1. Introduction
During the recent decades, there has been a significant surge of interest in the advancement of small-molecule fluorescent probes for imaging and detecting numerous biological targets
[1][2][3][4][5][6]. While a small subset of the probes may exhibit limitations in terms of target specificity, the majority of these pioneering probes have increasingly emerged as remarkably powerful tools for the real-time monitoring, in situ visualization, and quantitative analysis of various dynamic biological processes in live cells or tissues. This is attributed to their high specificity, exceptional sensitivity, non-destructive nature, and rapid analytical characteristics. Moreover, the modifiable chemical structures of small -molecule fluorescent probes enable the real-time visualization and detection of diverse ranges of enzymatic locations and activities in their native environments. The past few years have witnessed the rapid development of TrxR probes, and the arsenal of TrxR probes has become gradually enriched. These probes have contributed significantly to the investigation of TrxR’s biological functions and have been valuable tools in TrxR-related research.
2. The Mammalian TrxR Probes
2.1. Strained 1,2-Dithiolane as the Trigger
Enlightened by the early finding that the five-membered cyclic disulfide compounds lipoamide and lipoic acid could be efficiently opened by mammalian TrxR but not GSH
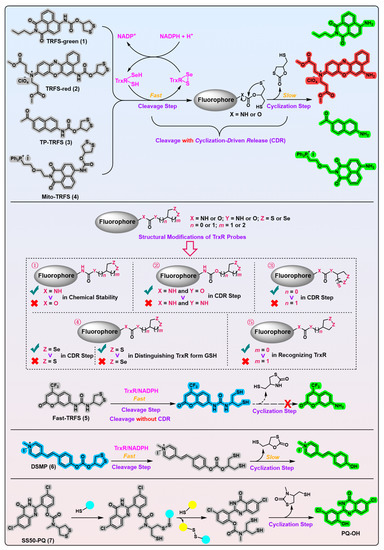
[7], the Fang group developed the first off–on fluorescent probe, called TRFS-green (
Figure 1, probe 1)
[8], for mammalian TrxR by linking the fluorophore naphthalimide to an artificial cyclic disulfide 1,2-dithiolane via a carbamate linker. With the reduction by NADPH under the catalysis of TrxR, TRFS-green emits a bright green fluorescence signal at λ = 538 nm when it is excited at λ = 438 nm. TRFS-green shows favorable selectivity to TrxR over other biologically related species, including small molecules like NADPH, GSH, ascorbic acid, and enzymes like lipoamide dehydrogenase and GR. Importantly, TRFS-green shows a trivial fluorescence signal upon U498C TrxR treatment, underlining the critical significance of TrxR’s Sec residue in recognizing TRFS-green. Further, lysates of different cell types with varying TrxR activity (determined by the classic Trx-mediated insulin reduction assay) show a corresponding ability to proportionally switch on the fluorescence signal of TRFS-green, whereas the TrxR inhibitor 2,4-dinitrochlorobenzene (DNCB) dose-dependently darkens the TRFS-green, highlighting the involvement of cellular TrxR in turning on the TRFS-green.
Figure 1. 1,2-Dithiolane-based TrxR probes. Probes 1–4 employ carbamate motif as the linker. Probes 5 and 6 use urea and carbonate motif as the linkers, respectively. The model probe 7 uses a stable phenolic carbamate motif as the linker. Following a two-step process, the free fluorophores are liberated in probes 1–4 and 6–7. Probe 5 is triggered only by cleavage of the disulfide bond in the recognition part. ①–⑤ indicate the influences of changing atoms in the linker unit and recognition part on the properties of probe 5. ①–⑤ also show the development process of probe 5. The cyan and yellow balls having sulfydryl on them shown in the working process of probe 7 indicate various biological thiols. Protein TrxR is shown in pink. The colors covering the chemical structures show their fluorescence colors after TrxR activation, and the gray indicates the fluorescence of the probes has been quenched.
