Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bingqian Xu | -- | 2468 | 2023-08-22 13:51:12 | | | |
| 2 | Rita Xu | Meta information modification | 2468 | 2023-08-23 03:46:50 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kandapal, S.; Xu, B. Atomic Force Microscopy in Biological Systems. Encyclopedia. Available online: https://encyclopedia.pub/entry/48328 (accessed on 10 January 2026).
Kandapal S, Xu B. Atomic Force Microscopy in Biological Systems. Encyclopedia. Available at: https://encyclopedia.pub/entry/48328. Accessed January 10, 2026.
Kandapal, Sneha, Bingqian Xu. "Atomic Force Microscopy in Biological Systems" Encyclopedia, https://encyclopedia.pub/entry/48328 (accessed January 10, 2026).
Kandapal, S., & Xu, B. (2023, August 22). Atomic Force Microscopy in Biological Systems. In Encyclopedia. https://encyclopedia.pub/entry/48328
Kandapal, Sneha and Bingqian Xu. "Atomic Force Microscopy in Biological Systems." Encyclopedia. Web. 22 August, 2023.
Copy Citation
Biological interactions often involve the transport of molecules, ions, or other species across biological membranes or between interacting proteins. The understanding of these transport phenomena is crucial for the development of therapies for various diseases. Atomic force microscopy is a powerful tool that has been increasingly used to study biological systems at the nano scale.
atomic force microscopy
biomarkers
transport
binding interactions
1. Introduction
Atomic force microscopy (AFM) [1][2] is a technique that is used not only for high-resolution imaging of a variety of materials, but also in the assessment of diverse physical characteristics at the nano scale. It finds extensive applications across various disciplines, including physics, chemistry, biology, and materials science, enabling the investigation of diverse physical and chemical properties at the molecular level. AFM [3] has equipped scientists with indispensable tools and resources to investigate the structure and properties of a wide range of samples, spanning from individual atoms to complex biological systems. These investigations hold broad applications, from fundamental research to industrial endeavors. Due to its remarkable capacity to deliver high-resolution images and precise measurements at the nano scale, it has emerged as an ever more crucial instrument in the examination of biosystems. This technology empowers researchers to visualize the surface of biological samples, enabling them to extract comprehensive data pertaining to topography, structure, and arrangement. Consequently, scientists can now delve into the intricate world of diverse biological molecules, such as proteins, DNA, RNA, and lipids [4][5][6][7][8], unraveling their structure–function relationships and gaining valuable insights. In addition it can serve as a valuable tool for investigating the mechanical characteristics of biological materials [9], such as their stiffness and elasticity, and provide insights into biomolecules and their role in biological processes such as cell adhesion [8][10], migration [11][12], and signaling [13][14]. With its remarkable ability to image and manipulate individual molecules, AFM has emerged as a potent instrument in the examination of isolated molecules, thereby offering invaluable insights into their behavior and function. This capability has revolutionized the study of biological molecules, including enzymes [15][16], receptors [17][18], and transporters [19][20].
AFM has proven highly valuable in the realm of investigating transport phenomena within biological systems. Transport phenomena involve the movement of particles or molecules across media like cell membranes or tissue barriers. In-depth exploration of transport phenomena within biological interactions, encompassing molecular binding and recognition processes involving proteins, nucleic acids, and lipids, sheds light on fundamental biological processes such as cell adhesion, protein–protein interactions, and drug delivery. Diverse mechanisms drive these interactions, including electrostatic interactions, hydrogen bonding, van der Waals interactions, and hydrophobic interactions. AFM finds noteworthy applications in the study of transport phenomena, with a specific focus on studying the blood–brain barrier (BBB) [21][22]. Acting as a complex barrier, the BBB separates blood vessels from brain tissue and plays a vital role in regulating the transport of nutrients, ions, and other molecules into the brain. AFM has facilitated investigations into the mechanical properties of the BBB and the interactions between the BBB and circulating cells and molecules. By quantifying the local mechanical properties of the BBB, AFM offers insights into the barrier’s permeability and transport properties. Similarly, AFM has proven to be instrumental in studying transport phenomena in ion channels [23][24][25][26]. These membrane proteins govern ion flow across cell membranes and are pivotal in various biological processes, such as nerve impulse transmission and muscle contraction. The exploration of biomolecular interactions through AFM has unveiled valuable insights into a wide array of biological processes, holding potential for the development of novel therapies and diagnostic tools.
2. Applications of AFM in the Study of Biomolecules
AFM enables the visualization of biomolecules in three dimensions, providing valuable insights into their shape and structure at the nano scale. It operates in diverse environments, making it suitable for studying biological systems. It plays a crucial role in investigating biomolecular interactions, including protein–protein, lipid–protein, and DNA–protein interactions. It allows direct, label-free measurements of molecular forces and interaction energies, facilitating the study of structural and mechanical properties.
