to infect and favor virus replication [72]. A similar observation has been reported indicating that viral population diversity remains higher in VNPs compared to standard progressors or RPs [146]. Persistent HIV-1 replication in the presence of supposedly efficient immune responses in VNPs is expected to lead to the accumulation of mutations to compensate viral fitness cost, which could result in a continuous Env escape from neutralizing Abs [147][148]. Furthermore, in the late AIDS phase of chronic infection in RPs, uncontrolled HIV-1 replication occurs together with the selection of the fittest variants [149]. Therefore, functional HIV-1 Envs are directly associated with infectious viruses of viremic and progressors patients in which HIV-1 infection evolves [70][71][72][106] (Figure 1, Figure 3 and Figure 4).
This efficient viral function of the HIV-1 Env, in a CD4 dependent manner, allows the virus to overcome cell barriers that limit HIV-1 Env-mediated pore fusion formation, viral entry to the cell, infection and replication (Figure 3a). A key restriction factor for HIV-1 infection that the researchers characterized is the cytoplasmic enzyme HDAC6 (histone deacetylase 6) [70][71][72][105][106][121][150], and more recently the transactive response of the DNA-binding protein (TARDBP or TDP-43) together with HDAC6 (i.e., the TDP-43/HDAC6 axis) [106]. An increase in the expression of functional TDP-43 concomitantly enhances the levels of mRNA and protein of HDAC6, leading to a diminution of the activity of functional HIV-1 Envs from viruses of VNP and RP individuals, reaching the levels of the inefficient Env from LTNP-EC individuals [106]. Silencing of the endogenous TDP-43 strongly reduces the levels of mRNA and of the HDAC6 enzyme [106], stabilizing acetylated MTs that favor the infection activity of primary HIV-1 Envs of VNP, progressors and even of non-functional Envs from LTNP-EC individuals [106]. This last observation suggests that defective viral features observed in a virus of LTNP-ECs [70][71][100][151] are possibly also modulated by the TDP-43/HDAC6 axis [106]. The TDP-43/HDAC6 axis therefore regulates cell permissivity to HIV-1 infection. This point may have negative consequences in HIV-1 LTNP-EC individuals, particularly if a negative regulation of TDP-43 occurs with a concomitant decrease in HDAC6 that would render cells more permissive against inefficient LTNP-EC Envs. Consistently, it has been reported that the ability of the viral Env to trigger signals that overcome the HDAC6 barrier is directly related to its fusion and infection activities [70][72][100][105][121]. The TDP-43/HDAC6 axis could be another factor that is worth exploring in EC individuals that lose the natural control of the infection [70][72][100][105][121][152].
References
- Simon, V.; Ho, D.D.; Abdool Karim, Q. HIV/AIDS epidemiology, pathogenesis, prevention, and treatment. Lancet 2006, 368, 489–504.
- Douek, D.C.; Picker, L.J.; Koup, R.A. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 2003, 21, 265–304.
- Moir, S.; Chun, T.W.; Fauci, A.S. Pathogenic mechanisms of HIV disease. Annu. Rev. Pathol. 2011, 6, 223–248.
- McCune, J.M. The dynamics of CD4+ T-cell depletion in HIV disease. Nature 2001, 410, 974–979.
- Deeks, S.G.; Overbaugh, J.; Phillips, A.; Buchbinder, S. HIV infection. Nat. Rev. Dis. Primers 2015, 1, 15035.
- Ramratnam, B.; Bonhoeffer, S.; Binley, J.; Hurley, A.; Zhang, L.; Mittler, J.E.; Markowitz, M.; Moore, J.P.; Perelson, A.S.; Ho, D.D. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 1999, 354, 1782–1785.
- Simon, V.; Ho, D.D. HIV-1 dynamics in vivo: Implications for therapy. Nat. Rev. Microbiol. 2003, 1, 181–190.
- Sepkowitz, K.A. AIDS—The first 20 years. N. Engl. J. Med. 2001, 344, 1764–1772.
- Hazenberg, M.D.; Hamann, D.; Schuitemaker, H.; Miedema, F. T cell depletion in HIV-1 infection: How CD4+ T cells go out of stock. Nat. Immunol. 2000, 1, 285–289.
- Borkowsky, W.; Rigaud, M.; Krasinski, K.; Moore, T.; Lawrence, R.; Pollack, H. Cell-mediated and humoral immune responses in children infected with human immunodeficiency virus during the first four years of life. J. Pediatr. 1992, 120, 371–375.
- Musey, L.K.; Krieger, J.N.; Hughes, J.P.; Schacker, T.W.; Corey, L.; McElrath, M.J. Early and persistent human immunodeficiency virus type 1 (HIV-1)-specific T helper dysfunction in blood and lymph nodes following acute HIV-1 infection. J. Infect. Dis. 1999, 180, 278–284.
- Kelley, C.F.; Barbour, J.D.; Hecht, F.M. The relation between symptoms, viral load, and viral load set point in primary HIV infection. J. Acquir. Immune Defic. Syndr. 2007, 45, 445–448.
- Katzenstein, T.L.; Pedersen, C.; Nielsen, C.; Lundgren, J.D.; Jakobsen, P.H.; Gerstoft, J. Longitudinal serum HIV RNA quantification: Correlation to viral phenotype at seroconversion and clinical outcome. Aids 1996, 10, 167–173.
- Sterling, T.R.; Vlahov, D.; Astemborski, J.; Hoover, D.R.; Margolick, J.B.; Quinn, T.C. Initial plasma HIV-1 RNA levels and progression to AIDS in women and men. N. Engl. J. Med. 2001, 344, 720–725.
- Prins, H.A.B.; Verbon, A.; Boucher, C.A.B.; Rokx, C. Ending the epidemic: Critical role of primary HIV infection. Neth J. Med. 2017, 75, 321–327.
- Andersson, S.; Norrgren, H.; da Silva, Z.; Biague, A.; Bamba, S.; Kwok, S.; Christopherson, C.; Biberfeld, G.; Albert, J. Plasma viral load in HIV-1 and HIV-2 singly and dually infected individuals in Guinea-Bissau, West Africa: Significantly lower plasma virus set point in HIV-2 infection than in HIV-1 infection. Arch. Intern. Med. 2000, 160, 3286–3293.
