Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Adrianna Magdalena Dzidek | -- | 2122 | 2023-08-25 20:24:19 | | | |
| 2 | Fanny Huang | Meta information modification | 2122 | 2023-08-28 07:25:36 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Dzidek, A.; Czerwińska-Ledwig, O.; Żychowska, M.; Pilch, W.; Piotrowska, A. Sirtuin 6 in Metabolic Activity. Encyclopedia. Available online: https://encyclopedia.pub/entry/48497 (accessed on 07 February 2026).
Dzidek A, Czerwińska-Ledwig O, Żychowska M, Pilch W, Piotrowska A. Sirtuin 6 in Metabolic Activity. Encyclopedia. Available at: https://encyclopedia.pub/entry/48497. Accessed February 07, 2026.
Dzidek, Adrianna, Olga Czerwińska-Ledwig, Małgorzata Żychowska, Wanda Pilch, Anna Piotrowska. "Sirtuin 6 in Metabolic Activity" Encyclopedia, https://encyclopedia.pub/entry/48497 (accessed February 07, 2026).
Dzidek, A., Czerwińska-Ledwig, O., Żychowska, M., Pilch, W., & Piotrowska, A. (2023, August 25). Sirtuin 6 in Metabolic Activity. In Encyclopedia. https://encyclopedia.pub/entry/48497
Dzidek, Adrianna, et al. "Sirtuin 6 in Metabolic Activity." Encyclopedia. Web. 25 August, 2023.
Copy Citation
Sirtuins, in mammals, are a group of seven enzymes (SIRT1–SIRT7) involved in the post-translational modification of proteins—they are considered longevity proteins. Sirtuin 6 (SIRT6), classified as class IV, is located on the cell nucleus. It affects many molecular pathways involved in aging: telomere maintenance, DNA repair, inflammatory processes or glycolysis. SIRT6 is involved in the regulation of homeostasis—an increase in the protein’s activity has been noted in calorie-restriction diets and with significant weight loss, among others. Expression of this protein is also elevated in people who regularly exercise.
sirtuins
SIRT6
aging
metabolism
1. Introduction
Silent information regulator (Sir) proteins belong to NAD+-dependent deacetylases, enzymes catalyzing the deacetylation reaction. Sirtuins were first discovered in the yeast Saccharomyces cerevisiae by virtue of their role in the establishment of transcriptional silencing of mating-type loci. Further studies have shown that Sir2 is also crucial in the partitioning of carbonylated proteins between mother and daughter cells, as well as for silencing at yeast telomeres and in the rDNA. The conserved enzymes called sirtuins, originally discovered in yeast, are produced by almost all organisms, from non-nucleated prokaryotes, to unicellular archaea and bacteria, to mammals [1][2].
Sirtuins play an important role in maintaining health and affect many pathways that increase the lifespan of organisms [1]. In mammals, they constitute a group of seven proteins (SIRT 1–7) belonging to class III histone deacetylases (HDACs). Sirtuins have a common catalytic domain of NAD+, consisting of about 260 amino acid residues [2]. Individual sirtuin isoforms differ in sequence and length in the N- and C-terminal domains, which affects their enzymatic activity, cellular localization and substrate specificity [3].
Initially, sirtuins were studied in the context of organism aging and gene silencing, but many other biological functions of proteins belonging to this group have been revealed in mammalian cells [4]. Studies on yeast aging showed a relationship between the Silent Information Regulator 2 (Sir2) gene and the viability of budding yeast [5].
Sirtuin 6 (SIRT6), belonging to class IV, is in the cell nucleus. SIRT6 promotes the repair of double-stranded DNA breaks by forming a complex with DNA-dependent protein kinase (DNA-PK) [6]. It has a hydrophobic pocket where the myristoylated group is located before being cleaved from the modified protein [7][8]. It has been suggested that this enzyme exhibits higher deacylase activity than deacetylase activity [4].
