Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alexis Murillo Carrasco | -- | 4737 | 2023-09-01 14:28:12 | | | |
| 2 | Fanny Huang | Meta information modification | 4737 | 2023-09-04 07:13:19 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Murillo Carrasco, A.G.; Otake, A.H.; Macedo-Da-Silva, J.; Feijoli Santiago, V.; Palmisano, G.; Andrade, L.N.D.S.; Chammas, R. Proteome of Extracellular Vesicles in Breast Cancer. Encyclopedia. Available online: https://encyclopedia.pub/entry/48742 (accessed on 08 February 2026).
Murillo Carrasco AG, Otake AH, Macedo-Da-Silva J, Feijoli Santiago V, Palmisano G, Andrade LNDS, et al. Proteome of Extracellular Vesicles in Breast Cancer. Encyclopedia. Available at: https://encyclopedia.pub/entry/48742. Accessed February 08, 2026.
Murillo Carrasco, Alexis Germán, Andreia Hanada Otake, Janaina Macedo-Da-Silva, Veronica Feijoli Santiago, Giuseppe Palmisano, Luciana Nogueira De Sousa Andrade, Roger Chammas. "Proteome of Extracellular Vesicles in Breast Cancer" Encyclopedia, https://encyclopedia.pub/entry/48742 (accessed February 08, 2026).
Murillo Carrasco, A.G., Otake, A.H., Macedo-Da-Silva, J., Feijoli Santiago, V., Palmisano, G., Andrade, L.N.D.S., & Chammas, R. (2023, September 01). Proteome of Extracellular Vesicles in Breast Cancer. In Encyclopedia. https://encyclopedia.pub/entry/48742
Murillo Carrasco, Alexis Germán, et al. "Proteome of Extracellular Vesicles in Breast Cancer." Encyclopedia. Web. 01 September, 2023.
Copy Citation
Breast cancer (BC) accounts for the highest incidence of tumor-related mortality among women worldwide, justifying the growing search for molecular tools for the early diagnosis and follow-up of BC patients under treatment. Circulating extracellular vesicles (EVs) are membranous nanocompartments produced by all human cells, including tumor cells.
extracellular vesicles
breast cancer
proteomes
1. Introduction
Breast cancer (BC) is currently the most commonly diagnosed type of cancer in the world. In 2020, this disease accounted for 25.5% of new cases among women, and it was responsible for 15.5% of the cases of death via cancer among women [1]. Due to these rates, it is important to continue searching for screening and therapeutic options. Cancer is a disease produced by specific tissue cells with uncontrolled growth. Tumor cells use cell-to-cell communication to spread through diverse strategies, invading lymph nodes or distant organs, evading growth suppressors, or inducing tumor-supporting angiogenesis [2].
Extracellular vesicles (EVs) represent one form of cell-to-cell communication through a range of different types of membranous nano- and microparticles (ranging from 30 nm to 5 µm in size), each with a specific biogenesis, cargo, and function [3]. EVs are involved in several biological processes such as cell signaling, cell proliferation, immune system modulation [4][5][6], and the production of amphisomes as a consequence of their fusion with autophagosomes [7]. EV proteins are markers for early diagnosis and are prognostic in several types of solid tumors. In lung cancer, EV leucine-rich alpha-2-glycoprotein 1 was reported to be up-regulated in urinary and lung tissue [8]. Moreover, ovarian cancer EVs are enriched in integrin, PI3 kinase, p53, Ras, and other proteins related to cancer development [9][10]. In gastrointestinal system cancers, the amount of exosome is increased in patients with colorectal cancer, and it is correlated with carcinoembryogenic antigen [11]. EVs also participate in cancer progression processes such as tumor invasion, growth, and metastasis [12][13]. For example, several studies have shown that EV protein content is associated with breast cancer cells, demonstrating its modulation during oncogenesis and tumor progression. EVs support cancer progression, signaling recipient cells’ motility and growth [14]. In breast cancer, increased EV biogenesis has been related to the activation of protease-activated receptor 2 (PAR2), which is induced by different factors, including coagulation factor-FVIIa and trypsin [15]. PAR2 is cleaved by trypsin, which triggers PIK3-dependent AKT phosphorylation. AKT’s phosphorylation induces Rab5a activation, resulting in actin polymerization, which contributes to microvesicle budding, cell migration, and invasion [16]. Moreover, three different signaling cascades contribute to EV production via PAR2: (1) actin polymerization via the sequential activation of P38, MK2, and HSP27; (2) ERK1/2 activation, which stimulates MLC2 via MLCK; and (3) MLC2 activation via ROCK-II independently of ERK1/2 activation. The activation of MLC2 and HSP27 is essential to actomyosin rearrangement and EV production [15].
The versatility attributed to EVs is due to their heterogeneity. EVs include at least three main groups of membranous particles: exosomes, ectosomes, and apoptotic bodies [3]. Exosomes (30–150 nm in size) are initially packaged by a cell membrane bud called a multivesicular body (MVB), whereas ectosomes (200 nm–5 µm in size) are produced by protrusions of the cell membrane shed by the cell. On the other hand, apoptotic bodies are formed via cell fragmentation during programmed cell death [3]. Therefore, these particles are classified according to their sizes and the expression of a signature of surface proteins, a consequence of their different biogenesis processes. Nevertheless, the classification of these EV subtypes neglects a fourth group of EVs with no expression of the minimal set of markers described for any of the three first groups. This group includes particles whose functions and surface proteins are still being discovered, for example, migrasomes [17]. Extracellular vesicle biology is a highly dynamic and relatively recent research field. The International Society of Extracellular Vesicles (ISEV) seeks to harmonize the studies in the field, and it continuously updates the markers, mechanisms, and technologies for classifying these EVs in documents published as Minimal Information for Studies of EVs (MISEV) [18].
Furthermore, EVs have arisen as a growing field of research due to their main characteristics, namely, (i) a wide of sizes from nano- to microscale (30 nm–5 µm) enabling their classification into groups (small or large EVs) [18][19][20]; (ii) a great diversity of proteins present on the surface of their lipid bilayer membranes, generally related to the molecular signature of the cell of origin [20][21]; and (iii) rhe mixed composition of their lumen or cargo, consisting of messenger RNA (mRNA) [22], microRNA (miRNA) [23], long non-coding RNA (lncRNA), circular RNA (circRNA) [24], double-stranded DNA (dsDNA) [25], mitochondrial DNA (mtDNA) [26], and proteins [27].
These characteristics allow EVs to be present in human fluids such as blood, urine, or saliva as a relevant form of cell-free circulating material helping to provide special protection for their luminal cargo, increasing the latter’s stability and half-life [28]. Furthermore, there is currently a need to understand more about the biology behind these vesicles and their potential applications, stimulating the creation of new EV-related databases [29][30][31]. These databases aim to organize the information on EV profiling being continuously produced worldwide despite the technical challenges surrounding their isolation and characterization [32].
2. Proteome of EVs in Breast Cancer
2.1. Analysis of BC-Related EV Proteomes Using High-Throughput Technologies
In order to elucidate the role of EV-related proteins during breast cancer pathogenesis, researchers reviewed the literature to determine the proteomes of EVs associated with breast cancer. A total of 21 studies [14][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52] that analyzed EVs from human breast cancer cell lines and biofluids using high-throughput and exploratory technologies were selected. A total of 8312 proteins were reported. The proteins were divided into core level 1 (protein identification in 50–100% of studies), core level 2 (protein identification in 40–50% of the studies), and core level 3 (protein identification in less than 40% of the studies) [14][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52]. Six and twenty-six proteins were assigned to cores 1 and 2, respectively. Although these data indicate high variability in the qualitative proteome content of EVs associated with breast cancer, researchers selected a few studies that evaluated methodological strategies for isolating EVs from breast cancer cells and biofluids and characterized their protein content. As many proteomic results were produced using BC cell lines, researchers used the classification system developed by Dai et al. (2017) to standardize the nomenclature and disease subtypes potentially represented by these cell lines [53].