- O’Brien, W.A.; Hartigan, P.M.; Martin, D.; Esinhart, J.; Hill, A.; Benoit, S.; Rubin, M.; Simberkoff, M.S.; Hamilton, J.D. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. Veterans Affairs Cooperative Study Group on AIDS. N. Engl. J. Med. 1996, 334, 426–431.
- McPhee, E.; Grabowski, M.K.; Gray, R.H.; Ndyanabo, A.; Ssekasanvu, J.; Kigozi, G.; Makumbi, F.; Serwadda, D.; Quinn, T.C.; Laeyendecker, O. Short Communication: The Interaction of HIV Set Point Viral Load and Subtype on Disease Progression. AIDS Res. Hum. Retroviruses 2019, 35, 49–51.
- Ananworanich, J.; Chomont, N.; Eller, L.A.; Kroon, E.; Tovanabutra, S.; Bose, M.; Nau, M.; Fletcher, J.L.K.; Tipsuk, S.; Vandergeeten, C.; et al. HIV DNA Set Point is Rapidly Established in Acute HIV Infection and Dramatically Reduced by Early ART. EBioMedicine 2016, 11, 68–72.
- Robb, M.L.; Ananworanich, J. Lessons from acute HIV infection. Curr. Opin. HIV AIDS 2016, 11, 555–560.
- Quinn, T.C.; Wawer, M.J.; Sewankambo, N.; Serwadda, D.; Li, C.; Wabwire-Mangen, F.; Meehan, M.O.; Lutalo, T.; Gray, R.H. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 2000, 342, 921–929.
- Wawer, M.J.; Gray, R.H.; Sewankambo, N.K.; Serwadda, D.; Li, X.; Laeyendecker, O.; Kiwanuka, N.; Kigozi, G.; Kiddugavu, M.; Lutalo, T.; et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J. Infect. Dis. 2005, 191, 1403–1409.
- Cohen, M.S.; Pilcher, C.D. Amplified HIV transmission and new approaches to HIV prevention. J. Infect. Dis. 2005, 191, 1391–1393.
- Mellors, J.W.; Rinaldo, C.R., Jr.; Gupta, P.; White, R.M.; Todd, J.A.; Kingsley, L.A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 1996, 272, 1167–1170.
- Gurdasani, D.; Iles, L.; Dillon, D.G.; Young, E.H.; Olson, A.D.; Naranbhai, V.; Fidler, S.; Gkrania-Klotsas, E.; Post, F.A.; Kellam, P.; et al. A systematic review of definitions of extreme phenotypes of HIV control and progression. AIDS 2014, 28, 149–162.
- Casado, C.; Colombo, S.; Rauch, A.; Martinez, R.; Gunthard, H.F.; Garcia, S.; Rodriguez, C.; Del Romero, J.; Telenti, A.; Lopez-Galindez, C. Host and viral genetic correlates of clinical definitions of HIV-1 disease progression. PLoS ONE 2010, 5, e11079.
- Beyrer, C.; Sullivan, P.; Sanchez, J.; Baral, S.D.; Collins, C.; Wirtz, A.L.; Altman, D.; Trapence, G.; Mayer, K. The increase in global HIV epidemics in MSM. AIDS 2013, 27, 2665–2678.
- Fontaine, J.; Coutlee, F.; Tremblay, C.; Routy, J.P.; Poudrier, J.; Roger, M.; Montreal Primary, H.I.V.I.; Long-Term Nonprogressor Study, G. HIV infection affects blood myeloid dendritic cells after successful therapy and despite nonprogressing clinical disease. J. Infect. Dis. 2009, 199, 1007–1018.
- Lajoie, J.; Fontaine, J.; Tremblay, C.; Routy, J.P.; Poudrier, J.; Roger, M. Persistence of high levels of blood soluble human leukocyte antigen-G is associated with rapid progression of HIV infection. AIDS 2009, 23, 1437–1440.
- Deeks, S.G.; Walker, B.D. Human immunodeficiency virus controllers: Mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 2007, 27, 406–416.
- Buchbinder, S.P.; Katz, M.H.; Hessol, N.A.; O’Malley, P.M.; Holmberg, S.D. Long-term HIV-1 infection without immunologic progression. AIDS 1994, 8, 1123–1128.
- Okulicz, J.F.; Marconi, V.C.; Landrum, M.L.; Wegner, S.; Weintrob, A.; Ganesan, A.; Hale, B.; Crum-Cianflone, N.; Delmar, J.; Barthel, V.; et al. Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV natural history study. J. Infect. Dis. 2009, 200, 1714–1723.
- Grabar, S.; Selinger-Leneman, H.; Abgrall, S.; Pialoux, G.; Weiss, L.; Costagliola, D. Prevalence and comparative characteristics of long-term nonprogressors and HIV controller patients in the French Hospital Database on HIV. AIDS 2009, 23, 1163–1169.
- Canducci, F.; Marinozzi, M.C.; Sampaolo, M.; Berre, S.; Bagnarelli, P.; Degano, M.; Gallotta, G.; Mazzi, B.; Lemey, P.; Burioni, R.; et al. Dynamic features of the selective pressure on the human immunodeficiency virus type 1 (HIV-1) gp120 CD4-binding site in a group of long term non progressor (LTNP) subjects. Retrovirology 2009, 6, 4.
- Diop, G.; Hirtzig, T.; Do, H.; Coulonges, C.; Vasilescu, A.; Labib, T.; Spadoni, J.L.; Therwath, A.; Lathrop, M.; Matsuda, F.; et al. Exhaustive genotyping of the interferon alpha receptor 1 (IFNAR1) gene and association of an IFNAR1 protein variant with AIDS progression or susceptibility to HIV-1 infection in a French AIDS cohort. Biomed. Pharmacother. 2006, 60, 569–577.
- Limou, S.; Le Clerc, S.; Coulonges, C.; Carpentier, W.; Dina, C.; Delaneau, O.; Labib, T.; Taing, L.; Sladek, R.; Deveau, C.; et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J. Infect. Dis. 2009, 199, 419–426.
