Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mohan Pammi | -- | 1449 | 2023-08-30 18:23:50 | | | |
| 2 | Wendy Huang | Meta information modification | 1449 | 2023-08-31 08:34:33 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Blutt, S.E.; Coarfa, C.; Neu, J.; Pammi, M. Multiomic Investigations into Lung Disease. Encyclopedia. Available online: https://encyclopedia.pub/entry/48657 (accessed on 07 February 2026).
Blutt SE, Coarfa C, Neu J, Pammi M. Multiomic Investigations into Lung Disease. Encyclopedia. Available at: https://encyclopedia.pub/entry/48657. Accessed February 07, 2026.
Blutt, Sarah E., Cristian Coarfa, Josef Neu, Mohan Pammi. "Multiomic Investigations into Lung Disease" Encyclopedia, https://encyclopedia.pub/entry/48657 (accessed February 07, 2026).
Blutt, S.E., Coarfa, C., Neu, J., & Pammi, M. (2023, August 30). Multiomic Investigations into Lung Disease. In Encyclopedia. https://encyclopedia.pub/entry/48657
Blutt, Sarah E., et al. "Multiomic Investigations into Lung Disease." Encyclopedia. Web. 30 August, 2023.
Copy Citation
Diseases of the lung account for more than 5 million deaths worldwide and are a healthcare burden. Improving clinical outcomes, including mortality and quality of life, involves a holistic understanding of the disease, which can be provided by the integration of lung multi-omics data. An enhanced understanding of comprehensive multiomic datasets provides opportunities to leverage those datasets to inform the treatment and prevention of lung diseases by classifying severity, prognostication, and discovery of biomarkers.
multiomics
lung
lung disease
lung cancer
machine learning
organoid models
animal models
1. Introduction
Respiratory diseases account for over 5 million deaths yearly, constitute a significant cause of morbidity and are a huge burden to healthcare systems worldwide [1][2]. Annually, about three million deaths are due to chronic obstructive pulmonary disease and asthma (the most common disease) accounts for half a million deaths. Other pulmonary diseases with chronic inflammation and obstruction and often exacerbated by infection include cystic fibrosis, idiopathic pulmonary fibrosis, ciliary dyskinesia, and pneumonia are also leading causes of death and lung cancer leads to the cancer-related deaths category [1][2].
With the ongoing global pandemic due to SARS-COVID-19 [3], respiratory diseases remain a leading cause of death and disability. Recent advances in high-throughput technologies have provided access to multiomics biological data, including genomics, epigenomics, transcriptomics, proteomics, metabolomics and immunomics, and provide a holistic view of pathophysiology in lung disease [4]. Multiomics data give a comprehensive overview of cellular processes (e.g., gene transcription, protein translation or epigenetic processes) associated with a disease and an insight into the complexity of the disease. Biological insights from multi-omics can be integrated with clinical and social data and applied in the clinical setting for improved health outcomes. Single omics are limited by providing associations, whereas multiomic integrations result in deriving a holistic insight that generates testable hypotheses about mechanisms.
State-of-the-art machine-learning methods can integrate high dimensional omics datasets resulting in the ability to predict short- and long-term health trajectories and enable early timely interventions that alter the health course towards better outcomes (precision medicine) [5]. Large datasets such as the omics dataset rely on ‘deep learning’ based on neural networks loosely modeled after neurons of the brain [6]. The insights gained by deep learning of multiomic datasets enhance personalized healthcare decision-making (precision medicine) and biomarker discovery [5][7].
2. Lung Multiomics Models
In almost all respiratory diseases, the epithelium, a monolayer of cells which comprises the lining of the conducting and respiratory airways, is damaged. This compromises the proximal airways’ ability to warm, humidify, and cleanse the inhaled air and distal air to facilitate gas exchange. As a result, health and quality of life are severely impacted by the impaired lung function that occurs in respiratory disease. Human models, such as primary cells and organoids, and animal studies involving integrated multi-omics will allow differences in markers and biological processes between disease and non-disease models to be elucidated (Figure 1). These differences will provide insight into lung disease, including pathways that result in regeneration and repair. Understanding these pathways will be critical in developing preventive treatments and therapeutic modalities to treat lung diseases. It can eventually be harnessed to develop a personalized approach to treating respiratory diseases.