Harris et al., 2015 investigated the subcellular localization of proteins found in the EVs of three BC cell lines with different types of metastatic profiles, namely, MDA-MB-231 (Triple-negative B subtype), MCF-7 overexpressing Rab27b, and MCF-7 (Luminal A subtype), and found several cellular localizations (the majority were from cytoplasm/cytoskeleton and integral/peripheral membrane proteins) and a minor proportion of EV proteins from the Golgi apparatus, ER, or mitochondria [14].
Regarding the molecular functions of the EV proteins, the majority displayed a protein-binding function and hydrolase activity. Comparing the EV protein expression between the three cell lines, 85 proteins were differentially expressed, with the non-invasive breast cancer cell line (MCF-7, Luminal A subtype) containing several up-regulated proteins with a tumor suppression function. Among the up-regulated proteins in MCF-7 compared to MDA-MB-231 were Tetraspanin (CD63, CD81, CD9, and Tetraspanin-14), Adhesion proteins (Neural cell adhesion molecule 2, Integrin alpha-V, Integrin beta-5, Epithelial cell adhesion molecule, Alpha-Parvin, Claudin-3, Cadherin-1, CD99, CD276, and Clusterin), stress response proteins (Heat shock protein beta-1), Small GTPase superfamily proteins (Rho-related GTP-binding protein RhoB, GTPase NRas, Ras-related protein Rap-2c, and RacGTPase-activating protein 1), and Endosome Trafficking/Transport proteins (Multivesicular body subunit 12B, Vacuolar protein sorting-associated protein 28 homolog, Vacuolar protein sorting-associated protein 37B, and Multivesicular body subunit 12A) [14]. The up-regulated proteins in MDA-MB-231 and MCF-7 overexpressing Rab27b compared to wild-type MCF-7 were adhesion/motility/cytoskeleton proteins (Vimentin, Galectin-3-binding protein, Annexin A1, Plectin, and Filamin-B), cell-surface receptor proteins (Ephrin type-A receptor 2), and stress response proteins (Protein NDRG1, Stress-70 protein, mitochondrial, and heat shock protein HSP 90-beta). Many of these proteins that were dysregulated in both cell lines are associated with tumorigenesis and metastasis [14]. Another protein of the Rab family, Rab5a, was associated with high migration and invasion rates in MDA-MB-231 cells along with the promotion of high rates of vesicle shedding. Interestingly, this process could be externally regulated by activating protease-activated receptor 2 (PAR2), using trypsin or coagulation factor-FVIIa to promote the accumulation of Rab5a [15][16].
Kruger et al., 2014 performed a molecular characterization of EVs from MCF-7 and MDA-MB-231 BC cells [52]. Using sucrose gradient ultracentrifugation and LC-MS/MS, the authors identified 59 and 88 proteins in MCF-7 and MDA-MB-231 EVs, respectively. The identified proteins were grouped based on molecular functions, such as catalytic activity, protein transport, adhesion, and extracellular matrix activity. Both EVs isolated from MCF-7 and MDA-MB-231 supernatants presented differences in their protein content. The MCF-7-derived EVs showed a greater abundance of biomolecule-binding and protein transport activity, while the MDA-MB-231-derived EVs contained proteins with catalytic activity. Moreover, 24% of the EV proteins of MDA-MB-231 were extracellular matrix proteins. These findings can be correlated with the higher metastatic potential of MDA-MB-231 (Triple-negative B subtype) compared to MCF-7 cells (Luminal A subtype) [52].
This study reported several proteins identified in both MDA-MB-231 and MCF-7 EVs, such as proteins from the Annexin family, Histone H4, and Calmodulin. Furthermore, the common EV proteins participate in cellular growth, signaling pathways, epigenomic alterations, DNA and histone methyltransferase, and Akt pathway regulation [54]. Moreover, these processes can be related to the malignancy of cancer cells and the poor prognosis of breast cancer [55]. Heinemann et al., 2014 used the MDA-MB-231 breast cancer cell line to develop a simple and efficient method for purifying cancer exosomes [47]. A three-step filtration method was implemented as the first step of a normal flow prefiltration using a 0.1 µm filter to remove larger constituents such as intact cells and cell debris. In the second step, tangential flow filtration using a 500kDa membrane filter was performed in order to remove proteins, and the retentate was passed to the third step, namely, track-etch filtration, where low pressure is applied using a 0.1 µm filter to isolate exosomes and remove microvesicles. A total of 60 unique proteins were identified sequential filtration, including the exosome marker CD63. In the exosome content, membrane trafficking proteins and proteins associated with transcription regulation, signal transduction, and the epigenetic modulation of nucleic acids were identified [47]. Regarding the heterogeneity of the vesicle entities, it is important to mention that filter-based techniques usually require the use of an automatized mechanism in order to avoid user interference during vesicle collection. Nevertheless, these techniques seem to support a relevant volume of EV proteomic data, so their application to EVs from different cell lines is recommended.
Smyth et al., 2014 examined the exosome proteomes from MCF-7 and PC3, cell lines of mammary gland/breast, and prostate cancers [49]. The identified exosome-delivered MCF-7 proteins were Heat Shock Proteins 70, 90, and 27; CD9; Ras-related protein 13 (RAB-13 and others); Annexins 1, 2, 5, and 7; Pyruvate kinase (PKM); Alpha-enolase (ENO1); Glyceraldehyde-3-phosphate dehydrogenase (GAPDH); Glucose-6-phosphate 1-dehydrogenase (G6PD); and actin. The functions of the exosomal proteins were related to amino acid transport, protease inhibition, the cytoskeleton, GTPase, ATPase, transcription regulation, transduction, adhesion, and chaperoning. Regarding subcellular localization, most of the proteins were in the membrane and cytoplasm; however, proteins localized in the Golgi and nucleus were also identified in MCF-7 exosomes. Moreover, it was found that the exosomal lipid composition also contributes to facilitating adherence/internalization in recipient cells [49]. The current stage of analytical methods, together with the optimization of pre-analytical factors and computational strategies, allowed for a better understanding of the EVs lipid compositions [56]. A comparison of the lipid compositions between EVs and cells of origin [57] has shown that, in vitro, EVs are more enriched in cholesterol, sphingomyelin, glycosphingolipids, and phosphatidylserine compared to their origin cells, while phosphatidylcholine and phosphatidylinositol are more enriched in cells compared to exosomes. Quantitative analyses of oxysterols in exosomes released from breast cancer cells revealed that levels of 27-Hydroxycholesterol were higher in exosomes from MCF-7 cells compared to MDA-MB-231 and non-cancerous cells, showing a dependence of the levels of this exosomal lipid on the ER status of the cell of origin [58]. Another study compared the lipid profiles of EVs isolated from TNBC cell lines, namely, D3H2LN and D3H1, with high and low metastatic potential, respectively [59]. The exosomal levels of unsaturated diacylglycerols isolated from the highly metastatic cells were higher and stimulated angiogenesis through the protein kinase D signaling pathway.
The EVs isolated from the MCF10 and MDA-MB231 cell lines shared proteins such as regulators of cell death, membrane components, adhesion, and cell motility proteins. The EVs from MDA-MB-231 were enriched in proteins associated with transcriptional regulation, proteolysis, EV formation (annexin, LAMP-1), cell cycle (NUMA1), and adherence to extracellular matrices (EDIL3, collagen, vitronectin). These results showed differences in the EV protein content of invasive and non-invasive breast cancer cells, which can contribute to the metastatic process [35].