In a healthy organism or with minor damage, this protein promotes cell proliferation and activation of repair processes, while in the case of serious damage, it supports the apoptosis of damaged cells. It is in this mechanism that SIRT6 prevents the proliferation of damaged cancer cells [9][10]. SIRT6 slows down the course of gluconeogenesis by inhibiting the action of the PGC-1α factor. The activity of PGC-1α depends on the degree of its acetylation. This process is controlled by the acetyltransferase GCN5. SIRT6 causes deacetylation and phosphorylation of GCN5, which increases its acetylase activity. Over-acetylating PGC-1α, the acetyltransferase leads to a decrease in its activity and, consequently, to the inhibition of gluconeogenesis [11][12].
The association of sirtuins with aging is a well-known topic. Imai et al. [13] pointed to several mechanisms, including: effects on metabolism and regulation of circadian clock mechanisms, and emphasized that sirtuin activity decreases with age, which should be associated with a decrease in NAD+. The most important sirtuin described by the paper’s authors was SIRT1. Furthermore, it has been the most extensively studied protein of the mammalian sirtuin family. However, it has been shown that not only a loss of SIRT1 activity enhances DNA damage, but also SIRT6 [14].
2. Structure
SIRT6 is a widely expressed protein [15]. Mahlknecht et al. [15] demonstrated that the human SIRT6 genomic sequence has one single genomic locus, which spans a region of 8427 bp. The human SIRT6 gene is located on chromosome 19p13.3 and consists of eight exons ranging in size from 60 bp (exon 4) to 838 bp (exon 8). The mRNA for human SIRT6 encodes a protein of 355 amino acids, with a predicted molecular weight of 39.1 kDa and an isoelectric point of 9.12 [9]. SIRT6 is composed of two main domains—a large domain containing a nucleotide binding and Rossmann fold (responsible for binding NAD+) and a small domain containing a Zn2+ binding loop. Zinc ions play a role in maintaining the integrity of the catalytic domain and stabilizing the enzyme structure [16].
Activation and Reactivation of Gene Expression
The DNA molecule, which is a long strand, is wound around histone proteins that constitute a structural element of chromatin. The modification reaction of histone proteins is histone deacetylation, which involves the detachment of an acetyl group from the N-terminus of the histone. This reaction is carried out by histone deacetylases [17].
SIRT6, as a histone deacetylase, inhibits the transcriptional activity of several transcription factors and deacetylates specific histone lysine residues, such as H3K9 and H3K56, depending on NAD+ [18].
The N-terminal extension of SIRT6 is important for chromatin association and the internal catalytic activity of the core domain [16]. As a non-histone protein deacetylase, SIRT6 deacetylates forkhead box O1 protein, pyruvate kinase M2, C-terminal binding protein and GATA-binding protein 3 [18]. The C-terminal extension is essential for proper nuclear localization [16].
Although SIRT6-mediated histone deacetylation generally correlates with chromatin condensation and gene silencing [19][20], there is also evidence that SIRT6 can activate certain genes by mediating histone deacetylation. For example, SIRT6 acts as a transcriptional coactivator of erythroid 2-related factor 2 (Nrf2) to protect against oxidative stress in human mesenchymal stem cells, and it has been found that SIRT6 is in a protein complex with Nrf2 and RNAP II [21][22]. Nrf2 is one of the master regulators of antioxidant responses—binds to the antioxidant response elements (AREs) and activates antioxidant genes, for example, heme oxygenase 1 (HMOX1), which is known for counteracting reactive oxygen species (ROS). In aging cells, a decrease in Nrf2-ARE activity is observed, which may be caused by oxidative stress-related tissue degeneration [21]. Nrf2 activation occurs during evasion from Keap1-mediated proteasomal degradation in the cytosol [23]. SIRT6 activates Nrf2 by inhibiting Keap1 transcription and directly interacting with Nrf2 [24]. It is indicated that SIRT6 is required for HOMX1 induction in response to oxidative stress and SIRT6 deficiency increases basal cellular ROS [21] (Figure 1).