In the further cellular evaluations of SS50-PQ and TRFS-green, the authors used SS00-PQ (a TrxR-independent linear disulfide probe) and RX1 (
Figure 2, probe 8; the most recently reported highly specific mammalian TrxR1 probe
[9]) as the controls. The results come that SS00-PQ, SS50-PQ, and TRFS-green are indeed almost independent of cellular TrxR modulation via chemical upregulation/knockdown, genetic knockout, and specific TrxR inhibitor treatment; however, the RX1 probe is significantly affected by these modulations. Although TRFS-green shows mild sensitivity to the genetic modulation of TrxR, it is important to highlight that TRFS-green retained its activation by 60% even in cells with aa complete knockout of TrxR1. Given the minimal response in TRFS-green to various TrxR modulations, the sensitivity of TRFS-green to Au-based treatment may be ascribed to the lipophilicity of Au compounds, such as auranofin, which enables them to effectively block cellular membrane thiols. Consequently, the diminished fluorescence of TRFS-green after a treatment of Au compounds may be a result of inhibited cellular uptake rather than the direct inhibition of TrxR by Au compounds. In combination with the aforementioned cell-free results, it is plausible to draw the inference that all existing carbamate-bridged 1,2-dithiolane probes may not possess selective cellular reporting capabilities for TrxR, irrespective of the observed partial reliance of TRFS-green on TrxR as evidenced by genetic studies
[8][10].
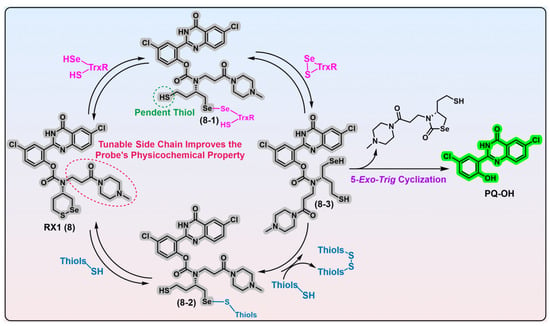
Figure 2. 1,2-Thiaselenane-based TrxR1 probe RX1. Probe RX1 interacts with the desired TrxR1 to liberate the fluorescent PQ-OH, during which process the key intermediates 8-1 and 8-3 are generated. Probe RX1 can also react with high concentrations of monothiols to give intermediate 8-2; however, this intermediate encounters significant kinetic and thermodynamic barriers to yield key intermediate 8-3. Thus, this unwanted parallel pathway induced by monothiols does not trigger the RX1. The bidirectional arrows indicate corresponding transformations are reversible. Protein TrxR1 is shown in pink and various biological thiols are shown in blue. Dashed ellipse in red shows the tunable side chain of probe 8, and the corresponding words in red explain the function of the side chain. Dashed cycle in green shows the potential reactive thiol in intermediate 8-1, and the corresponding words in green show the name of thiol appearing in the main text. The colors covering the chemical structures show their fluorescence colors after TrxR activation, and the gray indicates the fluorescence of the probes has been quenched.