- Rotger, M.; Dang, K.K.; Fellay, J.; Heinzen, E.L.; Feng, S.; Descombes, P.; Shianna, K.V.; Ge, D.; Gunthard, H.F.; Goldstein, D.B.; et al. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 2010, 6, e1000781.
- Kamya, P.; Boulet, S.; Tsoukas, C.M.; Routy, J.P.; Thomas, R.; Cote, P.; Boulassel, M.R.; Baril, J.G.; Kovacs, C.; Migueles, S.A.; et al. Receptor-ligand requirements for increased NK cell polyfunctional potential in slow progressors infected with HIV-1 coexpressing KIR3DL1*h/*y and HLA-B*57. J. Virol. 2011, 85, 5949–5960.
- Gillespie, G.M.; Kaul, R.; Dong, T.; Yang, H.B.; Rostron, T.; Bwayo, J.J.; Kiama, P.; Peto, T.; Plummer, F.A.; McMichael, A.J.; et al. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS 2002, 16, 961–972.
- Zhang, Z.; Jiang, Y.; Zhang, M.; Liu, J.; Sun, G.; Shi, W.; Wang, Y.; Shang, H. Alterations of CD4(+) CD25(+) Foxp3(+) regulatory T cells in HIV-infected slow progressors of former blood donors in China. Microbiol. Immunol. 2010, 54, 625–633.
- Lambotte, O.; Boufassa, F.; Madec, Y.; Nguyen, A.; Goujard, C.; Meyer, L.; Rouzioux, C.; Venet, A.; Delfraissy, J.F.; Group, S.-H.S. HIV controllers: A homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin. Infect. Dis. 2005, 41, 1053–1056.
- Blankson, J.N. Control of HIV-1 replication in elite suppressors. Discov. Med. 2010, 9, 261–266.
- Shaw, J.M.; Hunt, P.W.; Critchfield, J.W.; McConnell, D.H.; Garcia, J.C.; Pollard, R.B.; Somsouk, M.; Deeks, S.G.; Shacklett, B.L. Increased frequency of regulatory T cells accompanies increased immune activation in rectal mucosae of HIV-positive noncontrollers. J. Virol. 2011, 85, 11422–11434.
- Casado, C.; Galvez, C.; Pernas, M.; Tarancon-Diez, L.; Rodriguez, C.; Sanchez-Merino, V.; Vera, M.; Olivares, I.; De Pablo-Bernal, R.; Merino-Mansilla, A.; et al. Permanent control of HIV-1 pathogenesis in exceptional elite controllers: A model of spontaneous cure. Sci. Rep. 2020, 10, 1902.
- Samson, M.; Libert, F.; Doranz, B.J.; Rucker, J.; Liesnard, C.; Farber, C.M.; Saragosti, S.; Lapoumeroulie, C.; Cognaux, J.; Forceille, C.; et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 1996, 382, 722–725.
- Hutter, G.; Nowak, D.; Mossner, M.; Ganepola, S.; Mussig, A.; Allers, K.; Schneider, T.; Hofmann, J.; Kucherer, C.; Blau, O.; et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009, 360, 692–698.
- Gupta, R.K.; Abdul-Jawad, S.; McCoy, L.E.; Mok, H.P.; Peppa, D.; Salgado, M.; Martinez-Picado, J.; Nijhuis, M.; Wensing, A.M.J.; Lee, H.; et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature 2019, 568, 244–248.
- Dean, M.; Carrington, M.; Winkler, C.; Huttley, G.A.; Smith, M.W.; Allikmets, R.; Goedert, J.J.; Buchbinder, S.P.; Vittinghoff, E.; Gomperts, E.; et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 1996, 273, 1856–1862.
- Liu, R.; Paxton, W.A.; Choe, S.; Ceradini, D.; Martin, S.R.; Horuk, R.; MacDonald, M.E.; Stuhlmann, H.; Koup, R.A.; Landau, N.R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 1996, 86, 367–377.
- Masquelier, C.; Servais, J.Y.; Rusanganwa, E.; Roman, F.; Havuga, E.; Servais, J.; Tuyizere, S.; Omes, C.; Karasi, J.C.; Coruteille, O.; et al. A novel 24-base pair deletion in the coding region of CCR5 in an African population. AIDS 2007, 21, 111–113.
- Arendt, V.; Amand, M.; Iserentant, G.; Lemaire, M.; Masquelier, C.; Ndayisaba, G.F.; Verhofstede, C.; Karita, E.; Allen, S.; Chevigne, A.; et al. Predominance of the heterozygous CCR5 delta-24 deletion in African individuals resistant to HIV infection might be related to a defect in CCR5 addressing at the cell surface. J. Int. AIDS Soc. 2019, 22, e25384.
- Huang, Y.; Paxton, W.A.; Wolinsky, S.M.; Neumann, A.U.; Zhang, L.; He, T.; Kang, S.; Ceradini, D.; Jin, Z.; Yazdanbakhsh, K.; et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 1996, 2, 1240–1243.
- Gonzalez, E.; Bamshad, M.; Sato, N.; Mummidi, S.; Dhanda, R.; Catano, G.; Cabrera, S.; McBride, M.; Cao, X.H.; Merrill, G.; et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc. Natl. Acad. Sci. USA 1999, 96, 12004–12009.
- Ioannidis, J.P.; Rosenberg, P.S.; Goedert, J.J.; Ashton, L.J.; Benfield, T.L.; Buchbinder, S.P.; Coutinho, R.A.; Eugen-Olsen, J.; Gallart, T.; Katzenstein, T.L.; et al. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3′A alleles on HIV-1 disease progression: An international meta-analysis of individual-patient data. Ann. Intern. Med. 2001, 135, 782–795.
- Misrahi, M.; Teglas, J.P.; N’Go, N.; Burgard, M.; Mayaux, M.J.; Rouzioux, C.; Delfraissy, J.F.; Blanche, S. CCR5 chemokine receptor variant in HIV-1 mother-to-child transmission and disease progression in children. French Pediatric HIV Infection Study Group. JAMA 1998, 279, 277–280.