The EV proteomes of two tumorigenic breast cancer cell lines, invasive (SKBR3, Her2 subtype) and non-invasive (MCF-7), were compared to non-tumorigenic MCF-10a. In this study, the authors used a synthetic peptide (Vn96) with a high affinity for heat shock proteins (highly expressed in cancer cells) to isolate EVs. Heat shock proteins are present on the exosome surface, binding to Vn96 and facilitating EV recovery [60]. In total, 392 (SKBR3) and 301 (MCF-7) exosomal proteins were identified, and they were associated with membranes (19%) and the cell surface (12%). The functions of EV proteins from SKBR3 are related to metabolism (enolase, fatty acid synthase, phosphoglycerate kinase, fructose bisphosphatase 1, GAPDH, malate dehydrogenase, L-lactate dehydrogenase, aldehyde dehydrogenase, aldolase, triosephosphate isomerase, and glucosidase 2 subunit beta), binding (Selenium-binding protein 1, 60 kDa heat shock protein, Protein disulfide-isomerase, Lamin A/C, and Tumor protein D52), and assembly (Myosin-9, alpha-Actinin-4, Cytokeratin 16, Cytokeratin 18, Cytokeratin 8, and Cytokeratin 19). Although MCF-7 EV protein content displayed the same molecular function as SKBR3, different proteins were identified, namely, Aldolase, Pyruvate kinase, Tryptophan-tRNA ligase, Cathepsin D, Kynureninase, TER ATPase, Lactoferroxin-C, and Hexokinase-1, which are involved in metabolism, and HSP90-a, Agrin, and protein SET are related to binding. Cytokeratin 19, which was identified in both SKBR3 and MCF-7 EVs, has been suggested to be involved in more aggressive tumor proliferation, metastasis, and invasion [61]. Cytokeratin 8 and 18 were reported to be secreted cancer biomarkers when detected in the serum of breast cancer patients [62]. EV proteins from SKBR3 (Her2 subtype) and MCF-7 (Luminal A subtype) cells are involved in lipogenesis, glycolysis/gluconeogenesis, and tricarboxylic acid, corroborating the important role of altered metabolism in cancer. Moreover, glycolytic enzymes can protect cancer cells from stress by inhibiting apoptosis [63]. Taken together, these data show that EV cargo is associated with the progression, proliferation, metabolic control, and malignancy of breast cancer.
Koh et al., 2021 analyzed the modulation of the plasma EV proteome of breast cancer patients with and without cognitive impairment following anthracycline-based treatment [51]. Plasma from the early-stage breast cancer patients was collected longitudinally at three different time points (before the start of chemotherapy, 3 weeks after cycle 2 of chemotherapy, and 3 weeks of the last cycle of chemotherapy) from two groups (cognitive non-impaired and impaired). Circulating EVs were isolated through ultracentrifugation and analyzed using Tandem-Mass-Tag (TMT)-based quantitative proteomics. A total of 517 regulated proteins were identified in the cognitive impairment group, which was compared to a non-impairment group at three time points. Among these regulated proteins were EV markers, such as CD9, TSPAN14, and CD5L. Moreover, a down-regulation of galactosylceramidase from the plasma EV protein content was observed in the cognitive non-impaired group at timepoint T3 compared to T1. On the other hand, p2X purinoceptor and cofilin-1 were up-regulated at timepoint 3 compared to T1. In contrast, the EV content of the cognitive impairment group displayed a down-regulation of p2X purinoceptor, cofilin-1, nexilin, and ADAM10 at T3 compared to T1 [51]. In this study, the authors also analyzed N-glycosylation sites in the EV content from plasma samples. However, none of the regulated proteins were identified in the N-glycosylation analysis. However, CD5L, which is an EV marker, displayed two glycosylation sites in both groups (non-impaired and impaired). An altered N-glycosylation profile was identified in CD5L peptides, which presented decreased glycosylation in the non-impaired group. These findings contribute to researchers' understanding of how different omics characteristics interact between EVs, their cells of origin, and their destination. Nevertheless, due to the heterogeneity of the tumor itself as well as the intra and interpatient heterogeneity, further studies would need to include more available information about the clinical statuses of these patients (age at diagnosis, molecular subtype, immunohistochemical profiles, etc.) in order to find patterns in the EV-related markers using large-scale cohort analyses.
The EVs released from cancer cells can regulate specific biological processes in recipient cells and modulate the central nervous system, thereby impairing the cognitive abilities of breast cancer patients [51]. Jordan et al., analyzed the differences in the protein cargo of EVs isolated from breast cancer cell lines and the plasma of breast cancer patients and healthy donors to identify the structural features that contribute to altered functional activities [35]. EVs from peripheral plasma samples of patients presented an increased degree of invasion of non-invasive breast cancer cells. Breast cancer plasma EVs displayed a unique proteome profile, with typical EV markers, such as CD9, CD81, CD63, and HSP70; Rab proteins; and clathrin. Moreover, the data were used in a comparison between the gene expression of MCF-10 treated with EVs obtained from breast cancer plasma patients, healthy donors, MDA-MB-231 cells, and untreated cells. The results displayed altered gene expression associated with angiogenesis, cell adhesion, and proliferation. In particular, it was confirmed that EVs released from aggressive breast cancer cells could stimulate the invasive behavior of non-invasive breast cancer cells. Moreover, the EV content from breast cancer patients can be used as a biomarker once these EVs contain a specific set of proteins [35].
2.2. Protein Markers in EVs Associated with Breast Cancer Features
Cancer-delivered EVs are a potential source of markers due to the fact that the cargo of EVs is enriched with molecules associated with tumor progression, metastasis, and invasion [64][65][66]. Rontogianni et al., analyzed the proteome content of EVs to discriminate cancer types and subtypes [40]. For this analysis, the authors used ten BC cell lines to identify a protein signature in EVs for use as a biomarker for breast cancer monitoring, diagnosis, and subtyping. Among the 4676 identified proteins, 14-3-3 proteins, integrins, annexin proteins, and cytoskeletal proteins, which are required for intermediate EV formation, were identified in all ten cancer cell lines. To discriminate the breast cancer subtypes, a total of 64 proteins were differentially expressed in TNBC compared to HER2+; these proteins are involved in angiogenesis (PLAU, ADAM9, and EPHA2), integrin-binding (ITGA5 and TIMP2), and cell motility (VIM and AXL). HER2+ EVs presented proteins associated with translation (EIFs), axon guidance (DNM2 and PIK3R1), and ERBB signaling (GRB7 and SHC1). Periostin, an extracellular matrix component, was described as a metastatic breast cancer biomarker identified in exosomes. It was observed that this protein was up-regulated in both murine and human cancer cell lines. Periostin has been reported to be highly expressed in several cancer types, including breast cancer [67], and it has been associated with osteoblast adhesion and cancer cell migration and is also involved in tumor angiogenesis [68]. Increased abundance of the Transient Receptor Potential Channel 5 (TrpC5) in EVs was observed in adriamycin-resistant human breast cancer cells (MCF-7/ADM) and was correlated with EV formation; additionally, adriamycin was found in EVs. The transference of TrpC5 to recipient cells mediated by EVs allows recipient cells to acquire TrpC5 and to P-glycoprotein multidrug transporter expression, conferring chemoresistance to non-resistance cells [69]. Considering the relevance of studies involving several BC cell lines, there are two main factors that must be considered. First, previous studies on tumor-derived EVs have determined that culture conditions can affect the omics components of EV cargo [70]. Second, despite the above, it is necessary to have a reliable collection of vesicular omics data from BC cell lines to find consistent markers for further research.