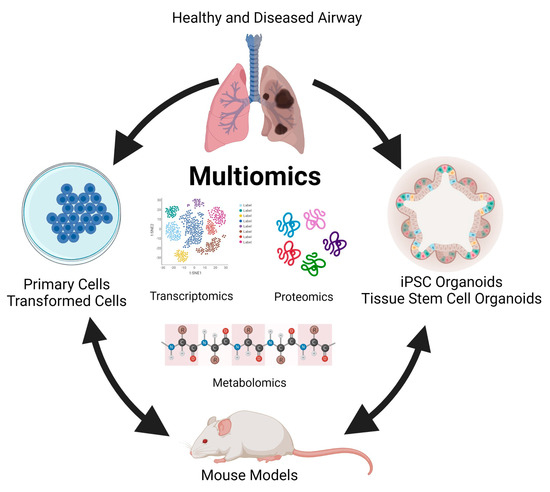
Figure 1. Modelling multi-omics studies in the study of human lung disease.
In recent decades, primary cells and transformed or tumor cell lines have been used to investigate lung diseases. The cells in these models retain many donor tissue characteristics and recapitulate markers and functions present in vivo [8][9][10]. These models have the advantage that they are amenable to genetic engineering, allowing the dissection of the role of individual molecules and pathways in disease [11]. Additionally, the ease of genetic engineering in these systems has allowed testing function via inducible gene expression [12]. Because of their wide use, many of these cell lines are well characterized, providing a foundation for multi-omics studies. Studies in cell lines are well suited for high throughput drug screening and evaluation of drug response [13] and are particularly valuable in studying lung cancer [14][15]. However, these models are not without limitations.
First and foremost, they fall short of replicating the complex nature of many respiratory diseases. Many lack the multiple cell types and cellular polarity present in the proximal and distal respiratory epithelium and exhibit an absence of morphology and structural features that play a significant role in lung biology. These cell lines also lack an immune cell component, which plays an important role in the etiology of many lung diseases [16]. Coupled with questions about the relevance of findings using these models, technical issues, including a requirement for tissue donors, a finite lifespan, and limited expansion capacity, have contributed to a reduced focus on using multi-omics in these models to study many respiratory diseases.
More recently, organoid models have come to the forefront of multi-omic studies of respiratory disorders. Organoids can be derived from either induced pluripotent stem cells or embryonic stem cells (hereafter referred to as iPSC organoids) or established from tissue-derived multipotent stem cells (referred to as ASC organoids) [17]. Differentiation of iPSC organoids occurs in a multistep process that involves a definitive endoderm stage, anterior foregut stage, and then into NKX2-1+ lung epithelial progenitors [18][19]. ASC organoids are established following mechanical and enzymatic isolation of conducting airway or respiratory epithelium stem cells from lavage, small amounts of native tissue, or biopsy specimen [20][21][22][23]. Both iPSC and ASC organoids rely heavily on manipulating exogenously added growth factors to induce differentiation of the mature polarized airway epithelium and the presence of extracellular matrix such as Matrigel®, synthetic matrices, or decellularized tissue scaffoldings. Both organoid models require cultivation on transwells under air-liquid interface conditions (ALI) where the basal side of the epithelium is in contact with media and the apical side is exposed to air to achieve maximum differentiation potential [22][24]. iPSC and ASC organoids can give rise to alveolar organoid models that recapitulate respiratory epithelium, nasal, trachea, or bronchial organoids that recapitulate conducting airway epithelium, and lung organoids that are a mixture [25][26][27]. Like primary and transformed cell models, organoids are amenable to genetic engineering and can be established from donors with genetic disorders that cause lung disease [28]. Organoids are well suited for drug screening and as models for infectious disease research [29][30][31][32] and recapitulate many aspects of other chronic lung diseases such as idiopathic pulmonary fibrosis [33] and cancer [22]. However, iPSC-derived airway cells do not seem to achieve the maturation levels observed in the human lung [34], although ASC organoids seem to contain mature epithelial cells, they lack stromal components such as the immune system that play a significant role in most lung diseases. However, the increased cellular complexity and the modeling of human epithelium combined with a forward-thinking multi-omics approach provide an area for advancement in information surrounding respiratory illness with significant translational potential.