- Ruiz-Mateos, E.; Tarancon-Diez, L.; Alvarez-Rios, A.I.; Dominguez-Molina, B.; Genebat, M.; Pulido, I.; Abad, M.A.; Munoz-Fernandez, M.A.; Leal, M. Association of heterozygous CCR5Delta32 deletion with survival in HIV-infection: A cohort study. Antiviral Res. 2018, 150, 15–19.
- Ding, J.; Liu, Y.; Lai, Y. Knowledge From London and Berlin: Finding Threads to a Functional HIV Cure. Front. Immunol. 2021, 12, 688747.
- Autran, B.; Descours, B.; Avettand-Fenoel, V.; Rouzioux, C. Elite controllers as a model of functional cure. Curr. Opin. HIV AIDS 2011, 6, 181–187.
- Canoui, E.; Lecuroux, C.; Avettand-Fenoel, V.; Gousset, M.; Rouzioux, C.; Saez-Cirion, A.; Meyer, L.; Boufassa, F.; Lambotte, O.; Noel, N.; et al. A Subset of Extreme Human Immunodeficiency Virus (HIV) Controllers Is Characterized by a Small HIV Blood Reservoir and a Weak T-Cell Activation Level. Open Forum Infect. Dis. 2017, 4, ofx064.
- Mendoza, D.; Johnson, S.A.; Peterson, B.A.; Natarajan, V.; Salgado, M.; Dewar, R.L.; Burbelo, P.D.; Doria-Rose, N.A.; Graf, E.H.; Greenwald, J.H.; et al. Comprehensive analysis of unique cases with extraordinary control over HIV replication. Blood 2012, 119, 4645–4655.
- International, H.I.V.C.S.; Pereyra, F.; Jia, X.; McLaren, P.J.; Telenti, A.; de Bakker, P.I.; Walker, B.D.; Ripke, S.; Brumme, C.J.; Pulit, S.L.; et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010, 330, 1551–1557.
- Fellay, J.; Shianna, K.V.; Ge, D.; Colombo, S.; Ledergerber, B.; Weale, M.; Zhang, K.; Gumbs, C.; Castagna, A.; Cossarizza, A.; et al. A whole-genome association study of major determinants for host control of HIV-1. Science 2007, 317, 944–947.
- Migueles, S.A.; Sabbaghian, M.S.; Shupert, W.L.; Bettinotti, M.P.; Marincola, F.M.; Martino, L.; Hallahan, C.W.; Selig, S.M.; Schwartz, D.; Sullivan, J.; et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 2000, 97, 2709–2714.
- Genovese, L.; Nebuloni, M.; Alfano, M. Cell-Mediated Immunity in Elite Controllers Naturally Controlling HIV Viral Load. Front. Immunol. 2013, 4, 86.
- Migueles, S.A.; Laborico, A.C.; Shupert, W.L.; Sabbaghian, M.S.; Rabin, R.; Hallahan, C.W.; Van Baarle, D.; Kostense, S.; Miedema, F.; McLaughlin, M.; et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 2002, 3, 1061–1068.
- Saez-Cirion, A.; Lacabaratz, C.; Lambotte, O.; Versmisse, P.; Urrutia, A.; Boufassa, F.; Barre-Sinoussi, F.; Delfraissy, J.F.; Sinet, M.; Pancino, G.; et al. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc. Natl. Acad. Sci. USA 2007, 104, 6776–6781.
- Martin, M.P.; Naranbhai, V.; Shea, P.R.; Qi, Y.; Ramsuran, V.; Vince, N.; Gao, X.; Thomas, R.; Brumme, Z.L.; Carlson, J.M.; et al. Killer cell immunoglobulin-like receptor 3DL1 variation modifies HLA-B*57 protection against HIV-1. J. Clin. Investig. 2018, 128, 1903–1912.
- Jiang, C.; Lian, X.; Gao, C.; Sun, X.; Einkauf, K.B.; Chevalier, J.M.; Chen, S.M.Y.; Hua, S.; Rhee, B.; Chang, K.; et al. Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature 2020, 585, 261–267.
- Abdel-Mohsen, M.; Raposo, R.A.; Deng, X.; Li, M.; Liegler, T.; Sinclair, E.; Salama, M.S.; Ghanem Hel, D.; Hoh, R.; Wong, J.K.; et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology 2013, 10, 106.
- Perez-Yanes, S.; Pernas, M.; Marfil, S.; Cabrera-Rodriguez, R.; Ortiz, R.; Urrea, V.; Rovirosa, C.; Estevez-Herrera, J.; Olivares, I.; Casado, C.; et al. The Characteristics of the HIV-1 Env Glycoprotein Are Linked With Viral Pathogenesis. Front. Microbiol. 2022, 13, 763039.
- Cabrera-Rodriguez, R.; Perez-Yanes, S.; Estevez-Herrera, J.; Marquez-Arce, D.; Cabrera, C.; Espert, L.; Blanco, J.; Valenzuela-Fernandez, A. The Interplay of HIV and Autophagy in Early Infection. Front. Microbiol. 2021, 12, 661446.
- Cabrera-Rodriguez, R.; Hebmann, V.; Marfil, S.; Pernas, M.; Marrero-Hernandez, S.; Cabrera, C.; Urrea, V.; Casado, C.; Olivares, I.; Marquez-Arce, D.; et al. HIV-1 envelope glycoproteins isolated from Viremic Non-Progressor individuals are fully functional and cytopathic. Sci. Rep. 2019, 9, 5544.
- Casado, C.; Pernas, M.; Sandonis, V.; Alvaro-Cifuentes, T.; Olivares, I.; Fuentes, R.; Martinez-Prats, L.; Grau, E.; Ruiz, L.; Delgado, R.; et al. Identification of a cluster of HIV-1 controllers infected with low replicating viruses. PLoS ONE 2013, 8, e77663.
- Murakoshi, H.; Koyanagi, M.; Akahoshi, T.; Chikata, T.; Kuse, N.; Gatanaga, H.; Rowland-Jones, S.L.; Oka, S.; Takiguchi, M. Impact of a single HLA-A*24:02-associated escape mutation on the detrimental effect of HLA-B*35:01 in HIV-1 control. EBioMedicine 2018, 36, 103–112.