Regarding the use of liquid biopsies in relation to metastatic breast cancer, in one study, EV proteins were monitored to identify potential biomarkers for prognostics using a thermophoretic aptasensor. A total of 286 plasma samples from metastatic patients, non-metastatic patients, and healthy donors were submitted to a thermophoretic aptasensor to obtain a profile of eight EV markers: CA 15-3, CA 125, carcinoembryonic antigen (CEA), human epidermal growth factor receptor 2 (HER2), epidermal growth factor receptor (EGFR), prostate-specific membrane antigen (PSMA), epithelial cell adhesion molecule (EpCAM), and vascular endothelial growth factor (VEGF). These markers are involved in survival and migration, cell proliferation, metastasis, invasion, cell stemness, vascular permeability, and angiogenesis [65]. In an independent study, sera-derived exosomes from breast cancer patients were used to characterize CD24 and EpCAM as markers for exosomes. Western-blotting analysis was performed to identify exosomal markers in breast cancer samples. EpCAM and CD24 were identified in exosomes isolated from ascites ovarian cancer patients. However, only CD24 was detected in the EVs isolated from the serum of breast cancer patients. The absence of EpCAM in the serum-derived exosomes from breast cancer can be explained by the cleavage of EpCAM by serum metalloproteinases and the fact that both CD24 and EpCAM are not present in the same EV populations, which was confirmed in this study after the immunoaffinity purification of exosomes using anti-EpCAM beads [71]. Taken together, these studies show the importance of EV proteins as breast cancer markers for clinical use in order to diagnose, stratify, and monitor breast cancer progression.
2.3. EV Protein Glycosylation in Breast Cancer
Glycoconjugates such as glycoproteins functionally decorate the cell surface and are released in the extracellular milieu, playing an essential role in cell communication with the extracellular environment in several pathophysiological conditions, including cancer [72][73]. Any alteration in glycosylation levels due to the variation in the expression of glycosyltransferases and glycosidases [74], the dysregulation of chaperone activity [75], different levels of substrates, and alterations in nucleotide sugar transporter levels are diagnostic signs in cancer [76]. Aberrant glycosylation in tumor cells influences angiogenesis, cell proliferation, invasion, and metastatic processes [77][78][79][80]. The surface of an EV is enriched with glycoproteins, such as the Tetraspanin CD63, which is known to regulate cancer malignancy, and glycolipids [81]. EV glycosylation has been reported in several cancer types, including breast cancer. A list of 15 studies on EV glycosylation in breast cancer was reported by Macedo-Silva et al. (2021) [82] [33][34][36][46][47][48][49][55][83][84][85][86][87][88][89].
Nishida-Aoki et al. (2020) developed an EV glycosylation profile from breast cancer cells with brain metastasis tropism (BMD2a) comparing lymph node-metastatic (MDA-MB-231-luc-D3H2LN) and primary tumor-derived TNBC cell lines (MDA-MB-231-luc-D3H1) using lectin blotting [90]. The data showed a reduced level of sialic acid and a specific glycosylation pattern associated with galactose, GalNAc, lactose, and GalNAcα1−3GalNAc in BMD2a EVs. This study concluded that the glycosylation of EVs exerted an inhibitory effect on EV uptake [90]. This evidence shows that EV uptake depends on glycosylation profiles, as the authors showed that the cessation of O-glycosylation increases tumor EV accumulation in the lungs; meanwhile, N-glycosylation inhibition does not alter tumor EV biodistribution. It appears that the specific glycosylation profile in tumor EVs highlights their importance in breast cancer aggressiveness. For instance, EVs secreted by TNBC cells present higher levels of sialylation when compared to parental cells [91], and this process could interfere with EVs uptake. Sialic acid is negatively charged; its removal from the surfaces of EVs can promote the interaction of EVs and recipient cells through the uncovering of carbohydrate ligands that bind to lectins and promote the uptake of EV by recipient cells. Additionally, EV cargo contains glycosylated proteins that can interfere with EV–cell interaction. Cytokines binding to surface glycosaminoglycan side chains of proteoglycans decorate EVs isolated from breast cancer patients, altering EV biodistribution and leading to a higher metastatic burden [92]. The structure of bisecting GlcNAc glycan (β1,4-linked GlcNAc at the β-mannose residue core) on target proteins (EGFR and integrins) plays a role in cell metastasis and adhesion. The role of bisecting GlcNAc of EVs in breast cancer cells was described by Tan et al. (2020) [93]. Some proteins were described as being markers of EVs from tumor cells in breast cancer, such as extracellular matrix metalloproteinase inducer (EMMPRIN). A higher abundance of glycosylated EMMPRIN was identified in EVs from metastatic breast cancer patients. PNGase F treatment resulted in EVs’ deglycosylation and inhibited EV-induced invasion [87]. Somehow, glycans being components of EV cargo seems to dictate cell function. ST6Gal1-mediated sialylation modulates cell surface receptor function, promoting cell proliferation and invasion. Hait et al., showed that ST6Gal1 RNA transcripts and protein expression were variable and heterogeneous in a panel of TNBC and ER+ cell lines. However, EVs containing ST6Gal1 potentiate aggressive cancer cell growth, proliferation, and invasion in cells containing low amounts of endogenous ST6Gal1 [94]. In a heterogeneous tumor microenvironment, EVs act as important mediators of tumor progression, dictating EV organ tropism and metastasis. CA15-3/CA27.29 epitopes from Mucin 1 were reported to be tumor markers for breast cancer diagnosis. Additionally, alterations in Mucin 1 glycosylation profiles were observed in several cancer types, including human breast cancer, and in exosomes derived from luminal A breast carcinoma cells MCF-7 [34][95]. Extracellular matrix protein nephronectin (NPNT) could be present in a truncated or a glycosylated form in EVs secreted by mouse breast carcinoma cells [96]. N-glycosylation sites in integrin β1 modulate cell migration and the adhesion of MCF7 cells (luminal A), activating focal adhesion kinase (FAK) signaling, and these sites are crucial to integrin activity in EVs [97]. Additionally, exosomes released by MCF-7 cells after irradiation (luminal A type) increase concentrations of the enzyme GalNAc-T6, which is important for promoting epithelial–mesenchymal transition [98]. Furthermore, there is still a gap concerning whether alterations in EV glycosylation are due to changes in glycoprotein levels or changes in the expression/activity levels of glycan-modifying enzymes.
2.4. EV Proteome during the Metastatic Process in Breast Cancer
A screening of the literature regarding EV proteins identified in human and other species’ breast cancer cells, tissues, and biofluids, focusing on the TNBC and metastatic cases, allowed us to retrieve 22 studies and a total of 7265 proteins [14][34][35][37][39][40][41][42][43][45][48][50][51][52][86][99][100][101][102][103][104][105]. Interestingly, researchers observed that one set of proteins was identified more frequently (Figure 1A), with HLA-A, HLA-B, A2M, ACTB, ALDOA, ANXA2, and FN1 identified in at least 13 of 22 studies that explored the vesicular proteome in breast cancer.