Further, the authors evaluated Fast-TRFS (
Figure 1, probe 5) and speculated that the probe may undergo a reduction-independent but concentration-dependent and strain-promoted
ring-
opening
polymerization (ROP) to emit fluorescence, which is quite different from the previously reported activation mechanism
[11]. The authors hypothesized that the hydrophobic Fast-TRFS would concentrate from the cellular aqueous phase to the apolar cellular lipid membrane of the high surface area, thus initiating ROP to form a polymer of Fast-TRFS. According to the activation mechanism of cleavage without a cyclization of Fast-TRFS (
Figure 1, probe 5)
[11], the polymer of Fast-TRFS would emit the fluorescence, and furthermore, this fluorescence intensity would be lipid concentration-dependent. The authors utilized small lipid/phospholipid vesicle suspensions to simulate the apolar environments of cellular membrane, and they then evaluated the performance of Fast-TRFS in this artificial medium. Indeed, the surface phenomena observed in this experiment are congruent with the proposed hypothesis. Nevertheless, there persist a number of essentially notable concerns regarding both the hypothesis and the experimental outcomes. First, Fast-TRFS is a superfast reporter of TrxR (~70-fold fluorescence intensity plateaus within 5 min in cellular conditions), whereas the ROP-mediated turn-on of Fast-TRFS plateaus after a 60 min treatment even in a high concentration of lecithin (10 mg/mL). Second, the liberated fluorescence of Fast-TRFS is evenly distributed in the cytosol
[11], whereas the ROP-induced fluorogenicity of Fast-TRFS is believed to be localized to the cellular membrane. Third, although the use of this artificial, cell-like environment devoid of TrxR may be more tractable, it fails to accurately emulate the inhomogeneous cellular context, as TrxR probably possesses superior kinetics when compared with other mechanisms responsible for lighting up Fast-TRFS. Fourth, for TRFS-green type probes working via the mechanism of cleavage with cyclization, the ROP-induced fluorescence emission wavelength (λ
em = ~490 nm for TRFS-green type probes when cleaving without cyclization
[11]) or any other intact probe- or membrane thiol exchange intermediates/adducts-induced environment-dependent fluorescence emission wavelength is different from the emission wavelength of free fluorophores (λ
em = 538 nm). This fact makes the hypothesis of ROP-mediated fluorogenicity non-universal, i.e., except for Fast-TRFS (
Figure 1, probe 5) with a unique activation mechanism, other TRFS probes all show the free fluorophores’ emission wavelength in cells but not the ROP-induced fluorescence emission wavelength.
While numerous fluorescent probes, including the majority of TrxR probes, may exhibit potentially interfering optical turn-on effects, such as the environment sensitivity of certain fluorophores, it remains obscure whether these effects competitively or comparably influence the fluorescence signal triggered by TrxR. On one hand, the existing 1,2-dithiolane probes have been found to lack sufficient specificity for TrxR thus far. On the other, it is noteworthy that 1,2-dithiolane probes incorporating different fluorophores and linkages display significantly distinct fluorogenic properties. These observations suggest that the reducible trigger of a trigger-cargo platform indeed executes a pivotal role in selectively recognizing the desired biological targets. Nevertheless, it is also important to acknowledge that the reducible trigger, the linker unit, and the fluorophore all have an impact on the ultimate fate of a probe. Therefore, substantial efforts are still required to delve deeper into the biochemical aspects of 1,2-dithiolane on multiple levels, with the anticipation that its new forthcoming derivatives may unexpectedly emerge to be validated as truly specific to TrxR.
Importantly, this work also possesses noteworthy merits that deserve significant recognition and advocacy
[10]. The modular reduction-triggered phenolic carbamate platform (
Figure 1, probe 7) developed by the authors has the following advantages: (1) the Haugland’s precipitating fluorophore PQ-OH (
Figure 1, probe 7) is featured with a large Stokes shift (λ
em/λ
ex = 530/360 nm) that ensures the platform has a high spatial resolution
[12][13]; (2) The PQ-OH-based platform switches on its ESIPT-based fluorescence signal only when cleavage with CDR takes place, indicating that the environment-dependent optical turn-on effects no longer interfere the signal generation. Furthermore, this phenomenon may contribute to the high signal-to-background fluorescence ratio observed; (3) The use of a phenol-based cargo release offers several attractive advantages, including a wider selection of potential cargos and more favorable leaving kinetics when compared with an aniline-based release. A key benefit of this approach is the ability to mask the hydroxyl group of many fluorophores, drugs, and agents, which can otherwise quench their fluorescence or activities. This allows for the construction of high-resolution probes, as well as the development of conceptually true prodrugs/theranostic agents.