- Kuse, N.; Murakoshi, H.; Akahoshi, T.; Chikata, T.; James, K.L.; Gatanaga, H.; Rowland-Jones, S.L.; Oka, S.; Takiguchi, M. Collaboration of a Detrimental HLA-B*35:01 Allele with HLA-A*24:02 in Coevolution of HIV-1 with T Cells Leading to Poorer Clinical Outcomes. J. Virol. 2021, 95, e0125921.
- Nehete, P.N.; Lewis, D.E.; Tang, D.N.; Pollack, M.S.; Sastry, K.J. Presence of HLA-C-restricted cytotoxic T-lymphocyte responses in long-term nonprogressors infected with human immunodeficiency virus. Viral Immunol. 1998, 11, 119–129.
- Malnati, M.S.; Ugolotti, E.; Monti, M.C.; Battista, D.; Vanni, I.; Bordo, D.; Sironi, F.; Larghero, P.; Marco, E.D.; Biswas, P.; et al. Activating Killer Immunoglobulin Receptors and HLA-C: A successful combination providing HIV-1 control. Sci. Rep. 2017, 7, 42470.
- Kyobe, S.; Mwesigwa, S.; Kisitu, G.P.; Farirai, J.; Katagirya, E.; Mirembe, A.N.; Ketumile, L.; Wayengera, M.; Katabazi, F.A.; Kigozi, E.; et al. Exome Sequencing Reveals a Putative Role for HLA-C*03:02 in Control of HIV-1 in African Pediatric Populations. Front. Genet. 2021, 12, 720213.
- Alexander, L.; Weiskopf, E.; Greenough, T.C.; Gaddis, N.C.; Auerbach, M.R.; Malim, M.H.; O’Brien, S.J.; Walker, B.D.; Sullivan, J.L.; Desrosiers, R.C. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J. Virol. 2000, 74, 4361–4376.
- Lassen, K.G.; Lobritz, M.A.; Bailey, J.R.; Johnston, S.; Nguyen, S.; Lee, B.; Chou, T.; Siliciano, R.F.; Markowitz, M.; Arts, E.J. Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog. 2009, 5, e1000377.
- Curriu, M.; Fausther-Bovendo, H.; Pernas, M.; Massanella, M.; Carrillo, J.; Cabrera, C.; López-Galíndez, C.; Clotet, B.; Debré, P.; Vieillard, V.; et al. Viremic HIV infected individuals with high CD4 T cells and functional envelope proteins show anti-gp41 antibodies with unique specificity and function. PLoS ONE 2012, 7, e30330.
- Miura, T.; Brumme, Z.L.; Brockman, M.A.; Rosato, P.; Sela, J.; Brumme, C.J.; Pereyra, F.; Kaufmann, D.E.; Trocha, A.; Block, B.L.; et al. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J. Virol. 2010, 84, 7581–7591.
- Sandonis, V.; Casado, C.; Alvaro, T.; Pernas, M.; Olivares, I.; Garcia, S.; Rodriguez, C.; del Romero, J.; Lopez-Galindez, C. A combination of defective DNA and protective host factors are found in a set of HIV-1 ancestral LTNPs. Virology 2009, 391, 73–82.
- Sáez-Cirión, A.; Bacchus, C.; Hocqueloux, L.; Avettand-Fenoel, V.; Girault, I.; Lecuroux, C.; Potard, V.; Versmisse, P.; Melard, A.; Prazuck, T.; et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 2013, 9, e1003211.
- Quillay, H.; El Costa, H.; Duriez, M.; Marlin, R.; Cannou, C.; Madec, Y.; de Truchis, C.; Rahmati, M.; Barre-Sinoussi, F.; Nugeyre, M.T.; et al. NK cells control HIV-1 infection of macrophages through soluble factors and cellular contacts in the human decidua. Retrovirology 2016, 13, 39.
- Quaranta, M.G.; Napolitano, A.; Sanchez, M.; Giordani, L.; Mattioli, B.; Viora, M. HIV-1 Nef impairs the dynamic of DC/NK crosstalk: Different outcome of CD56(dim) and CD56(bright) NK cell subsets. FASEB J. 2007, 21, 2323–2334.
- Rydyznski, C.; Daniels, K.A.; Karmele, E.P.; Brooks, T.R.; Mahl, S.E.; Moran, M.T.; Li, C.; Sutiwisesak, R.; Welsh, R.M.; Waggoner, S.N. Generation of cellular immune memory and B-cell immunity is impaired by natural killer cells. Nat. Commun. 2015, 6, 6375.
- Castellano, P.; Prevedel, L.; Eugenin, E.A. HIV-infected macrophages and microglia that survive acute infection become viral reservoirs by a mechanism involving Bim. Sci. Rep. 2017, 7, 12866.
- Moanna, A.; Dunham, R.; Paiardini, M.; Silvestri, G. CD4+ T-cell depletion in HIV infection: Killed by friendly fire? Curr. HIV/AIDS Rep. 2005, 2, 16–23.
- Okoye, A.A.; Picker, L.J. CD4(+) T-cell depletion in HIV infection: Mechanisms of immunological failure. Immunol. Rev. 2013, 254, 54–64.
- Denizot, M.; Varbanov, M.; Espert, L.; Robert-Hebmann, V.; Sagnier, S.; Garcia, E.; Curriu, M.; Mamoun, R.; Blanco, J.; Biard-Piechaczyk, M. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy 2008, 4, 998–1008.
- Cunyat, F.; Curriu, M.; Marfil, S.; Garcia, E.; Clotet, B.; Blanco, J.; Cabrera, C. Evaluation of the cytopathicity (fusion/hemifusion) of patient-derived HIV-1 envelope glycoproteins comparing two effector cell lines. J. Biomol. Screen. 2012, 17, 727–737.