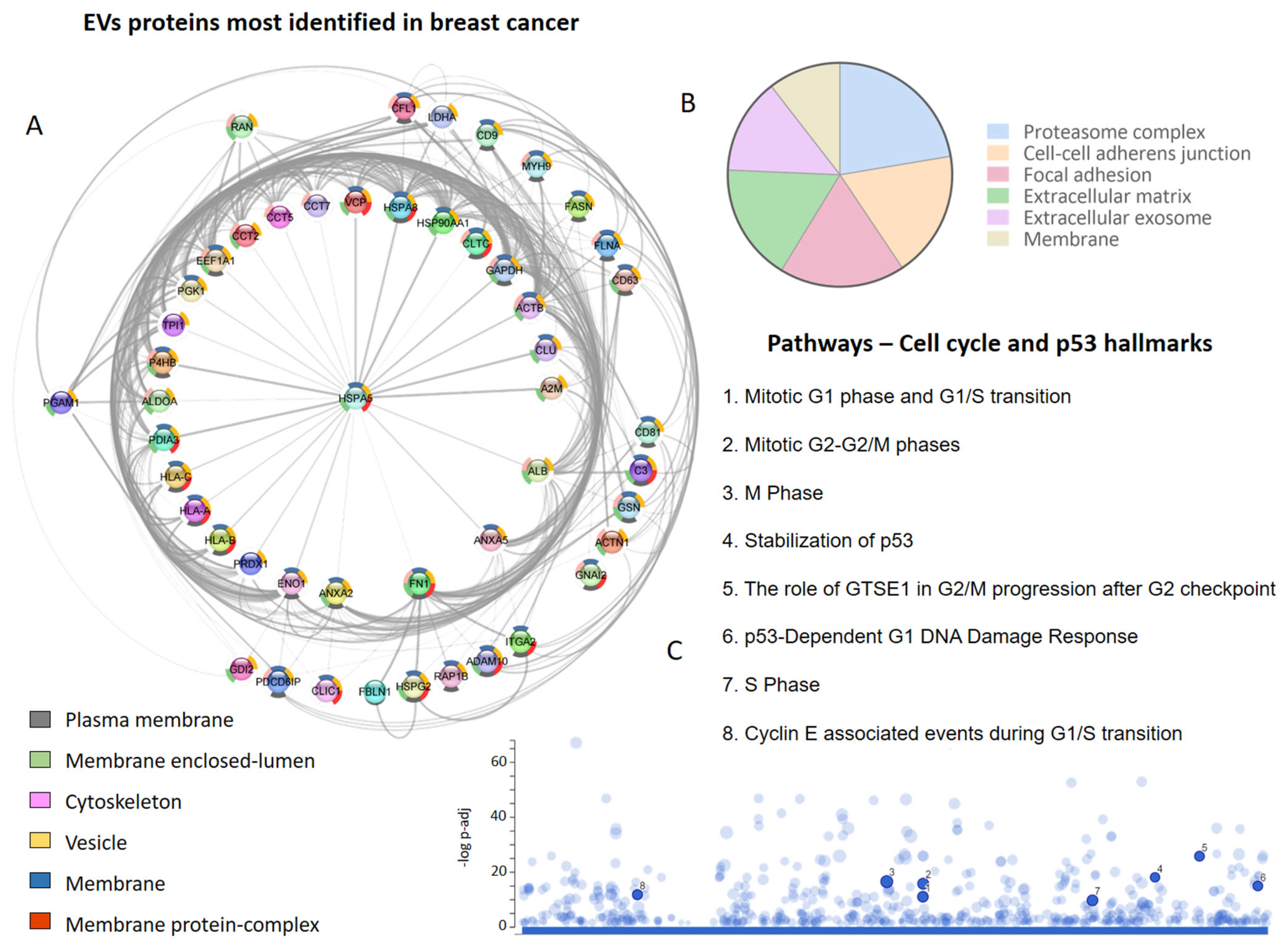
Figure 1. Extracellular vesicle (EVs) proteins in breast cancer proteome studies. (A) The most frequently identified proteins in the evaluated studies. The donut graph shows the corresponding subcellular locations. (B) Subcellular localization of proteins identified in EVs in at least two studies. (C) Cell-cycle-related and p53 pathways in which EV proteins participate.
Among the most frequent proteins, those located in membranes (plasma and organellar) and the cytoskeleton were particularly prominent (Figure 1A). On the other hand, after evaluating the subcellular localization of all the proteins identified at least twice in the EVs derived from breast cancer cells, it was determined that the proteasome complex, cell–cell adhesion junctions, focal adhesions, extracellular matrix, extracellular exosome, and membranes were also relevant (Figure 1B). It is possible to identify vesicular proteins that are involved in different phases of the cell cycle and the p53-linked response (Figure 1C), reinforcing the role of EVs in metastatic processes and breast cancer progression.
In fact, studies have demonstrated the crucial role of EVs in pro-metastatic processes, wherein a cancer cell’s communication with its local environment is necessary to initiate growth and invasion through the transfer of molecules, which include mRNAs, microRNAs, and proteins [100]. Different studies have revealed the vesicular proteins associated with metastatic events, and among those listed, ADAM10 (four studies), VIM (three studies), and ANXA6 (two studies) stand out. An important finding was that exosomes expressing these proteins play important roles in TNBC.
It has been identified that the levels of the CD151 protein are increased in serum exosomes from TNBC patients, and this protein has been shown to promote proliferation, migration, and invasion by modulating the activities of laminin-binding integrins α3β1, α6β4, and α6β1 [39]. An increase in CD151 abundance was also verified in a TNBC cell model (MDA-MB-231) [45]. These results indicate that exosomal CD151 can be used as a biomarker to distinguish TNBC patients from healthy individuals.
The data gathered reveal the importance of other vesicular proteins in TNBC, such as VIM (three studies) and ANXA6, EDIL3, EPHA2, FLNA, FLNB, and PLAU (two studies). It has been observed that the VIM protein and its gene play important roles in controlling migration and invasion in different types of cancer [106][107]; here, it is highlighted that its location in EVs can be associated with the aggressiveness of cancer. Based on the collected data, it is possible to select possible markers of EVs in breast cancer. In addition to conventional markers such as CD9, CD81, and CD83, the proteins HLA-A, HLA-B, HSPA5, A2M, ACTB, ALDOA, ANXA2, FN1, ANXA5, GSN, MYH9, PGK1, and ACTN1 are also often identified in EVs in cells and breast cancer patients at different stages. Therefore, these markers could be used to purify EVs potentially related to highly aggressive tumor subpopulations and study them separately.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674.
- Dixson, A.C.; Dawson, T.R.; Di Vizio, D.; Weaver, A.M. Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nat. Rev. Mol. Cell Biol. 2023, 24, 454–476.
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome Secretion: Molecular Mechanisms and Roles in Immune Responses. Traffic 2011, 12, 1659–1668.
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920.
- Théry, C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3, 15.
- Gross, J.C.; Parbin, S. Crossroads of the endosomal machinery: Multivesicular bodies, small extracellular vesicles and autophagy. Trillium. Exctracellular. Vesicles 2020, 2, 48–53.
- Li, Y.; Zhang, Y.; Qiu, F.; Qiu, Z. Proteomic identification of exosomal LRG1: A potential urinary biomarker for detecting NSCLC. Electrophoresis 2011, 32, 1976–1983.
- Liang, B.; Peng, P.; Chen, S.; Li, L.; Zhang, M.; Cao, D.; Yang, J.; Li, H.; Gui, T.; Li, X.; et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J. Proteom. 2013, 80, 171–182.
- Sinha, A.; Ignatchenko, V.; Ignatchenko, A.; Mejia-Guerrero, S.; Kislinger, T. In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochem. Biophys. Res. Commun. 2014, 445, 694–701.
- Silva, J.; Garcia, V.; Rodriguez, M.; Compte, M.; Cisneros, E.; Veguillas, P.; Garcia, J.M.; Dominguez, G.; Campos-Martin, Y.; Cuevas, J.; et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosom. Cancer 2012, 51, 409–418.
- Park, J.E.; Sen Tan, H.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteom. 2010, 9, 1085–1099.
- Zhang, X.; Yuan, X.; Shi, H.; Wu, L.; Qian, H.; Xu, W. Exosomes in cancer: Small particle, big player. J. Hematol. Oncol. 2015, 8, 83.
- Harris, D.A.; Patel, S.H.; Gucek, M.; Hendrix, A.; Westbroek, W.; Taraska, J.W. Exosomes Released from Breast Cancer Carcinomas Stimulate Cell Movement. PLoS ONE 2015, 10, e0117495.
- Das, K.; Prasad, R.; Singh, A.; Bhattacharya, A.; Roy, A.; Mallik, S.; Mukherjee, A.; Sen, P. Protease-activated receptor 2 promotes actomyosin dependent transforming microvesicles generation from human breast cancer. Mol. Carcinog. 2018, 57, 1707–1722.
- Das, K.; Prasad, R.; Roy, S.; Mukherjee, A.; Sen, P. The Protease Activated Receptor2 Promotes Rab5a Mediated Generation of Pro-metastatic Microvesicles. Sci. Rep. 2018, 8, 7357.