The diversification of recognition parts to create novel redox substrates that are specific to diverse oxidoreductases is a challenging yet highly promising research field. The incorporation of selective triggers for various oxidoreductases such as GPx, GR, Trx, Msr, Prx, etc., into this modular platform presents an exciting opportunity to expedite the identification of high-quality probes. This approach can greatly facilitate and accelerate the study of redox biology. Furthermore, expanding this modular platform would have additional benefits, including the construction of oxidoreductase-targeted prodrugs and theranostic agents.
2.2. 1,2-Thiaselenane as the Trigger
Previous studies revealed that both the topology (linear or cyclic) and geometry (strained or stabilized) of disulfide exert profound effects on the cellular performance of disulfide-based probes, as both characteristics significantly influence the sensitivity of the probes to background monothiols
[14]. Theoretically, disulfide probes with linear topology exhibit kinetic irreversibility against cellular monothiols in high concentrations (~1–10 mM). Disulfide probes with cyclic topology appear to demonstrate varying degrees of resistance to background monothiols
[11][15]. Theoretically, strained cyclic probes are susceptible to interference from monothiols due to the inherent kinetic lability of strained cyclic disulfides. Conversely, non-tensile cyclic probes typically remain stable against monothiols attack via reversible thiol–disulfide exchange reactions. Interestingly, alicyclic six-membered disulfides were recently reported as selective substrates of Trx by the Thorn-Seshold group
[14][16]. In this sense, 1,2-dithianes may be tuned and harnessed for probe designs, acting as specific triggers for various oxidoreductases.
Most recently, based on their recently developed modular reduction-triggered phenolic carbamate platform
[10], the Thorn-Seshold group successfully developed an exceptionally selective mammalian TrxR1 fluorescent probe (
Figure 2, RX1, probe 8) that employs a novel stereoisomeric 1,2-thiaselenane as the recognition moiety. Given the catalytic principles of TrxR and monothiols, the replacement of a sulfur atom in 1,2-dithiane with a selenium atom may be advantageous for facilitating the selective recognition of TrxR as well as strengthening resistance against monothiols. Experimental evidence suggests that it is Sec in TrxR1 and a selenium atom in the 1,2-thiaselenane of RX1 probe that are both kinetically and thermodynamically preferred in the first recognizing exchange reaction step, which gives the key intermediate 8-1 (
Figure 2). Despite the presence of a pendent thiol, the intermediate 8-1 displays significant chemical resistance to undergo a 6-
exo-
trig “on-reductant” cyclization. Instead, it exhibits a strong preference for a kinetically smooth reaction, whereby the nascent diselenide bond of intermediate 8-1 is attacked by the thiolate in the active site of TrxR, ultimately resulting in the completion of the catalytic cycle of TrxR and the generation of the fully reduced intermediate 8-3. While the reductive intermediate 8-3 is capable of reducing the disulfide biomolecules accessible within cells, there appears no fluorescent signal in this process. Furthermore, due to the inherent nucleophilicity of its selenolate moiety, intermediate 8-3 can readily undergo a chemically favored 5-
exo-
trig cyclization, resulting in the release of the fluorescent PQ-OH molecule. Alternatively, although the reaction of RX1 with high levels of background monothiols can generate intermediate 8-2, the subsequent extrusion of the fully reduced intermediate 8-3, propelled by a second molecule of monothiol, is extremely disfavored in both kinetics and thermodynamics. Based on these observations, it can be inferred that RX1 exhibits strong kinetic and thermodynamic selectivity for TrxR, effectively precluding the interference from high levels of monothiols present within cells. In other words, the selenium present in the trigger of RX1 serves two distinct functions: (1) facilitating the recruitment of the desired TrxR in cells and (2) acting as a sink for the monothiols-induced parallel pathway, thereby preventing unwanted interference. Further LC-MS analysis provides additional confirmation of this underlying mechanistic process. In vitro evaluation of RX1 reveals its strong selectivity towards the TrxR1 isoform, as evidenced by the inability of the Sec-deficient U498C TrxR1 to trigger RX1. This observation highlights the essential role of selenium in TrxR1 for the recognition and cleavage of the selenenylsulfide bond present within RX1. The cellular modulation of TrxR1 via genetic knockout/knock-in, chemical inhibition, and selenium supplementation/starvation further underscores the high selectivity of RX1 towards cellular TrxR1. Importantly, high concentrations of monothiols such as GSH, as well as protein thiols such as GR, Grx, and TRP14, have been shown to have a minimal impact on the efficacy of RX1, further accentuating its exceptional selectivity for TrxR1.