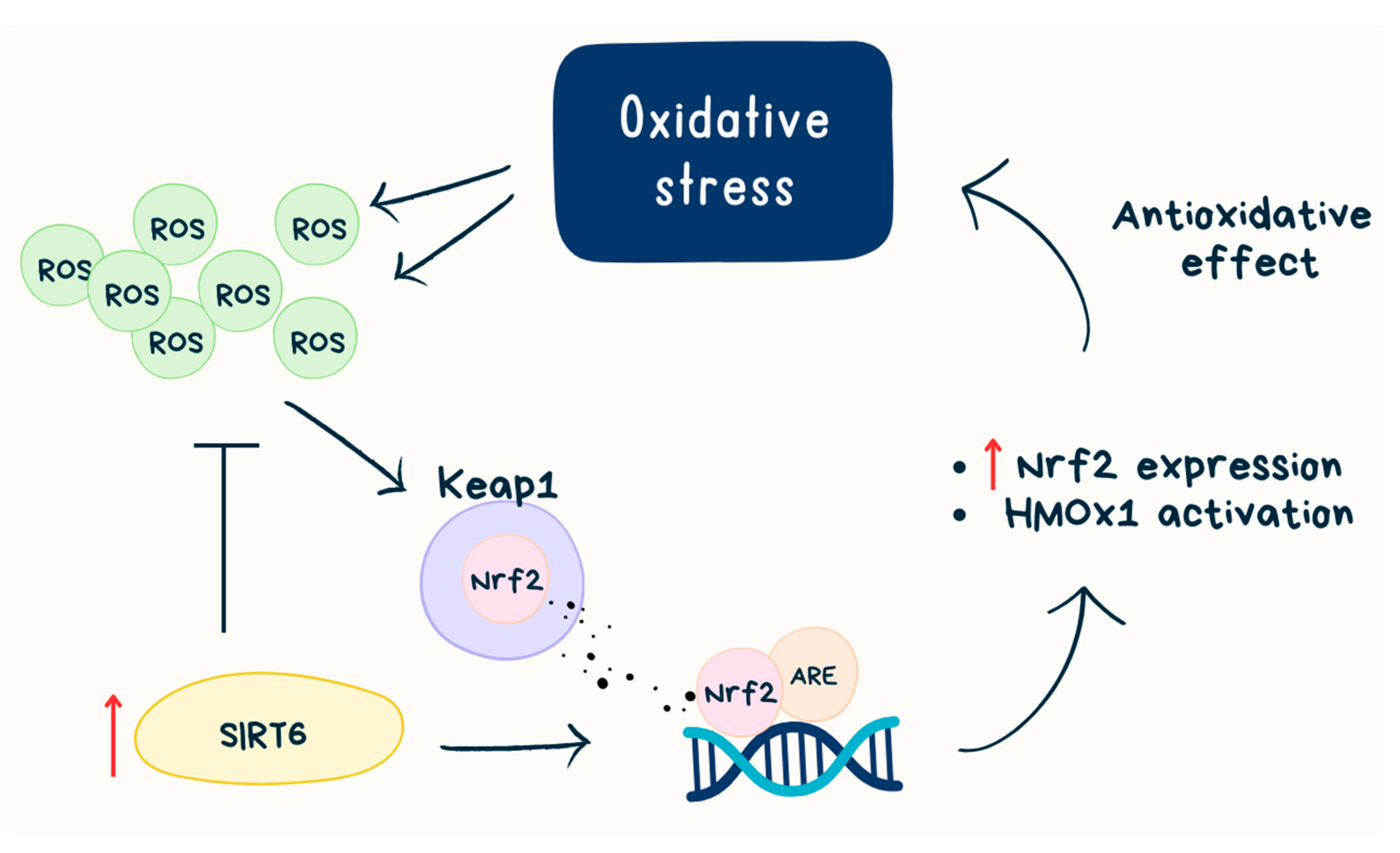
Figure 1. SIRT6 and Nrf2 path. ROS—reactive oxygen species; SIRT6—sirtuin 6, Keap1—Kelch-like ECH-associated protein 1, Nrf2—erythroid 2-related factor 2; ARE—antioxidant response elements; HMOX1—heme oxygenase 1.