- Ma, L.; Li, Y.; Peng, J.; Wu, D.; Zhao, X.; Cui, Y.; Chen, L.; Yan, X.; Du, Y.; Yu, L. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 2015, 25, 24–38.
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750.
- Alberro, A.; Iparraguirre, L.; Fernandes, A.; Otaegui, D. Extracellular Vesicles in Blood: Sources, Effects, and Applications. Int. J. Mol. Sci. 2021, 22, 8163.
- Martins, Á.M.; Ramos, C.C.; Freitas, D.; Reis, C.A. Glycosylation of Cancer Extracellular Vesicles: Capture Strategies, Functional Roles and Potential Clinical Applications. Cells 2021, 10, 109.
- Li, Y.; He, X.; Li, Q.; Lai, H.; Zhang, H.; Hu, Z.; Li, Y.; Huang, S. EV-origin: Enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput. Struct. Biotechnol. J. 2020, 18, 2851–2859.
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606.
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659.
- Li, Y.; Zhao, J.; Yu, S.; Wang, Z.; He, X.; Su, Y.; Guo, T.; Sheng, H.; Chen, J.; Zheng, Q.; et al. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin. Chem. 2019, 65, 798–808.
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769.
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. 2009, 117, 1–4.
- Belov, L.; Matic, K.J.; Hallal, S.; Best, O.G.; Mulligan, S.P.; Christopherson, R.I. Extensive surface protein profiles of extracellular vesicles from cancer cells may provide diagnostic signatures from blood samples. J. Extracell. Vesicles 2016, 5, 25355.
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom.—Clin. Appl. 2015, 9, 358–367.
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112.
- Liu, C.-J.; Xie, G.-Y.; Miao, Y.-R.; Xia, M.; Wang, Y.; Lei, Q.; Zhang, Q.; Guo, A.-Y. EVAtlas: A comprehensive database for ncRNA expression in human extracellular vesicles. Nucleic Acids Res. 2022, 50, D111–D117.
- Liu, T.; Zhang, Q.; Zhang, J.; Li, C.; Miao, Y.R.; Lei, Q.; Li, Q.; Guo, A.Y. EVmiRNA: A database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019, 47, D89–D93.
- Melling, G.E.; Carollo, E.; Conlon, R.; Simpson, J.C.; Carter, D.R.F. The Challenges and Possibilities of Extracellular Vesicles as Therapeutic Vehicles. Eur. J. Pharm. Biopharm. 2019, 144, 50–56.
- Hurwitz, S.N.; Rider, M.A.; Bundy, J.L.; Liu, X.; Singh, R.K.; Meckes, D.G., Jr. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget 2016, 7, 86999–87015.
- Staubach, S.; Razawi, H.; Hanisch, F.G. Proteomics of MUC1-containing lipid rafts from plasma membranes and exosomes of human breast carcinoma cells MCF-7. Proteomics 2009, 9, 2820–2835.
- Jordan, K.R.; Hall, J.K.; Schedin, T.; Borakove, M.; Xian, J.J.; Dzieciatkowska, M.; Lyons, T.R.; Schedin, P.; Hansen, K.C.; Borges, V.F. Extracellular vesicles from young women’s breast cancer patients drive increased invasion of non-malignant cells via the Focal Adhesion Kinase pathway: A proteomic approach. Breast Cancer Res. 2020, 22, 128.
- Antonyak, M.A.; Li, B.; Boroughs, L.K.; Johnson, J.L.; Druso, J.E.; Bryant, K.L.; Holowka, D.A.; Cerione, R.A. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. USA 2011, 108, 4852–4857.
- Tutanov, O.; Orlova, E.; Proskura, K.; Grigor’eva, A.; Yunusova, N.; Tsentalovich, Y.; Alexandrova, A.; Tamkovich, S. Proteomic Analysis of Blood Exosomes from Healthy Females and Breast Cancer Patients Reveals an Association between Different Exosomal Bioactivity on Non-tumorigenic Epithelial Cell and Breast Cancer Cell Migration in Vitro. Biomolecules 2020, 10, 495.
- Vinik, Y.; Ortega, F.G.; Mills, G.B.; Lu, Y.; Jurkowicz, M.; Halperin, S.; Aharoni, M.; Gutman, M.; Lev, S. Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci. Adv. 2020, 6, eaba5714.
- Li, S.; Li, X.; Yang, S.; Pi, H.; Li, Z.; Yao, P.; Zhang, Q.; Wang, Q.; Shen, P.; Li, X.; et al. Proteomic landscape of exosomes reveals the functional contributions of CD151 in triple-negative breast cancer. Mol. Cell. Proteom. 2021, 20, 100121.
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun. Biol. 2019, 2, 325.
- Tutanov, O.; Proskura, K.; Kamyshinsky, R.; Shtam, T.; Tsentalovich, Y.; Tamkovich, S. Proteomic Profiling of Plasma and Total Blood Exosomes in Breast Cancer: A Potential Role in Tumor Progression, Diagnosis, and Prognosis. Front. Oncol. 2020, 10, 2173.
- Martínez-Greene, J.A.; Hernández-Ortega, K.; Quiroz-Baez, R.; Resendis-Antonio, O.; Pichardo-Casas, I.; Sinclair, D.A.; Budnik, B.; Hidalgo-Miranda, A.; Uribe-Querol, E.; del Pilar Ramos-Godínez, M.; et al. Quantitative proteomic analysis of extracellular vesicle subgroups isolated by an optimized method combining polymer-based precipitation and size exclusion chromatography. J. Extracell. Vesicles 2021, 10, e12087.
- Dalla, P.V.; Santos, J.; Milthorpe, B.K.; Padula, M.P. Selectively-Packaged Proteins in Breast Cancer Extracellular Vesicles Involved in Metastasis. Int. J. Mol. Sci. 2020, 21, 4990.
- Vardaki, I.; Ceder, S.; Rutishauser, D.; Baltatzis, G.; Foukakis, T.; Panaretakis, T.; Vardaki, I.; Ceder, S.; Rutishauser, D.; Baltatzis, G.; et al. Periostin is identified as a putative metastatic marker in breast cancer-derived exosomes. Oncotarget 2016, 7, 74966–74978.
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers. Sci. Rep. 2020, 10, 13572.
- Palazzolo, G.; Albanese, N.N.; Di Cara, G.; Gygax, D.; Vittorelli, M.L.; Pucci-Minafra, I. Proteomic analysis of exosome-like vesicles derived from breast cancer cells. Anticancer Res. 2012, 32, 847–860.
- Heinemann, M.L.; Ilmer, M.; Silva, L.P.; Hawke, D.H.; Recio, A.; Vorontsova, M.A.; Alt, E.; Vykoukal, J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A 2014, 1371, 125–135.
- Clark, D.J.; Fondrie, W.E.; Liao, Z.; Hanson, P.I.; Fulton, A.; Mao, L.; Yang, A.J. Redefining the Breast Cancer Exosome Proteome by Tandem Mass Tag Quantitative Proteomics and Multivariate Cluster Analysis. Anal. Chem. 2015, 87, 10462–10469.
- Smyth, T.J.; Redzic, J.S.; Graner, M.W.; Anchordoquy, T.J. Examination of the specificity of tumor cell derived exosomes with tumor cells in vitro. Biochim. Biophys. Acta-Biomembr. 2014, 1838, 2954–2965.
- Griffiths, S.G.; Cormier, M.T.; Clayton, A.; Doucette, A.A. Differential Proteome Analysis of Extracellular Vesicles from Breast Cancer Cell Lines by Chaperone Affinity Enrichment. Proteomes 2017, 5, 25.