2.3. Linear Diselenide as the Trigger
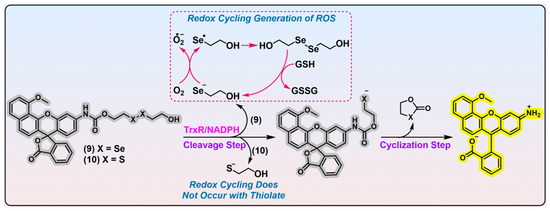
In 2020, Strongin et al. reported an off–on fluorescent probe specific for mammalian TrxR (
Figure 3, probe 9; λ
em/λ
ex = 580/531 nm). Probe 9 consists of seminaphthorhodafluor as the fluorophore, linear diselenide as the recognition part, and carbamate as the linker unit
[17]. Docking simulations and kinetic studies (
Km = 15.89 μM) both indicate a favorable binding affinity of probe 9 with TrxR, in comparison with its disulfide counterpart probe 10. LC-MS analysis confirmed the proposed releasing mechanism shown in
Figure 3, and a further imaging application of probe 9 in HCC827 cells with TrxR overexpressed shows its potential in TrxR detection. Notwithstanding its purported selectivity for TrxR, the confidence in its specificity may be significantly undermined by the following essential issues. First, previous studies unraveled that a diselenide bond in strained geometry is sensitive to GSH challenge
[11][18], and linear diselenide is kinetically labile to background monothiols in theory
[19]. According to this present study, probe 9 has a merely ~5-fold selectivity for TrxR (0.12 units) over the cellular abundant GSH (1 mM). Moreover, the applied concentration of GSH is a little low compared with physiological concentrations (~5 mM; it is even higher in cancer cells). This result was obtained upon a 20 min treatment, the incubation time of which is too short when considering the plateau time of probe 9 is ~30 min. Second, the research design fails to consider several critical interfering species, namely U498C TrxR, Trx, GR, GPx, Sec, and NADPH. Neglecting these species poses a significant challenge to claiming the selectivity of probe 9 for TrxR, as they exhibit either an overlapping biological functionality with TrxR or reactivity towards linear diselenide. Hence, it is imperative to verify whether the activation of probe 9 would be elicited by these particular species. Other disadvantages including a limited fluorescence increment and short Stokes shift of probe 9 also plague its further application. In addition, it may be deemed more appropriate to designate probe 9 as a theranostic agent, rather than a fluorescent probe for TrxR, given the substantial cytotoxicity exhibited by probe 9 towards SK-MEL-5 (IC
50 = 3.5 μM) and A375 (IC
50 = 4.8 μM) melanoma cell lines. Prior studies have unveiled that compounds containing diselenide bonds exhibit superior cytotoxicity over their disulfide counterparts
[18][19]. The remarkable cytotoxicity of probe 9 may be attributed to the generation of a free selenolate upon reaction with TrxR (
Figure 3, shown in red dashed box), and the selenolate is amenable to redox cycling in the presence of reducing species. This process leads to a rapid accumulation of excess ROS, which induces oxidative stress and cell death.
Figure 3. Linear diselenide-based TrxR probe. Probes 9 and 10 are diselenide- and disulfide-based TrxR probes. Experimental evidence indicates probe 9 is better than probe 10 in recognizing TrxR. During the activation process of probe 9 by TrxR, free selenolate is generated, which leads to the production of ROS. The dashed box in red shows the process of generation of ROS through free selenolate byproduct, and the arrows indicate the transformation of the substances. The curved arrows in black show the generated side products after activation by TrxR. Protein TrxR is shown in pink. The colors covering the chemical structures show their fluorescence colors after TrxR activation, and the gray indicates the fluorescence of the probes has been quenched.