2.1. Protein
Proteins play a vital role in biological systems, participating in essential cellular processes such as protein synthesis, signal transduction, and enzymatic reactions. They are also implicated in the development of various diseases, including neurodegenerative disorders like Parkinson’s and Alzheimer’s, as well as localized conditions such as type 2 diabetes and cataracts. Understanding protein aggregation is crucial in clinical settings for comprehending disease pathology and developing diagnostics and therapies. One example of protein aggregation is pseudoexfoliation syndrome (PEX), a disorder characterized by extracellular matrix protein aggregation that obstructs the eye’s aqueous outflow, posing a significant risk of glaucoma. Creasey et al. [27] utilized AFM-based antibody recognition imaging (Figure 1a) to investigate the molecular nature of PEX aggregates on human lens capsules. Their findings revealed an association between lysyl oxidase-like 1 (LOXL1) and an increased susceptibility to the syndrome. Another notable example is Alzheimer’s disease, which is characterized by the self-assembly and accumulation of fibrillar amyloid-β (Aβ) peptides in the brain. Biophysical studies, including AFM, have significantly advanced the understanding of the mechanisms underlying amyloid formation and its role in the pathogenesis of Alzheimer’s.
AFM plays a crucial role in studying protein folding and unfolding processes, which are fundamental to cellular function. Proteins transition from an unfolded state to well-defined three-dimensional structures, unique to each protein, for their biological activity. AFM enables direct visualization of the topographic structures of individual proteins and provides mechanical insights into their unfolding dynamics. Protein misfolding can lead to the formation of aggregated protofibrils, contributing to various diseases, such as Alzheimer’s, prion diseases, cystic fibrosis, and amyotrophic lateral sclerosis. AFM studies on protein folding have focused on proteins with tandem repeats of similar modules, such as tenascin, spectrin, and titin, which exhibit mechanical strength that is essential for their physiological functions. Best et al. [28] aimed to investigate the resistance of proteins to force and discovered that proteins that are not specifically designed for tensile strength may not exhibit the same resistance to force as those that are. They also observed that proteins with similar unfolding rates in solution may exhibit different unfolding properties under force, emphasizing the importance of considering protein-specific characteristics when studying their response to mechanical forces using AFM. Kawakami and Smith [29] introduced a novel force-ramp modification that enables precise control of multiple unfolding events in a multi-modular protein. This advancement utilizes software-based digital force feedback control to maintain a constant force-loading rate, regardless of the length of the soft elastic linkage or the number of unfolded polypeptide domains. By applying this technique, one can observe distinct pathways of unfolding in a controlled manner. Peng and Li. [30] conducted an experimental study that provided empirical evidence for the kinetic partitioning mechanism involved in the mechanical unfolding of T4 lysozyme—a compact protein composed of two subdomains. By subjecting T4 lysozyme to applied stretching forces from its N and C termini, they observed multiple distinct pathways of unfolding (Figure 1b,c). The majority of T4 lysozyme molecules exhibited an all-or-none unfolding behavior, surpassing a dominant kinetic barrier. However, a minority fraction of T4 lysozyme molecules followed a three-state unfolding pathway involving intermediate states. Notably, these three-state unfolding pathways displayed variability and diversity, deviating from well-defined routes. These findings provide direct evidence for the presence of kinetic partitioning in the mechanical unfolding pathways of T4 lysozyme. Moreover, the complex unfolding behaviors observed highlight the stochastic nature of kinetic barrier rupture during mechanical unfolding processes. In a study by Mahmood, Moheimani, and Bhikkaji [31], the effectiveness of integrating genetic manipulation with the AFM technique was investigated as a robust method to study the responses of proteins to forces within living cells. The findings underscore the potential of this approach for unraveling the intricate mechanisms underlying protein mechanics in a cellular context. Thus, AFM studies have significantly contributed to the understanding of proteins’ aggregation, folding, and unfolding processes. They shed light on the structural and mechanical properties of proteins, offering insights into their biological functions and their roles in disease development.