Reports of machine learning integrated multiomics in organoid models and animal models have been published. One organoid model investigated the tumorigenic potential of alveolar type 2 cells (cells of origin of lung adenocarcinoma) using a multiomics and unsupervised machine learning approach [35]. The findings highlight the utility of understanding chromatin regulation in the early oncogenic versions of epithelial cells, which may reveal more effective means to intervene in the progression of Kras-driven lung cancer. In a murine model, researchers investigated the interplay of the gut microbiome, metabolism, and host inflammation in obesity-associated asthma using a multiomics approach (microbiome, metabolomics and proteomics) to profile the gut-lung axis in the setting of allergic airway disease and diet-induced obesity [36]. The researchers reported that changes to structural proteins in the lung airways and parenchyma may contribute to heightened lung elastance and serve as a potential therapeutic target for obese allergic asthma.
There are many animal models of lung diseases, including chronic diseases such as cystic fibrosis [37], idiopathic pulmonary fibrosis [38][39], viral and bacterial infections [40], and cancer [41]. Animal models of respiratory diseases offer several advantages including reproducibility, control of environmental factors, unlimited numbers of replicates, genetic phenotyping, and accessibility to lung tissue. Multi-omics approaches can be easily used to provide insight into the relationship between environmental stressors and the effect of the stressor on respiratory disease. The information gained can lead to detailed physiologic and pathologic pathways contributing to disease pathogenesis. Animal models of lung disease are instrumental in assessing the predisposition of genetic mutations in causing a specific disease and provide a model in which interactions between components of the whole system can also be examined [42]. Animal models are limited because, in most cases, there are significant differences between human lung tissue and animal lung tissue [43][44]. In addition, many human respiratory diseases are not recapitulated in animal models, and clinical manifestations are difficult to assess. However, comparisons between human and animal multi-omics analyses can validate animal models. Together, multi-omic-based approaches combining data collected from human in vitro and animal in vivo models will provide robustness, rigor, and reproducibility to support drawn conclusions.
References
- Humbert, M.V.; Spalluto, C.M.; Bell, J.; Blume, C.; Conforti, F.; Davies, E.R.; Dean, L.S.N.; Elkington, P.; Haitchi, H.M.; Jackson, C.; et al. Towards an artificial human lung: Modelling organ-like complexity to aid mechanistic understanding. Eur. Respir. J. 2022, 60, 2200455.
- GBD Chronic Respiratory Disease Collaborators. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med. 2020, 8, 585–596.
- Atzrodt, C.L.; Maknojia, I.; McCarthy, R.D.P.; Oldfield, T.M.; Po, J.; Ta, K.T.L.; Stepp, H.E.; Clements, T.P. A Guide to COVID-19: A global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020, 287, 3633–3650.
- Lee, A.J.; Einarsson, G.G.; Gilpin, D.F.; Tunney, M.M. Multi-Omics Approaches: The Key to Improving Respiratory Health in People with Cystic Fibrosis? Front. Pharmacol. 2020, 11, 569821.
- MacEachern, S.J.; Forkert, N.D. Machine learning for precision medicine. Genome 2021, 64, 416–425.
- Mathema, V.B.; Sen, P.; Lamichhane, S.; Orešič, M.; Khoomrung, S. Deep learning facilitates multi-data type analysis and predictive biomarker discovery in cancer precision medicine. Comput. Struct. Biotechnol. J. 2023, 21, 1372–1382.
- Bellman, R. Dynamic programming. Science 1966, 153, 34–37.
- Gruenert, D.C.; Willems, M.; Cassiman, J.J.; Frizzell, R.A. Established cell lines used in cystic fibrosis research. J. Cyst. Fibros. 2004, 3 (Suppl. S2), 191–196.
- Ren, H.; Birch, N.P.; Suresh, V. An Optimised Human Cell Culture Model for Alveolar Epithelial Transport. PLoS ONE 2016, 11, e0165225.
- Hermanns, M.I.; Unger, R.E.; Kehe, K.; Peters, K.; Kirkpatrick, C.J. Lung epithelial cell lines in coculture with human pulmonary microvascular endothelial cells: Development of an alveolo-capillary barrier in vitro. Lab. Investig. 2004, 84, 736–752.
- Kiełbus, M.; Czapiński, J.; Kałafut, J.; Woś, J.; Stepulak, A.; Rivero-Müller, A. Genetically Engineered Lung Cancer Cells for Analyzing Epithelial-Mesenchymal Transition. Cells 2019, 8, 1644.
- Kallunki, T.; Barisic, M.; Jäättelä, M.; Liu, B. How to Choose the Right Inducible Gene Expression System for Mammalian Studies? Cells 2019, 8, 796.