Recently, a benzoselenadiazole-based theranostic agent was developed by Lin et al. for cancer treatment and cancer cell tracing/imaging
[20], and mitochondrial TrxR may play a role in triggering this agent. However, the intrinsically unstable benzoselenadiazole moiety is widely known to be sensitive to many biological species including GSH, Cys, selenol, H
2Se, homocysteine, protein kinases, and others
[21][22][23][24][25][26][27]. Thus, it is imperative that the theranostic agent contrived by Lin et al. should be subjected to further improvement and reevaluation to ascertain its selectivity towards TrxR2.
2.4. α,β-Unsaturated Ketone Moieties as the Triggers for Labelling/Imaging Agents
Compounds containing α,β-unsaturated ketone moieties have long been documented as TrxR inhibitors, due to their potential to undergo the Michael addition with reactive Sec residue in the C-terminal active site of TrxR under physiological conditions
[28][29][30][31][32][33][34][35][36][37][38][39]. Inspired by this interesting chemistry, the Bu group developed an α,β-unsaturated ketone-based fluorogenic probe for the covalent labelling of TrxR (
Figure 4, TR-green, probe 12)
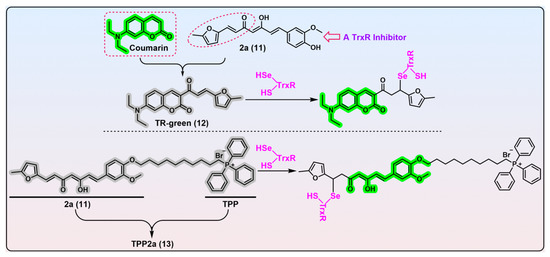
[40]. Prior to this work, Bu et al. identified compound 2a (
Figure 4, compound 11) as a highly selective TrxR inhibitor from a library of synthesized compounds and proved the furanyl–acryl moiety is the structural determinant responsible for its reactivity with TrxR
[41][42]. Further, the conjugation of the fluorophore coumarin with the furanyl–acryl moiety as the recognition part generates the off–on probe TR-green (λ
em/λ
ex = 500/440 nm). A biological evaluation revealed TR-green has great potential in the gel-imaging of TrxR, determining the concentrations of TrxR in the nanomolar level, and live-cell imaging of TrxR.
Figure 4. α,β-Unsaturated ketone-based TrxR probes. Probe 12, TR-green, is constructed from a specific TrxR inhibitor 2a (compound 11) and fluorescent coumarin. TR-green undergoes the Michael addition with the nucleophilic TrxR’s Sec residue. TR-green can be used in gel-imaging of TrxR and live-cell imaging of TrxR. Probe 13, TPP2a, is built by the integration of compound 2a with TPP. TPP2a can be used as a labelling agent for TrxR2, or a theranostic agent for cancers overexpressing TrxR2. The dashed box in red shows the structure of fluorophore coumarin, and the dashed ellipse in red show the structural determinant of compound 2a for inhibition of TrxR activity. Protein TrxR is shown in pink. The colors covering the chemical structures show their fluorescence colors after TrxR activation, and the gray indicates the fluorescence of the probes has been quenched.