- Cunyat, F.; Marfil, S.; García, E.; Svicher, V.; Pérez-Alvárez, N.; Curriu, M.; Perno, C.F.; Clotet, B.; Blanco, J.; Cabrera, C. The HR2 polymorphism N140I in the HIV-1 gp41 combined with the HR1 V38A mutation is associated with a less cytopathic phenotype. Retrovirology 2012, 9, 15.
- Blanco, J.; Barretina, J.; Ferri, K.F.; Jacotot, E.; Gutierrez, A.; Armand-Ugon, M.; Cabrera, C.; Kroemer, G.; Clotet, B.; Este, J.A. Cell-surface-expressed HIV-1 envelope induces the death of CD4 T cells during GP41-mediated hemifusion-like events. Virology 2003, 305, 318–329.
- Barretina, J.; Blanco, J.; Armand-Ugon, M.; Gutierrez, A.; Clotet, B.; Este, J.A. Anti-HIV-1 activity of enfuvirtide (T-20) by inhibition of bystander cell death. Antivir. Ther. 2003, 8, 155–161.
- Blanco, J.; Barretina, J.; Clotet, B.; Este, J.A. R5 HIV gp120-mediated cellular contacts induce the death of single CCR5-expressing CD4 T cells by a gp41-dependent mechanism. J. Leukoc. Biol. 2004, 76, 804–811.
- Pitcher, C.J.; Quittner, C.; Peterson, D.M.; Connors, M.; Koup, R.A.; Maino, V.C.; Picker, L.J. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat. Med. 1999, 5, 518–525.
- Rosenberg, E.S.; Billingsley, J.M.; Caliendo, A.M.; Boswell, S.L.; Sax, P.E.; Kalams, S.A.; Walker, B.D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 1997, 278, 1447–1450.
- Valentine, F.T.; Paolino, A.; Saito, A.; Holzman, R.S. Lymphocyte-proliferative responses to HIV antigens as a potential measure of immunological reconstitution in HIV disease. AIDS Res. Hum. Retroviruses 1998, 14 (Suppl. 2), S161–S166.
- Casado, C.; Marrero-Hernandez, S.; Marquez-Arce, D.; Pernas, M.; Marfil, S.; Borras-Granana, F.; Olivares, I.; Cabrera-Rodriguez, R.; Valera, M.S.; de Armas-Rillo, L.; et al. Viral Characteristics Associated with the Clinical Nonprogressor Phenotype Are Inherited by Viruses from a Cluster of HIV-1 Elite Controllers. mBio 2018, 9, e02338-17.
- Serena, M.; Parolini, F.; Biswas, P.; Sironi, F.; Blanco Miranda, A.; Zoratti, E.; Scupoli, M.T.; Ziglio, S.; Valenzuela-Fernandez, A.; Gibellini, D.; et al. HIV-1 Env associates with HLA-C free-chains at the cell membrane modulating viral infectivity. Sci. Rep. 2017, 7, 40037.
- Garcia-Exposito, L.; Ziglio, S.; Barroso-Gonzalez, J.; de Armas-Rillo, L.; Valera, M.S.; Zipeto, D.; Machado, J.D.; Valenzuela-Fernandez, A. Gelsolin activity controls efficient early HIV-1 infection. Retrovirology 2013, 10, 39.
- Garcia-Exposito, L.; Barroso-Gonzalez, J.; Puigdomenech, I.; Machado, J.D.; Blanco, J.; Valenzuela-Fernandez, A. HIV-1 requires Arf6-mediated membrane dynamics to efficiently enter and infect T lymphocytes. Mol. Biol. Cell 2011, 22, 1148–1166.
- Barrero-Villar, M.; Cabrero, J.R.; Gordon-Alonso, M.; Barroso-Gonzalez, J.; Alvarez-Losada, S.; Munoz-Fernandez, M.A.; Sanchez-Madrid, F.; Valenzuela-Fernandez, A. Moesin is required for HIV-1-induced CD4-CXCR4 interaction, F-actin redistribution, membrane fusion and viral infection in lymphocytes. J. Cell Sci. 2009, 122, 103–113.
- Valenzuela-Fernandez, A.; Cabrero, J.R.; Serrador, J.M.; Sanchez-Madrid, F. HDAC6: A key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 2008, 18, 291–297.
- Cabrera-Rodríguez, R.; Pérez-Yanes, S.; Montelongo, R.; Lorenzo-Salazar, J.M.; Estévez-Herrera, J.; García-Luis, J.; Íñigo-Campos, A.; Rubio-Rodríguez, L.A.; Muñoz-Barrera, A.; Trujillo-González, R.; et al. Transactive Response DNA-Binding Protein (TARDBP/TDP-43) Regulates Cell Permissivity to HIV-1 Infection by Acting on HDAC6. Int. J. Mol. Sci. 2022, 23, 6180.
- McDougal, J.S.; Kennedy, M.S.; Sligh, J.M.; Cort, S.P.; Mawle, A.; Nicholson, J.K. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science 1986, 231, 382–385.
- Dalgleish, A.G.; Beverley, P.C.; Clapham, P.R.; Crawford, D.H.; Greaves, M.F.; Weiss, R.A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 1984, 312, 763–767.
- Klatzmann, D.; Barre-Sinoussi, F.; Nugeyre, M.T.; Danquet, C.; Vilmer, E.; Griscelli, C.; Brun-Veziret, F.; Rouzioux, C.; Gluckman, J.C.; Chermann, J.C.; et al. Selective tropism of lymphadenopathy associated virus (LAV) for helper-inducer T lymphocytes. Science 1984, 225, 59–63.
- Klatzmann, D.; Champagne, E.; Chamaret, S.; Gruest, J.; Guetard, D.; Hercend, T.; Gluckman, J.C.; Montagnier, L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 1984, 312, 767–768.
- Maddon, P.J.; Dalgleish, A.G.; McDougal, J.S.; Clapham, P.R.; Weiss, R.A.; Axel, R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 1986, 47, 333–348.
- Kwong, P.D.; Wyatt, R.; Robinson, J.; Sweet, R.W.; Sodroski, J.; Hendrickson, W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998, 393, 648–659.
- Bleul, C.C.; Farzan, M.; Choe, H.; Parolin, C.; Clark-Lewis, I.; Sodroski, J.; Springer, T.A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 1996, 382, 829–833.