- Ling, A.; Gruener, R.F.; Fessler, J.; Huang, R.S. More than fishing for a cure: The promises and pitfalls of high throughput cancer cell line screens. Pharmacol. Ther. 2018, 191, 178–189.
- Kitaeva, K.V.; Rutland, C.S.; Rizvanov, A.A.; Solovyeva, V.V. Cell Culture Based in vitro Test Systems for Anticancer Drug Screening. Front. Bioeng. Biotechnol. 2020, 8, 322.
- Wong, A.H.; Li, H.; Jia, Y.; Mak, P.I.; Martins, R.; Liu, Y.; Vong, C.M.; Wong, H.C.; Wong, P.K.; Wang, H.; et al. Drug screening of cancer cell lines and human primary tumors using droplet microfluidics. Sci. Rep. 2017, 7, 9109.
- Wilson, J.D.K. The lungs at the frontlines of immunity. Nat. Immunol. 2015, 16, 17.
- van der Vaart, J.; Clevers, H. Airway organoids as models of human disease. J. Intern. Med. 2021, 289, 604–613.
- McCauley, K.B.; Hawkins, F.; Serra, M.; Thomas, D.C.; Jacob, A.; Kotton, D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 2017, 20, 844–857.e6.
- McCauley, K.B.; Hawkins, F.; Kotton, D.N. Derivation of Epithelial-Only Airway Organoids from Human Pluripotent Stem Cells. Curr. Protoc. Stem Cell Biol. 2018, 45, e51.
- Kumar, P.A.; Hu, Y.; Yamamoto, Y.; Hoe, N.B.; Wei, T.S.; Mu, D.; Sun, Y.; Joo, L.S.; Dagher, R.; Zielonka, E.M.; et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 2011, 147, 525–538.
- Usui, S.; Shimizu, T.; Kishioka, C.; Fujita, K.; Sakakura, Y. Secretory cell differentiation and mucus secretion in cultures of human nasal epithelial cells: Use of a monoclonal antibody to study human nasal mucin. Ann. Otol. Rhinol. Laryngol. 2000, 109, 271–277.
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38, e100300.
- Chiu, M.C.; Li, C.; Liu, X.; Yu, Y.; Huang, J.; Wan, Z.; Xiao, D.; Chu, H.; Cai, J.P.; Zhou, B.; et al. A bipotential organoid model of respiratory epithelium recapitulates high infectivity of SARS-CoV-2 Omicron variant. Cell Discov. 2022, 8, 57.
- Bluhmki, T.; Traub, S.; Müller, A.K.; Bitzer, S.; Schruf, E.; Bammert, M.T.; Leist, M.; Gantner, F.; Garnett, J.P.; Heilker, R. Functional human iPSC-derived alveolar-like cells cultured in a miniaturized 96-Transwell air-liquid interface model. Sci. Rep. 2021, 11, 17028.
- Yang, W.; Li, Y.; Shi, F.; Liu, H. Human lung organoid: Models for respiratory biology and diseases. Dev. Biol. 2023, 494, 26–34.
- Yu, F.; Liu, F.; Liang, X.; Duan, L.; Li, Q.; Pan, G.; Ma, C.; Liu, M.; Li, M.; Wang, P.; et al. iPSC-Derived Airway Epithelial Cells: Progress, Promise, and Challenges. Stem Cells 2023, 41, 1–10.
- Lu, T.; Cao, Y.; Zhao, P.; Shen, S.; Xi, Y. Organoid: A powerful tool to study lung regeneration and disease. Cell Regen. 2021, 10, 21.
- Chen, J.; Na, F. Organoid technology and applications in lung diseases: Models, mechanism research and therapy opportunities. Front. Bioeng. Biotechnol. 2022, 10, 1066869.
- van der Sanden, S.M.G.; Sachs, N.; Koekkoek, S.M.; Koen, G.; Pajkrt, D.; Clevers, H.; Wolthers, K.C. Enterovirus 71 infection of human airway organoids reveals VP1-145 as a viral infectivity determinant. Emerg. Microbes Infect. 2018, 7, 84.
- Zhou, J.; Li, C.; Sachs, N.; Chiu, M.C.; Wong, B.H.; Chu, H.; Poon, V.K.; Wang, D.; Zhao, X.; Wen, L.; et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. USA 2018, 115, 6822–6827.