2.5. Linear Disulfide as the Trigger
Compared with traditional imaging agents, carbon dots have several advantages such as good aqueous solubility, wide pH tolerance, high photostability, and easy surface functionalization. Thus, fluorescent probes based on carbon dots have been finding various applications in recent years
[43][44]. Singh et al. developed two carbon dots-based analytical tools for detecting mammalian TrxR (
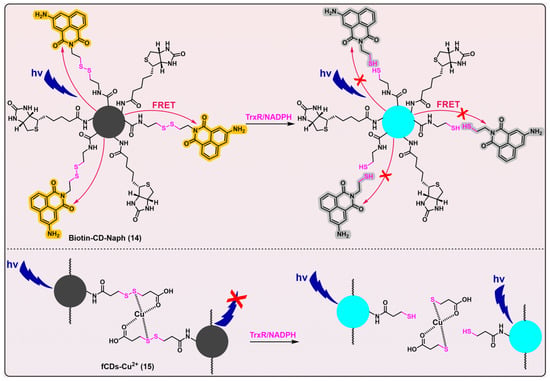
Figure 5, probes 14
[45] and 15
[46]). Both Biotin-CD-Naph (λ
em/λ
ex = 450/360 nm) and fCDs-Cu
2+ (λ
em/λ
ex = 446/340 nm) have linear disulfides as their triggers, that are designed to be cleaved by TrxR. Biotin-CD-Naph is a ratiometric probe, constructed on the basis of a fluorescence resonance energy transfer (FRET) mechanism. Whilst Biotin-CD-Naph shows a discernible level of TrxR selectivity and demonstrates potential applicability in cancer cell imaging and screening, the specificity of the probe still needs refinement because the employment of a disulfide bond as the trigger likely makes the probe susceptible to other disulfide-reducing species, e.g., Trx. Fortunately, Biotin-CD-Naph was additionally attached with biotin as a supplementation moiety, thereby enabling the preferential and selective recruitment and imaging of cancer cells that overexpress biotin receptors such as avidin and streptavidin on their cellular surfaces. Given the potential cytotoxic effects of 3-aminonaphthalimide, as well as the ROS-generating capabilities of carbon dots under photic conditions
[47][48][49], Biotin-CD-Naph holds immense promise to be applied as a theranostic agent in cancer treatment.
Figure 5. Linear disulfide-based TrxR probes. Probes 14 and 15 are based on carbon dots, and both have linear disulfide as their recognition parts for TrxR. Biotin-CD-Naph is a ratiometric probe that uses biotin to selectively target cancer cells. When the disulfide bond in Biotin-CD-Naph is cleaved by TrxR, the FRET process is blocked, and carbon dots emit fluorescence. fCDs-Cu2+ is quenched because of the chelation of Cu2+ within this probe. When the disulfide bond in fCDs-Cu2+ is cleaved by TrxR, Cu2+ is removed from the surface of carbon dots, and the fluorescence generates. Both probes 14 and 15 are able to image cancer cells that overexpress TrxR. The curved arrows in red indicate the FRET process, which leads to the turning-on of naphthalimide fluorophore (shown in yellow) and quench of carbon dots (shown in grey). The red × in the upper panel indicates the blockage of FRET process, which results in turning-on of carbon dots (shown in blue) and quench of naphthalimide fluorophore (shown in grey). The red × in the bottom panel indicates the quench process by chelation of Cu2+. Protein TrxR is shown in pink.
Another carbon dots probe fCDs-Cu
2+ (
Figure 5, probe 15) has been developed as an off–on fluorogenic type, with demonstrated selectivity for TrxR over a series of other biological species. Additionally, this probe has been successfully utilized for the imaging of cancer cells
[46]. However, the same problem suffered by Biotin-CD-Naph (
Figure 5, probe 14) also pertains to fCDs-Cu
2+, i.e., a possible insufficient specificity of disulfide bond in fCDs-Cu
2+ for TrxR. Thus, the potential lack of selectivity of Biotin-CD-Naph and fCDs-Cu
2+ towards TrxR underscores the pressing need for significant enhancements in their specificity.
Drug delivery systems based on disulfide bond usually target reducing the environment in cells. However, conferring individualized selectivity upon a given target through the utilization of the disulfide moiety presents a formidable challenge, given the presence of multiple cellular disulfide-reducing enzymes and a pronounced background of monothiols that exhibit different degrees of potential for the irreversible cleavage of the moiety. Perhaps exploiting the kinetic differences in various disulfide compounds towards TrxR and other biological species may contribute to the solution of this intractable selectivity
[50].