3. Aging
Aging of the body is a physiological, dynamic and inevitable process. It is believed that the aging process begins at different times in different systems (the skin is one of the first organs to age). However, aging usually begins in the fourth decade of life and ends at the end of biological life. Important factors influencing aging are extrinsic, biological and psychosocial factors. However, the basic determinant of organism aging is the individual’s genotype [25]. Lopez-Otin et al. [26] proposed nine so-called hallmarks of aging: telomere attrition, genomic instability, loss of proteostasis, epigenetic alterations, deregulated nutrient sensing, mitochondrial dysfunction, stem cell exhaustion, cellular senescence and altered intercellular communication. There are some symptoms that occur in psychological aging, but at a younger age [27]. It is associated with accelerated or premature aging, where interrelated molecular and cellular phenomena are intensified.
Nowadays, it is hypothesized that SIRT6 regulates lifespan by influencing a series of processes that control aging, such as genome stability, transcriptional processes of DNA repair elements, the ability to regulate telomere length and metabolic homeostasis (including carbohydrate metabolism) [9][28][29] (Figure 2).

Figure 2. The role of the SIRT6 in aging.
4. Metabolic Activity
SIRT6 is involved in regulating glucose homeostasis in the body. It has been shown that SIRT6 levels are increased during fasting. Additionally, by increasing the expression of genes involved in gluconeogenesis, it controls this process in the liver. Zhong et al. [30] discovered that the expression of gluconeogenic genes was increased in livers with SIRT6 deficiency. Researchers also identified the role of SIRT6 as a corepressor of Hif1α (Hypoxia-inducible factor 1α), a critical regulator of the response to nutritional stress. Cells deficient in SIRT6 exhibit increased Hif1α activity, thus showing increased glucose uptake. SIRT6, therefore, acts as a corepressor of the transcription factor Hif1α, reducing glycolysis during normal nutrition and stimulating mitochondrial fatty acid oxidation [30].
Kim et al. [31] pointed out that liver-specific deletion of SIRT6 in mice leads to profound changes in gene expression, causing increased glycolysis, intensified triglyceride synthesis, decreased β-oxidation and liver steatosis. Other authors suggest that SIRT6 may therefore be a potential target in the treatment of liver diseases characterized by lipid accumulation [32].
Dominy et al. [11] indicated the usefulness of activating liver SIRT6 in the therapeutic treatment of insulin-resistant diabetes. Xiong et al. [33] demonstrated a significant decrease (50%) in glucose-stimulated insulin secretion in mice subjected to SIRT6 knockdown in pancreatic beta cells. The mice also had lower levels of ATP in the studied cells compared to the wild-type control group. An increased number of damaged mitochondria was also identified. Based on the obtained results, the authors suggested that SIRT6 regulates proper insulin secretion through the regulation of mitochondrial glucose oxidation. Therefore, activating the protein may be helpful in improving insulin secretion in diabetic states. The process described here takes place in the mitochondria. This underscores that SIRT6 acts in many cellular organelles, so it can be said that it acts throughout the cell.
Further experiments have indicated that mice with SIRT6 knockout were more susceptible to high-fat diet-induced obesity, attributed to adipocyte hypertrophy. Moreover, increased macrophage infiltration into the examined adipose tissue was observed, indicating an intensified inflammatory process in these mice. It was found that SIRT6 regulates energy homeostasis by modulating the activity and expression of lipase in adipose tissue [34]. Kanfi et al. [35] noticed increased glucose tolerance and insulin secretion stimulated by glucose in mice with SIRT6 overexpression. Their study indicated that SIRT6 overexpression increases triglyceride clearance in the blood and reduces triglyceride production in adipose tissue. Mice with SIRT6 overexpression subjected to a high-fat diet accumulated significantly less visceral fat, LDL cholesterol and triglycerides compared to wild-type mice. This suggests a protective role of SIRT6 against metabolic consequences of obesity caused by an improper diet [35]. Tang et al. [36] came to similar conclusions—SIRT6 overexpression in the arcuate nucleus of the hypothalamus in mice reduced their body weight induced by a high-fat diet. A decrease in the weight of eWAT (epididymal white adipose tissue) and iWAT (inguinal white adipose tissue) and adipocyte size was also observed. Furthermore, the impairment of leptin activity in the POMC neurons of mice subjected to SIRT6 neuron-specific knockout was demonstrated. These mice exhibited a predisposition to obesity and increased food consumption.