- Heo, I.; Dutta, D.; Schaefer, D.A.; Iakobachvili, N.; Artegiani, B.; Sachs, N.; Boonekamp, K.E.; Bowden, G.; Hendrickx, A.P.A.; Willems, R.J.L.; et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018, 3, 814–823.
- Hui, K.P.Y.; Ching, R.H.H.; Chan, S.K.H.; Nicholls, J.M.; Sachs, N.; Clevers, H.; Peiris, J.S.M.; Chan, M.C.W. Tropism, replication competence, and innate immune responses of influenza virus: An analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir. Med. 2018, 6, 846–854.
- Wilkinson, D.C.; Alva-Ornelas, J.A.; Sucre, J.M.; Vijayaraj, P.; Durra, A.; Richardson, W.; Jonas, S.J.; Paul, M.K.; Karumbayaram, S.; Dunn, B.; et al. Development of a Three-Dimensional Bioengineering Technology to Generate Lung Tissue for Personalized Disease Modeling. Stem Cells Transl. Med. 2017, 6, 622–633.
- Miller, A.J.; Dye, B.R.; Ferrer-Torres, D.; Hill, D.R.; Overeem, A.W.; Shea, L.D.; Spence, J.R. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019, 14, 518–540.
- Kim, C.; Li, J.; Dang, S.; Schurmann, P.; Dost, A.; Moye, A.; Paschini, M.; Bhetariya, P.; Bronson, R.; Sui, S.H. Organoid modeling reveals the tumorigenic potential of the alveolar progenitor cell state. Res. Sq. 2023.
- Heinrich, V.A.; Uvalle, C.; Manni, M.L.; Li, K.; Mullett, S.J.; Donepudi, S.R.; Clader, J.; Fitch, A.; Ellgass, M.; Cechova, V.; et al. Meta-omics profiling of the gut-lung axis illuminates metabolic networks and host-microbial interactions associated with elevated lung elastance in a murine model of obese allergic asthma. Front. Microbiomes 2023, 2, 1153691.
- Wilke, M.; Buijs-Offerman, R.M.; Aarbiou, J.; Colledge, W.H.; Sheppard, D.N.; Touqui, L.; Bot, A.; Jorna, H.; de Jonge, H.R.; Scholte, B.J. Mouse models of cystic fibrosis: Phenotypic analysis and research applications. J. Cyst. Fibros. 2011, 10 (Suppl. S2), S152–S171.
- Walters, D.M.; Kleeberger, S.R. Mouse models of bleomycin-induced pulmonary fibrosis. Curr. Protoc. Pharmacol. 2008, 40, 5–46.
- Tashiro, J.; Rubio, G.A.; Limper, A.H.; Williams, K.; Elliot, S.J.; Ninou, I.; Aidinis, V.; Tzouvelekis, A.; Glassberg, M.K. Exploring Animal Models that Resemble Idiopathic Pulmonary Fibrosis. Front. Med. 2017, 4, 118.
- Lemaitre, J.; Naninck, T.; Delache, B.; Creppy, J.; Huber, P.; Holzapfel, M.; Bouillier, C.; Contreras, V.; Martinon, F.; Kahlaoui, N.; et al. Non-human primate models of human respiratory infections. Mol. Immunol. 2021, 135, 147–164.
- Kwon, M.C.; Berns, A. Mouse models for lung cancer. Mol. Oncol. 2013, 7, 165–177.
- Baron, R.M.; Choi, A.J.; Owen, C.A.; Choi, A.M. Genetically manipulated mouse models of lung disease: Potential and pitfalls. Am. J. Physiol. Lung Cell Mol. Physiol. 2012, 302, L485–L497.
- Ardini-Poleske, M.E.; Clark, R.F.; Ansong, C.; Carson, J.P.; Corley, R.A.; Deutsch, G.H.; Hagood, J.S.; Kaminski, N.; Mariani, T.J.; Potter, S.S.; et al. LungMAP: The Molecular Atlas of Lung Development Program. Am. J. Physiol. Lung Cell Mol. Physiol. 2017, 313, L733–L740.
- Pan, H.; Deutsch, G.H.; Wert, S.E. Comprehensive anatomic ontologies for lung development: A comparison of alveolar formation and maturation within mouse and human lung. J. Biomed. Semant. 2019, 10, 18.
More
Information
Subjects:
Respiratory System
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
526
Revisions:
2 times
(View History)
Update Date:
31 Aug 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