- Koh, Y.Q.; Ng, D.Q.; Ng, C.C.; Boey, A.; Wei, M.; Sze, S.K.; Ho, H.K.; Acharya, M.; Limoli, C.L.; Chan, A. Extracellular Vesicle Proteome of Breast Cancer Patients with and without Cognitive Impairment Following Anthracycline-based Chemotherapy: An Exploratory Study. Biomark. Insights 2021, 16, 11772719211018204.
- Kruger, S.; Elmageed, Z.Y.A.; Hawke, D.H.; Wörner, P.M.; Jansen, D.A.; Abdel-Mageed, A.B.; Alt, E.U.; Izadpanah, R. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer 2014, 14, 1–10.
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141.
- Tryndyak, V.P.; Kovalchuk, O.; Pogribny, I.P. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol. Ther. 2006, 5, 65–70.
- Coticchia, C.M.; Revankar, C.M.; Deb, T.B.; Dickson, R.B.; Johnson, M.D. Calmodulin modulates Akt activity in human breast cancer cell lines. Breast Cancer Res. Treat. 2008, 115, 545–560.
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321.
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18.
- Roberg-Larsen, H.; Lund, K.; Seterdal, K.E.; Solheim, S.; Vehus, T.; Solberg, N.; Krauss, S.; Lundanes, E.; Wilson, S.R. Mass spectrometric detection of 27-hydroxycholesterol in breast cancer exosomes. J. Steroid Biochem. Mol. Biol. 2017, 169, 22–28.
- Nishida-Aoki, N.; Izumi, Y.; Takeda, H.; Takahashi, M.; Ochiya, T.; Bamba, T. Lipidomic Analysis of Cells and Extracellular Vesicles from High- and Low-Metastatic Triple-Negative Breast Cancer. Metabolites 2020, 10, 67.
- Góss Santos, T.; Regina Martins, V.; Noeli Maroso Hajj, G.; Di Sansebastiano, G.-P.; Gaballo, A. Unconventional Secretion of Heat Shock Proteins in Cancer. Int. J. Mol. Sci. 2017, 18, 946.
- Zhang, D.H.; Tai, L.K.; Wong, L.L.; Sethi, S.K.; Koay, E.S.C. Proteomics of breast cancer: Enhanced expression of cytokeratin19 in human epidermal growth factor receptor type 2 positive breast tumors. Proteomics 2005, 5, 1797–1805.
- Olofsson, M.H.; Ueno, T.; Pan, Y.; Xu, R.; Cai, F.; Van Der Kuip, H.; Muerdter, T.E.; Sonnenberg, M.; Aulitzky, W.E.; Schwarz, S.; et al. Cytokeratin-18 Is a Useful Serum Biomarker for Early Determination of Response of Breast Carcinomas to Chemotherapy. Clin. Cancer Res. 2007, 13, 3198–3206.
- Muñoz-Pinedo, C.; El Mjiyad, N.; Ricci, J.E. Cancer metabolism: Current perspectives and future directions. Cell Death Dis. 2012, 3, e248.
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848.
- Tian, F.; Zhang, S.; Liu, C.; Han, Z.; Liu, Y.; Deng, J.; Li, Y.; Wu, X.; Cai, L.; Qin, L.; et al. Protein analysis of extracellular vesicles to monitor and predict therapeutic response in metastatic breast cancer. Nat. Commun. 2021, 12, 2536.
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232.
- Horiuchi, K.; Amizuka, N.; Takeshita, S.; Takamatsu, H.; Katsuura, M.; Ozawa, H.; Toyama, Y.; Bonewald, L.F.; Kudo, A. Identification and Characterization of a Novel Protein, Periostin, with Restricted Expression to Periosteum and Periodontal Ligament and Increased Expression by Transforming Growth Factor β. J. Bone Miner. Res. 1999, 14, 1239–1249.
- Shao, R.; Bao, S.; Bai, X.; Blanchette, C.; Anderson, R.M.; Dang, T.; Gishizky, M.L.; Marks, J.R.; Wang, X.-F. Acquired Expression of Periostin by Human Breast Cancers Promotes Tumor Angiogenesis through Up-Regulation of Vascular Endothelial Growth Factor Receptor 2 Expression. Mol. Cell. Biol. 2004, 24, 3992–4003.
- Ma, X.; Chen, Z.; Hua, D.; He, D.; Wang, L.; Zhang, P.; Wang, J.; Cai, Y.; Gao, C.; Zhang, X.; et al. Essential role for TrpC5-containing extracellular vesicles in breast cancer with chemotherapeutic resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 6389–6394.
- Palviainen, M.; Saari, H.; Kärkkäinen, O.; Pekkinen, J.; Auriola, S.; Yliperttula, M.; Puhka, M.; Hanhineva, K.; Siljander, P.R.-M. Metabolic signature of extracellular vesicles depends on the cell culture conditions. J. Extracell. Vesicles 2019, 8, 1596669.
- Rupp, A.K.; Rupp, C.; Keller, S.; Brase, J.C.; Ehehalt, R.; Fogel, M.; Moldenhauer, G.; Marmé, F.; Sültmann, H.; Altevogt, P. Loss of EpCAM expression in breast cancer derived serum exosomes: Role of proteolytic cleavage. Gynecol. Oncol. 2011, 122, 437–446.
- Ilver, D.; Johansson, P.; Miller-Podraza, H.; Nyholm, P.G.; Teneberg, S.; Karlsson, K.A. Bacterium–Host Protein–Carbohydrate Interactions. Methods Enzymol. 2003, 363, 134–157.
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555.
- Marcos, N.T.; Bennett, E.P.; Gomes, J.; Magalhaes, A.; Gomes, C.; David, L.; Dar, I.; Jeanneau, C.; DeFrees, S.; Krustrup, D.; et al. ST6GalNAc-I controls expression of sialyl-Tn antigen in gastrointestinal tissues. Front. Biosci.-Elit. 2011, 3, 1443–1455.
- Wang, Y.; Ju, T.; Ding, X.; Xia, B.; Wang, W.; Xia, L.; He, M.; Cummings, R.D. Cosmc is an essential chaperone for correct protein O-glycosylation. Proc. Natl. Acad. Sci. USA 2010, 107, 9228–9233.
- Kumamoto, K.; Goto, Y.; Sekikawa, K.; Takenoshita, S.; Ishida, N.; Kawakita, M.; Kannagi, R. Increased Expression of UDP-Galactose Transporter Messenger RNA in Human Colon Cancer Tissues and Its Implication in Synthesis of Thomsen-Friedenreich Antigen and Sialyl Lewis A/X Determinants1. Cancer Res. 2001, 61, 4620–4627.
- Croci, D.O.; Cerliani, J.P.; Pinto, N.A.; GMorosi, L.; Rabinovich, G.A. Regulatory role of glycans in the control of hypoxia-driven angiogenesis and sensitivity to anti-angiogenic treatment. Glycobiology 2014, 24, 1283–1290.
- Freitas, D.; Campos, D.; Gomes, J.; Pinto, F.; Macedo, J.A.; Matos, R.; Mereiter, S.; Pinto, M.T.; Polónia, A.; Gartner, F.; et al. O-glycans truncation modulates gastric cancer cell signaling and transcription leading to a more aggressive phenotype. EBioMedicine 2019, 40, 349–362.
- Häuselmann, I.; Borsig, L. Altered tumor-cell glycosylation promotes metastasis. Front. Oncol. 2014, 4, 28.
- Lau, K.S.; Partridge, E.A.; Grigorian, A.; Silvescu, C.I.; Reinhold, V.N.; Demetriou, M.; Dennis, J.W. Complex N-Glycan Number and Degree of Branching Cooperate to Regulate Cell Proliferation and Differentiation. Cell 2007, 129, 123–134.
- Tominaga, N.; Hagiwara, K.; Kosaka, N.; Honma, K.; Nakagama, H.; Ochiya, T. RPN2-mediated glycosylation of tetraspanin CD63 regulates breast cancer cell malignancy. Mol. Cancer 2014, 13, 134.