Increased SIRT6 levels were observed in in vivo models—in mice after fasting, rats after caloric restriction and in vitro—in cell cultures in a medium deprived of nutrients. The authors indicated that the increase in SIRT6 levels is caused by the stabilization of the SIRT6 protein, not an increase in SIRT6 transcription. Moreover, p53 positively regulates SIRT6 protein levels under standard growth conditions but does not play a role in regulating SIRT6 under caloric restriction [37]. It is widely known that calorie restriction diets (CR) slow down aging processes and may contribute to extending life [38]. In light of the above research, it can be suggested that the beneficial effects of a calorie-restricted diet are strongly related to increased SIRT6 stability, and the increased expression of the protein generated by factors other than starvation may exhibit similar effects to CR [37]. The role of SIRT6 in metabolism has been summarized in Table 1.
Table 1. Role of the SIRT6 in metabolic activity regulation.
| SIRT6 and Metabolic Activity | |
|---|---|
| Obesity | SIRT6 plays a protective role against the metabolic consequences of diet-induced obesity, which suggests a potentially beneficial effect of SIRT6 activation on age-related metabolic diseases [35]. In obese patients, the expression of SIRT6 is reduced. It suggests that SIRT6 is an attractive therapeutic target for treating obesity and obesity-related metabolic disorders [34]. |
| Fat metabolism | SIRT6 plays a critical role in fat metabolism, and may therefore be a potential target in the treatment of liver diseases characterized by lipid accumulation [31]. |
| Carbohydrate metabolism | Activation of hepatic by SIRT6 may be therapeutically useful for treating insulin-resistant diabetes [11]. SIRT6 may be useful to improve insulin secretion in diabetes [33]. |
| Energy balance | SIRT6 is an important molecular regulator for POMC neurons to promote negative energy balance [36]. |
| Other | SIRT6 appears to function as a corepressor of the Hif1α—a critical regulator of nutrient stress responses [30]. Expression of SIRT6 increased upon nutrient deprivation in cultured cells. The increase in SIRT6 levels is due to the stabilization of the SIRT6 protein, and not via an increase in SIRT6 transcription [37]. |
The “sirtfoods’’ diet, combing sirtuin-activating foods belonging to both Mediterranean and Asian diets, may be a promising dietary strategy in preventing chronic diseases, thereby ensuring healthy aging [39]. “Sirtfoods’’ can be found in: olive oil [40]; red wine [41]; grapes [42][43]; apple, strawberries, onion and cabbage [43]; soybeans [44]; tofu [45]; licorice [46]; shallot [46]. Pallauf et al. [39] suggested that omega-3 fatty acids, vitamins and antioxidants do not work in isolation. They should synergistically work to prevent chronic diseases.
References
- Haigis, M.C.; Sinclair, D.A. Mammalian Sirtuins: Biological Insights and Disease Relevance. Annu. Rev. Pathol. Mech. Dis. 2010, 5, 253–295.
- Carafa, V.; Rotili, D.; Forgione, M.; Cuomo, F.; Serretiello, E.; Hailu, G.S.; Jarho, E.; Lahtela-Kakkonen, M.; Mai, A.; Altucci, L. Sirtuin Functions and Modulation: From Chemistry to the Clinic. Clin. Epigenetics 2016, 8, 61.
- Frydzińska, Z.; Owczarek, A.; Winiarska, K. Sirtuiny i Ich Rola w Regulacji Metabolizmu. Postep. Biochem. 2019, 65, 31–40.