- Oberlin, E.; Amara, A.; Bachelerie, F.; Bessia, C.; Virelizier, J.L.; Arenzana-Seisdedos, F.; Schwartz, O.; Heard, J.M.; Clark-Lewis, I.; Legler, D.F.; et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 1996, 382, 833–835.
- Choe, H.; Farzan, M.; Sun, Y.; Sullivan, N.; Rollins, B.; Ponath, P.D.; Wu, L.; Mackay, C.R.; LaRosa, G.; Newman, W.; et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 1996, 85, 1135–1148.
- Chanel, C.; Staropoli, I.; Baleux, F.; Amara, A.; Valenzuela-Fernandez, A.; Virelizier, J.L.; Arenzana-Seisdedos, F.; Altmeyer, R. Low levels of co-receptor CCR5 are sufficient to permit HIV envelope-mediated fusion with resting CD4 T cells. AIDS 2002, 16, 2337–2340.
- Valenzuela-Fernández, A.; Palanche, T.; Amara, A.; Magerus, A.; Altmeyer, R.; Delaunay, T.; Virelizier, J.L.; Baleux, F.; Galzi, J.L.; Arenzana-Seisdedos, F. Optimal inhibition of X4 HIV isolates by the CXC chemokine stromal cell-derived factor 1 alpha requires interaction with cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2001, 276, 26550–26558.
- Puigdomènech, I.; Massanella, M.; Cabrera, C.; Clotet, B.; Blanco, J. On the steps of cell-to-cell HIV transmission between CD4 T cells. Retrovirology 2009, 6, 89.
- Santos, G.; Valenzuela-Fernandez, A.; Torres, N.V. Quantitative analysis of the processes and signaling events involved in early HIV-1 infection of T cells. PLoS ONE 2014, 9, e103845.
- Liu, Y.; Belkina, N.V.; Shaw, S. HIV infection of T cells: Actin-in and actin-out. Sci. Signal. 2009, 2, pe23.
- Valenzuela-Fernandez, A.; Alvarez, S.; Gordon-Alonso, M.; Barrero, M.; Ursa, A.; Cabrero, J.R.; Fernandez, G.; Naranjo-Suarez, S.; Yanez-Mo, M.; Serrador, J.M.; et al. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol. Biol. Cell 2005, 16, 5445–5454.
- Liu, Y.; Woodward, A.; Zhu, H.; Andrus, T.; McNevin, J.; Lee, J.; Mullins, J.I.; Corey, L.; McElrath, M.J.; Zhu, T. Preinfection human immunodeficiency virus (HIV)-specific cytotoxic T lymphocytes failed to prevent HIV type 1 infection from strains genetically unrelated to viruses in long-term exposed partners. J. Virol. 2009, 83, 10821–10829.
- Jimenez-Baranda, S.; Gomez-Mouton, C.; Rojas, A.; Martinez-Prats, L.; Mira, E.; Ana Lacalle, R.; Valencia, A.; Dimitrov, D.S.; Viola, A.; Delgado, R.; et al. Filamin-A regulates actin-dependent clustering of HIV receptors. Nat. Cell Biol. 2007, 9, 838–846.
- Yoder, A.; Yu, D.; Dong, L.; Iyer, S.R.; Xu, X.; Kelly, J.; Liu, J.; Wang, W.; Vorster, P.J.; Agulto, L.; et al. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell 2008, 134, 782–792.
- Liu, Y.; Curlin, M.E.; Diem, K.; Zhao, H.; Ghosh, A.K.; Zhu, H.; Woodward, A.S.; Maenza, J.; Stevens, C.E.; Stekler, J.; et al. Env length and N-linked glycosylation following transmission of human immunodeficiency virus Type 1 subtype B viruses. Virology 2008, 374, 229–233.
- Keele, B.F.; Giorgi, E.E.; Salazar-Gonzalez, J.F.; Decker, J.M.; Pham, K.T.; Salazar, M.G.; Sun, C.; Grayson, T.; Wang, S.; Li, H.; et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 2008, 105, 7552–7557.
- Quan, Y.; Liang, C.; Brenner, B.G.; Wainberg, M.A. Multidrug-resistant variants of HIV type 1 (HIV-1) can exist in cells as defective quasispecies and be rescued by superinfection with other defective HIV-1 variants. J. Infect. Dis. 2009, 200, 1479–1483.
- Fischer, W.; Ganusov, V.V.; Giorgi, E.E.; Hraber, P.T.; Keele, B.F.; Leitner, T.; Han, C.S.; Gleasner, C.D.; Green, L.; Lo, C.C.; et al. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS ONE 2010, 5, e12303.
- Roche, M.; Jakobsen, M.R.; Sterjovski, J.; Ellett, A.; Posta, F.; Lee, B.; Jubb, B.; Westby, M.; Lewin, S.R.; Ramsland, P.A.; et al. HIV-1 escape from the CCR5 antagonist maraviroc associated with an altered and less-efficient mechanism of gp120-CCR5 engagement that attenuates macrophage tropism. J. Virol. 2011, 85, 4330–4342.
- Fraser, C.; Lythgoe, K.; Leventhal, G.E.; Shirreff, G.; Hollingsworth, T.D.; Alizon, S.; Bonhoeffer, S. Virulence and pathogenesis of HIV-1 infection: An evolutionary perspective. Science 2014, 343, 1243727.
- Dang, L.V.P.; Pham, H.V.; Dinh, T.T.; Nguyen, T.H.; Vu, Q.T.H.; Vu, N.T.P.; Le, P.T.B.; Nguyen, L.V.; Le, H.T.; Vu, P.T.; et al. Characterization of envelope sequence of HIV virus in children infected with HIV in Vietnam. SAGE Open Med. 2020, 8, 2050312120937198.
- Koot, M.; Keet, I.P.; Vos, A.H.; de Goede, R.E.; Roos, M.T.; Coutinho, R.A.; Miedema, F.; Schellekens, P.T.; Tersmette, M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 1993, 118, 681–688.