- Macedo-da-Silva, J.; Santiago, V.F.; Rosa-Fernandes, L.; Marinho, C.R.F.; Palmisano, G. Protein glycosylation in extracellular vesicles: Structural characterization and biological functions. Mol. Immunol. 2021, 135, 226–246.
- Battke, C.; Ruiss, R.; Welsch, U.; Wimberger, P.; Lang, S.; Jochum, S.; Zeidler, R. Tumour exosomes inhibit binding of tumour-reactive antibodies to tumour cells and reduce ADCC. Cancer Immunol. Immunother. 2011, 60, 639–648.
- Pigati, L.; Yaddanapudi, S.C.S.; Iyengar, R.; Kim, D.J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 2010, 5, e13515.
- Smyth, T.; Petrova, K.; Payton, N.M.; Persaud, I.; Redzic, J.S.; Graner, M.W.; Smith-Jones, P.; Anchordoquy, T.J. Surface functionalization of exosomes using click chemistry. Bioconjug. Chem. 2014, 25, 1777–1784.
- Gangoda, L.; Liem, M.; Ang, C.S.; Keerthikumar, S.; Adda, C.G.; Parker, B.S.; Mathivanan, S. Proteomic Profiling of Exosomes Secreted by Breast Cancer Cells with Varying Metastatic Potential. Proteomics 2017, 17, 1600370.
- Menck, K.; Scharf, C.; Bleckmann, A.; Dyck, L.; Rost, U.; Wenzel, D.; Dhople, V.M.; Siam, L.; Pukrop, T.; Binder, C.; et al. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J. Mol. Cell Biol. 2015, 7, 143–153.
- Koga, K.; Matsumoto, K.; Akiyoshi, T.; Kubo, M.; Yamanaka, N.; Tasaki, A.; Nakashima, H.; Nakamura, M.; Kuroki, S.; Tanaka, M.; et al. Purification, Characterization and Biological Significance of Tumor-derived Exosomes. Anticancer Res. 2005, 25, 3703–3707.
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Théry, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303.
- Nishida-Aoki, N.; Tominaga, N.; Kosaka, N.; Ochiya, T. Altered biodistribution of deglycosylated extracellular vesicles through enhanced cellular uptake. J. Extracell. Vesicles 2020, 9, 1713527.
- Kavanagh, E.L.; Halasz, M.; Dowling, P.; Withers, J.; Lindsay, S.; Higgins, M.J.; Irwin, J.A.; Rudd, P.M.; Saldova, R.; McCann, A. N -Linked glycosylation profiles of therapeutic induced senescent (TIS) triple negative breast cancer cells (TNBC) and their extracellular vesicle (EV) progeny. Mol. Omics 2021, 17, 72–85.
- Lima, L.G.; Ham, S.; Shin, H.; Chai, E.P.Z.; Lek, E.S.H.; Lobb, R.J.; Müller, A.F.; Mathivanan, S.; Yeo, B.; Choi, Y.; et al. Tumor microenvironmental cytokines bound to cancer exosomes determine uptake by cytokine receptor-expressing cells and biodistribution. Nat. Commun. 2021, 12, 3543.
- Tan, Z.; Cao, L.; Wu, Y.; Wang, B.; Song, Z.; Yang, J.; Cheng, L.; Yang, X.; Zhou, X.; Dai, Z. Bisecting GlcNAc modification diminishes the pro-metastatic functions of small extracellular vesicles from breast cancer cells. J. Extracell. Vesicles 2020, 10, e12005.
- Hait, N.C.; Maiti, A.; Wu, R.; Andersen, V.L.; Hsu, C.-C.; Wu, Y.; Chapla, D.G.; Takabe, K.; Rusiniak, M.E.; Bshara, W.; et al. Extracellular sialyltransferase st6gal1 in breast tumor cell growth and invasiveness. Cancer Gene Ther. 2022, 29, 1662–1675.
- Stowell, S.R.; Ju, T.; Cummings, R.D. Protein glycosylation in cancer. Annu. Rev. Pathol. 2015, 10, 473–510.
- Toraskar, J.; Magnussen, S.N.; Hagen, L.; Sharma, A.; Hoang, L.; Bjørkøy, G.; Svineng, G.; Steigedal, T.S. A novel truncated form of nephronectin is present in small extracellular vesicles isolated from 66cl4 cells. J. Proteome Res. 2019, 18, 1237–1247.
- Cao, L.; Wu, Y.; Wang, X.; Li, X.; Tan, Z.; Guan, F. Role of Site-Specific Glycosylation in the I-Like Domain of Integrin β1 in Small Extracellular Vesicle-Mediated Malignant Behavior and FAK Activation. Int. J. Mol. Sci. 2021, 22, 1770.
- AL-Abedi, R.; Tuncay Cagatay, S.; Mayah, A.; Brooks, S.A.; Kadhim, M. Ionising Radiation Promotes Invasive Potential of Breast Cancer Cells: The Role of Exosomes in the Process. Int. J. Mol. Sci. 2021, 22, 11570.
- Bard, M.P.; Hegmans, J.P.; Hemmes, A.; Luider, T.M.; Willemsen, R.; Severijnen, L.A.A.; Van Meerbeeck, J.P.; Burgers, S.A.; Hoogsteden, H.C.; Lambrecht, B.N. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol. 2004, 31, 114–121.
- Keklikoglou, I.; Cianciaruso, C.; Güç, E.; Squadrito, M.L.; Spring, L.M.; Tazzyman, S.; Lambein, L.; Poissonnier, A.; Ferraro, G.B.; Baer, C.; et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat. Cell Biol. 2018, 21, 190–202.
- Vergauwen, G.; Tulkens, J.; Pinheiro, C.; Avila Cobos, F.; Dedeyne, S.; De Scheerder, M.A.; Vandekerckhove, L.; Impens, F.; Miinalainen, I.; Braems, G.; et al. Robust sequential biophysical fractionation of blood plasma to study variations in the biomolecular landscape of systemically circulating extracellular vesicles across clinical conditions. J. Extracell. Vesicles 2021, 10, e12122.
- Gonzalez, E.; Piva, M.; Rodriguez-Suarez, E.; Gil, D.; Royo, F.; Elortza, F.; Falcon-Perez, J.M.; Vivanco, M.D.M. Human Mammospheres Secrete Hormone-Regulated Active Extracellular Vesicles. PLoS ONE 2014, 9, e83955.
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012, 151, 1542–1556.
- Shimoda, M.; Principe, S.; Jackson, H.W.; Luga, V.; Fang, H.; Molyneux, S.D.; Shao, Y.W.; Aiken, A.; Waterhouse, P.D.; Karamboulas, C.; et al. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat. Cell Biol. 2014, 16, 889–901.
- Moon, P.G.; Lee, J.E.; Cho, Y.E.; Lee, S.J.; Jung, J.H.; Chae, Y.S.; Bae, H.I.; Kim, Y.B.; Kim, I.S.; Park, H.Y.; et al. Identification of Developmental Endothelial Locus-1 on Circulating Extracellular Vesicles as a Novel Biomarker for Early Breast Cancer Detection. Clin. Cancer Res. 2016, 22, 1757–1766.
- SUN, J.G.; LI, X.B.; YIN, R.H.; LI, X.F. lncRNA VIM-AS1 promotes cell proliferation, metastasis and epithelial–mesenchymal transition by activating the Wnt/β-catenin pathway in gastric cancer. Mol. Med. Rep. 2020, 22, 4567–4578.
- Wu, S.; Du, Y.; Beckford, J.; Alachkar, H. Upregulation of the EMT marker vimentin is associated with poor clinical outcome in acute myeloid leukemia. J. Transl. Med. 2018, 16, 170.
More
Information
Subjects:
Biochemistry & Molecular Biology; Oncology; Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
473
Revisions:
2 times
(View History)
Update Date:
04 Sep 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