- Jing, H.; Lin, H. Sirtuins in Epigenetic Regulation. Chem. Rev. 2015, 115, 2350–2375.
- Sinclair, D.A.; Guarente, L. Extrachromosomal RDNA Circles-A Cause of Aging in Yeast. Cell 1997, 91, 1033–1042.
- Sharma, A.; Diecke, S.; Zhang, W.Y.; Lan, F.; He, C.; Mordwinkin, N.M.; Chua, K.F.; Wu, J.C. The Role of SIRT6 Protein in Aging and Reprogramming of Human Induced Pluripotent Stem Cells. J. Biol. Chem. 2013, 288, 18439–18447.
- Madsen, A.S.; Andersen, C.; Daoud, M.; Anderson, K.A.; Laursen, J.S.; Chakladar, S.; Huynh, F.K.; Colaço, A.R.; Backos, D.S.; Fristrup, P.; et al. Investigating the Sensitivity of NAD+-Dependent Sirtuin Deacylation Activities to NADH. J. Biol. Chem. 2016, 291, 7128–7141.
- Kanfi, Y.; Naiman, S.; Amir, G.; Peshti, V.; Zinman, G.; Nahum, L.; Bar-Joseph, Z.; Cohen, H.Y. The Sirtuin SIRT6 Regulates Lifespan in Male Mice. Nature 2012, 483, 218–221.
- Vitiello, M.; Zullo, A.; Servillo, L.; Mancini, F.P.; Borriello, A.; Giovane, A.; Della Ragione, F.; D’Onofrio, N.; Balestrieri, M.L. Multiple Pathways of SIRT6 at the Crossroads in the Control of Longevity, Cancer, and Cardiovascular Diseases. Ageing Res. Rev. 2017, 35, 301–311.
- Ran, L.K.; Chen, Y.; Zhang, Z.Z.; Tao, N.N.; Ren, J.H.; Zhou, L.; Tang, H.; Chen, X.; Chen, K.; Li, W.Y.; et al. SIRT6 Overexpression Potentiates Apoptosis Evasion in Hepatocellular Carcinoma via BCL2-Associated X Protein-Dependent Apoptotic Pathway. Clin. Cancer Res. 2016, 22, 3372–3382.
- Dominy, J.E.; Lee, Y.; Jedrychowski, M.P.; Chim, H.; Jurczak, M.J.; Camporez, J.P.; Ruan, H.B.; Feldman, J.; Pierce, K.; Mostoslavsky, R.; et al. The Deacetylase Sirt6 Activates the Acetyltransferase GCN5 and Suppresses Hepatic Gluconeogenesis. Mol. Cell 2012, 48, 900–913.
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as Regulators of Gene Expression. Science 2016, 351, 6274.
- Imai, S.i.; Guarente, L. NAD+ and Sirtuins in Aging and Disease. Trends Cell Biol. 2014, 24, 464–471.
- Tennen, R.I.; Chua, K.F. Chromatin Regulation and Genome Maintenance by Mammalian SIRT6. Trends Biochem. Sci. 2011, 36, 39–46.
- Mahlknecht, U.; Ho, A.; Volter-Mahlknecht, S. Chromosomal Organization and Fluorescence in Situhybridization of the Human Sirtuin 6 Gene. Int. J. Oncol. 2006, 28, 447–456.
- Gertler, A.A.; Cohen, H.Y. SIRT6, a Protein with Many Faces. Biogerontology 2013, 14, 629–639.
- Bielawski, A.; Nalepa, I. Sirtuins—Intriguing multi-tasking "keepers" of life processes. Wszechświat 2019, 20, 4–6.
- Koo, J.H.; Jang, H.Y.; Lee, Y.; Moon, Y.J.; Bae, E.J.; Yun, S.K.; Park, B.H. Myeloid Cell-Specific Sirtuin 6 Deficiency Delays Wound Healing in Mice by Modulating Inflammation and Macrophage Phenotypes. Exp. Mol. Med. 2019, 51, 1–10.