- Kitrinos, K.M.; Nelson, J.A.; Resch, W.; Swanstrom, R. Effect of a protease inhibitor-induced genetic bottleneck on human immunodeficiency virus type 1 env gene populations. J. Virol. 2005, 79, 10627–10637.
- Kitchen, C.M.; Philpott, S.; Burger, H.; Weiser, B.; Anastos, K.; Suchard, M.A. Evolution of human immunodeficiency virus type 1 coreceptor usage during antiretroviral Therapy: A Bayesian approach. J. Virol. 2004, 78, 11296–11302.
- Hunt, P.W.; Harrigan, P.R.; Huang, W.; Bates, M.; Williamson, D.W.; McCune, J.M.; Price, R.W.; Spudich, S.S.; Lampiris, H.; Hoh, R.; et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J. Infect. Dis. 2006, 194, 926–930.
- Duenas-Decamp, M.J.; Peters, P.; Burton, D.; Clapham, P.R. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J. Virol. 2008, 82, 5807–5814.
- Salazar-Gonzalez, J.F.; Bailes, E.; Pham, K.T.; Salazar, M.G.; Guffey, M.B.; Keele, B.F.; Derdeyn, C.A.; Farmer, P.; Hunter, E.; Allen, S.; et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 2008, 82, 3952–3970.
- Kassaye, S.; Johnston, E.; McColgan, B.; Kantor, R.; Zijenah, L.; Katzenstein, D. Envelope coreceptor tropism, drug resistance, and viral evolution among subtype C HIV-1-infected individuals receiving nonsuppressive antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 2009, 50, 9–18.
- Moore, P.L.; Gray, E.S.; Morris, L. Specificity of the autologous neutralizing antibody response. Curr. Opin. HIV AIDS 2009, 4, 358–363.
- Shi, B.; Kitchen, C.; Weiser, B.; Mayers, D.; Foley, B.; Kemal, K.; Anastos, K.; Suchard, M.; Parker, M.; Brunner, C.; et al. Evolution and recombination of genes encoding HIV-1 drug resistance and tropism during antiretroviral therapy. Virology 2010, 404, 5–20.
- Sterjovski, J.; Churchill, M.J.; Ellett, A.; Gray, L.R.; Roche, M.J.; Dunfee, R.L.; Purcell, D.F.; Saksena, N.; Wang, B.; Sonza, S.; et al. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology 2007, 4, 89.
- Coffin, J.; Swanstrom, R. HIV pathogenesis: Dynamics and genetics of viral populations and infected cells. Cold Spring Harb. Perspect. Med. 2013, 3, a012526.
- Mishra, N.; Makhdoomi, M.A.; Sharma, S.; Kumar, S.; Dobhal, A.; Kumar, D.; Chawla, H.; Singh, R.; Kanga, U.; Das, B.K.; et al. Viral Characteristics Associated with Maintenance of Elite Neutralizing Activity in Chronically HIV-1 Clade C-Infected Monozygotic Pediatric Twins. J. Virol. 2019, 93, e00654-19.
- Turk, G.; Seiger, K.; Lian, X.; Sun, W.; Parsons, E.M.; Gao, C.; Rassadkina, Y.; Polo, M.L.; Czernikier, A.; Ghiglione, Y.; et al. A Possible Sterilizing Cure of HIV-1 Infection Without Stem Cell Transplantation. Ann. Intern. Med. 2022, 175, 95–100.
- Kirchhoff, F.; Greenough, T.C.; Brettler, D.B.; Sullivan, J.L.; Desrosiers, R.C. Brief report: Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 1995, 332, 228–232.
- Weber, J.; Gibson, R.M.; Sacka, L.; Strunin, D.; Hodek, J.; Weberova, J.; Pavova, M.; Alouani, D.J.; Asaad, R.; Rodriguez, B.; et al. Impaired human immunodeficiency virus type 1 replicative fitness in atypical viremic non-progressor individuals. AIDS Res. Ther. 2017, 14, 15.
- Armand-Ugon, M.; Quinones-Mateu, M.E.; Gutierez, A.; Barretina, J.; Blanco, J.; Schols, D.; De Clercq, E.; Clotet, B.; Este, J.A. Reduced fitness of HIV-1 resistant to CXCR4 antagonists. Antivir. Ther. 2003, 8, 1–8.
- Pietzsch, J.; Scheid, J.F.; Mouquet, H.; Klein, F.; Seaman, M.S.; Jankovic, M.; Corti, D.; Lanzavecchia, A.; Nussenzweig, M.C. Human anti-HIV-neutralizing antibodies frequently target a conserved epitope essential for viral fitness. J. Exp. Med. 2010, 207, 1995–2002.
- Shankarappa, R.; Margolick, J.B.; Gange, S.J.; Rodrigo, A.G.; Upchurch, D.; Farzadegan, H.; Gupta, P.; Rinaldo, C.R.; Learn, G.H.; He, X.; et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 1999, 73, 10489–10502.
- Valera, M.S.; de Armas-Rillo, L.; Barroso-Gonzalez, J.; Ziglio, S.; Batisse, J.; Dubois, N.; Marrero-Hernandez, S.; Borel, S.; Garcia-Exposito, L.; Biard-Piechaczyk, M.; et al. The HDAC6/APOBEC3G complex regulates HIV-1 infectiveness by inducing Vif autophagic degradation. Retrovirology 2015, 12, 53.
- Rosas-Umbert, M.; Llano, A.; Bellido, R.; Olvera, A.; Ruiz-Riol, M.; Rocafort, M.; Fernandez, M.A.; Cobarsi, P.; Crespo, M.; Dorrell, L.; et al. Mechanisms of Abrupt Loss of Virus Control in a Cohort of Previous HIV Controllers. J. Virol. 2019, 93, e01436-18.
- Borrell, M.; Fernandez, I.; Etcheverrry, F.; Ugarte, A.; Plana, M.; Leal, L.; Garcia, F. High rates of long-term progression in HIV-1-positive elite controllers. J. Int. AIDS Soc. 2021, 24, e25675.
More
Information
Subjects:
Infectious Diseases
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
706
Revisions:
2 times
(View History)
Update Date:
04 Sep 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