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 Polarization. Eur. J. Pharmacol. 2020, 877, 173090.
- Chang, A.R.; Ferrer, C.M.; Mostoslavsky, R. SIRT6, a Mammalian Deacylase with Multitasking Abilities. Physiol. Rev. 2020, 100, 145–169.
- Pan, H.; Guan, D.; Liu, X.; Li, J.; Wang, L.; Wu, J.; Zhou, J.; Zhang, W.; Ren, R.; Zhang, W.; et al. SIRT6 Safeguards Human Mesenchymal Stem Cells from Oxidative Stress by Coactivating NRF2. Cell Res. 2016, 26, 190–205.
- Thandavarayan, R.A.; Garikipati, V.N.S.; Joladarashi, D.; Suresh Babu, S.; Jeyabal, P.; Verma, S.K.; Mackie, A.R.; Khan, M.; Arumugam, S.; Watanabe, K.; et al. Sirtuin-6 Deficiency Exacerbates Diabetes-Induced Impairment of Wound Healing. Exp. Dermatol. 2015, 24, 773–778.
- Itoh, K.; Tong, K.I.; Yamamoto, M. Molecular Mechanism Activating Nrf2-Keap1 Pathway in Regulation of Adaptive Response to Electrophiles. Free. Radic. Biol. Med. 2004, 36, 1208–1213.
- Kanwal, A.; Pillai, V.B.; Gupta, M.P. The Nuclear and Mitochondrial Sirtuins, Sirt6 and Sirt3, Regulate each others’ Activity and Protect the Heart from Developing Obesity-Mediated Diabeticcardiomyopathy. J. Fed. Am. Soc. Exp. Biol. 2020, 34, 14057.
- Dziechciaż, M.; Filip, R. Biological Psychological and Social Determinants of Old Age: Bio-Psycho-Social Aspects of Human Aging. Ann. Agric. Environ. Med. 2014, 21, 835–838.
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194.
- Franco, A.C.; Aveleira, C.; Cavadas, C. Skin Senescence: Mechanisms and Impact on Whole-Body Aging. Trends Mol. Med. 2022, 28, 97–109.
- Mao, Z.; Hine, C.; Tian, X.; Van Meter, M.; Au, M.; Vaidya, A.; Seluanov, A.; Gorbunova, V. SIRT6 Promotes DNA Repair under Stress by Activating PARP1. Science 2011, 332, 1443–1446.
- Cardus, A.; Uryga, A.K.; Walters, G.; Erusalimsky, J.D. SIRT6 Protects Human Endothelial Cells from DNA Damage, Telomere Dysfunction, and Senescence. Cardiovasc. Res. 2013, 97, 571–579.
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1α. Cell 2010, 140, 280–293.
- Kim, H.S.; Xiao, C.; Wang, R.H.; Lahusen, T.; Xu, X.; Vassilopoulos, A.; Vazquez-Ortiz, G.; Jeong, W.I.; Park, O.; Ki, S.H.; et al. Hepatic-Specific Disruption of SIRT6 in Mice Results in Fatty Liver Formation Due to Enhanced Glycolysis and Triglyceride Synthesis. Cell Metab. 2010, 12, 224–236.
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as Regulators of Metabolism and Healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238.
- Xiong, X.; Wang, G.; Tao, R.; Wu, P.; Kono, T.; Li, K.; Ding, W.X.; Tong, X.; Tersey, S.A.; Harris, R.A.; et al. Sirtuin 6 Regulates Glucose-Stimulated Insulin Secretion in Mouse Pancreatic Beta Cells. Diabetologia 2016, 59, 151–160.
- Kuang, J.; Zhang, Y.; Liu, Q.; Shen, J.; Pu, S.; Cheng, S.; Chen, L.; Li, H.; Wu, T.; Li, R.; et al. Fat-Specific Sirt6 Ablation Sensitizes Mice to High-Fat Diet-Induced Obesity and Insulin Resistance by Inhibiting Lipolysis. Diabetes 2017, 66, 1159–1171.
- Kanfi, Y.; Peshti, V.; Gil, R.; Naiman, S.; Nahum, L.; Levin, E.; Kronfeld-Schor, N.; Cohen, H.Y. SIRT6 Protects against Pathological Damage Caused by Diet-Induced Obesity. Aging Cell 2010, 9, 162–173.
- Tang, Q.; Gao, Y.; Liu, Q.; Yang, X.; Wu, T.; Huang, C.; Huang, Y.; Zhang, J.; Zhang, Z.; Li, R.; et al. Sirt6 in Pro-Opiomelanocortin Neurons Controls Energy Metabolism by Modulating Leptin Signaling. Mol. Metab. 2020, 37, 100994.
- Kanfi, Y.; Shalman, R.; Peshti, V.; Pilosof, S.N.; Gozlan, Y.M.; Pearson, K.J.; Lerrer, B.; Moazed, D.; Marine, J.C.; de Cabo, R.; et al. Regulation of SIRT6 Protein Levels by Nutrient Availability. FEBS Lett. 2008, 582, 543–548.
- Mccay, C.M.; Crowell, M.F.; Maynard, L.A. The Effect of Retarded Growth upon the Length of Life Span and upon the Ultimate Body Size. 1935. Nutrition 1989, 5, 155–171.
- Pallauf, K.; Giller, K.; Huebbe, P.; Rimbach, G. Nutrition and Healthy Ageing: Calorie Restriction or Polyphenol-Rich “MediterrAsian” Diet? Oxid. Med. Cell Longev. 2013, 2013, 707421.
- Menendez, J.A.; Joven, J.; Aragonès, G.; Barrajón-Catalán, E.; Beltrán-Debón, R.; Borrás-Linares, I.; Camps, J.; Corominas-Faja, B.; Cufí, S.; Fernández-Arroyo, S.; et al. Xenohormetic and Anti-Aging Activity of Secoiridoid Polyphenols Present in Extra Virgin Olive Oil: A New Family of Gerosuppressant Agents. Cell Cycle 2013, 12, 555–578.
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.-L.; et al. Small Molecule Activators of Sirtuins Extend Saccharomyces Cerevisiae Lifespan. Nature 2003, 425, 191–196.
- Guerrero, R.F.; Puertas, B.; Fernández, M.I.; Palma, M.; Cantos-Villar, E. Induction of Stilbenes in Grapes by UV-C: Comparison of Different Subspecies of Vitis. Innov. Food Sci. Emerg. Technol. 2010, 11, 231–238.
- Arai, Y.; Watanabe, S.; Kimira, M.; Shimoi, K.; Mochizuki, R.; Kinae, N. Human Nutrition and Metabolism Dietary Intakes of Flavonols, Flavones and Isoflavones by Japanese Women and the Inverse Correlation between Quercetin Intake and Plasma LDL Cholesterol Concentration 1. J. Nutr. 2000, 130, 2243–2250.
- Rasbach, K.A.; Schnellmann, R.G. Isoflavones Promote Mitochondrial Biogenesis. J. Pharmacol. Exp. Ther. 2008, 325, 536–543.
- Song, T.; Barua, K.; Buseman, G.; Murphy, P. Soy Isoflavone Analysis: Quality Control and a New Internalstandard. Am. J. Clin. Nutr. 1998, 68, 1474–1479.
- Hsu, Y.L.; Chia, C.C.; Chen, P.J.; Huang, S.E.; Huang, S.C.; Kuo, P.L. Shallot and Licorice Constituent Isoliquiritigenin Arrests Cell Cycle Progression and Induces Apoptosis through the Induction of ATM/P53 and Initiation of the Mitochondrial System in Human Cervical Carcinoma HeLa Cells. Mol. Nutr. Food Res. 2009, 53, 826–835.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
704
Revisions:
2 times
(View History)
Update Date:
28 Aug 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