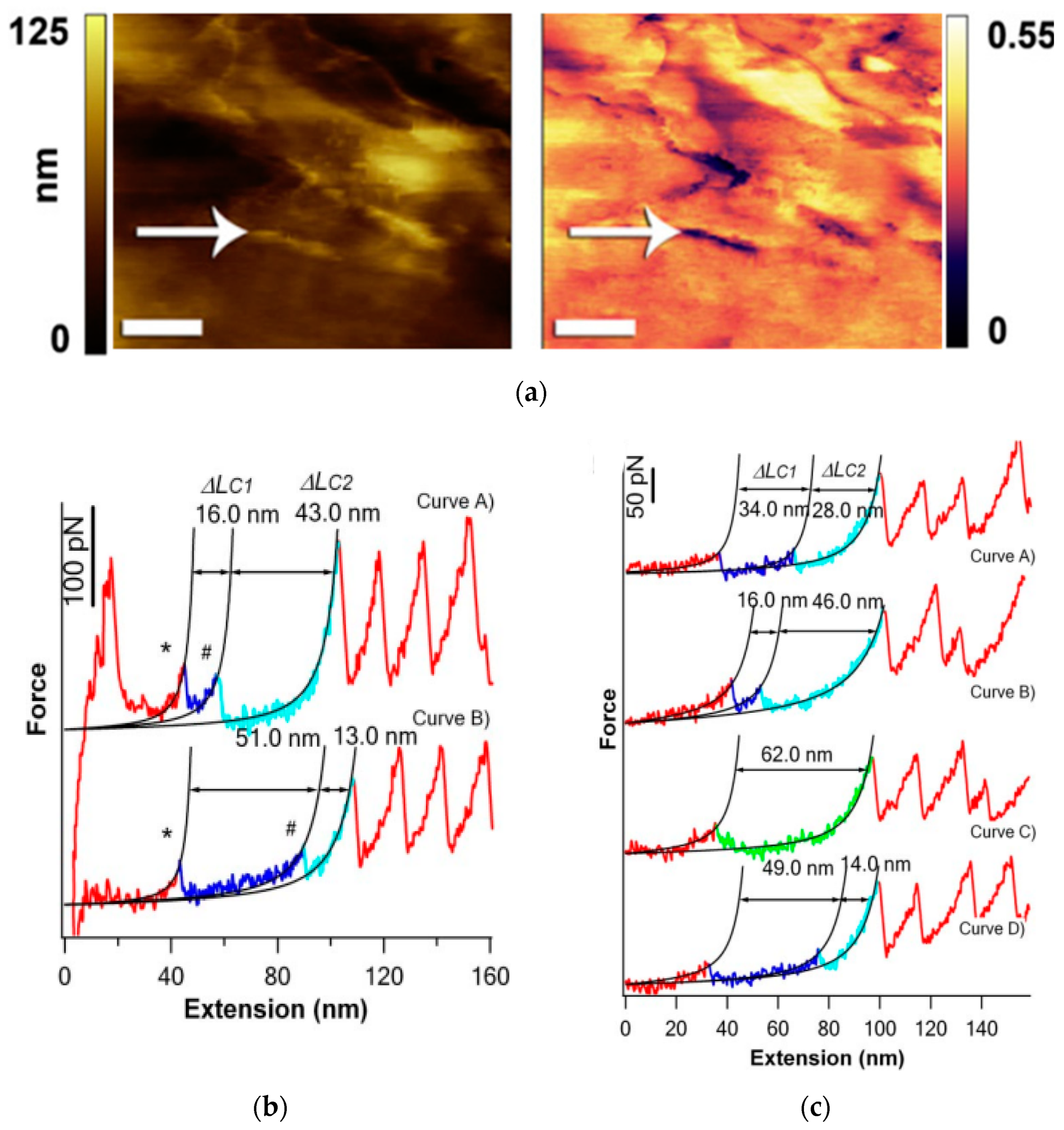
Figure 1. (a) AFM images of a lens capsule from a PEX patient acquired using an anti-LOXL1-antibody-functionalized tip, from an area near the center of the capsule. (b) Typical force–extension curves of cysteine-free pseudo wild-type T4 lysozyme (T4L*) showing three-state unfolding behaviors. (#-second unfolding force peaks) (c) A series of force–extension curves of the same T4L* measured during repeated stretching–relaxation experiments.
2.2. Nucleic Acids
The direct imaging of nucleic acids [32] in aqueous solutions has long been a challenging task, primarily because of the relatively weak interactions between nucleic acids and substrates. However, advancements in substrate modifications and detection methods have improved the quality and resolution of nucleic acid imaging. Zhu et al. [33] provided a method for studying DNA methylation that cannot be detected using conventional DNA probes. They used an antibody-tethered AFM cantilever to measure the distances between 5-methylcytidine bases in DNA strands, with a resolution of 4 Å. By binding to two 5-methylcytidine bases, the cantilever retraction produced a unique rupture signature reflecting the spacing between the tagged bases. Shim et al. [34] developed a force mapping method to directly quantify low-abundance DNA containing methyl-CpG at a specific site. Their approach utilized methyl-CpG-binding (MBD) proteins to distinguish methylation states. The MBD protein was positioned on the probe opposite the target DNA site of interest. If the target site was methylated, the MBD protein would identify the methylated CpG, while it would disregard the site if it was not methylated. This method allowed for the detection of specific methylation patterns without the need for chemical treatment or amplification of the target DNA. Strobl et al. [35] investigated the factors influencing nucleic acid condensation and release in individual parvovirus particles. A single virus particle was attached to a cantilever and exposed to various physicochemical conditions, where the pH and salt concentration were manipulated to induce the condensation or release of nucleic acids. The study revealed that parvovirus particles are highly responsive to changes in physicochemical conditions for nucleic acid condensation and release.
2.3. Polysaccharides
The application of AFM in the study of polysaccharides [36] is not as extensive as it is for other biomolecules, despite the crucial biological roles that they play. Nevertheless, there have been several AFM investigations that have aimed to unravel the structural characteristics of polysaccharides found in bacteria, plants, and fungi, as well as densely glycosylated peptides and proteoglycans. Furthermore, AFM proves valuable in exploring the molecular interactions between polysaccharides and other biomolecules, such as proteins and lipids. In addition to these efforts, recent advancements in AFM techniques offer opportunities to delve deeper into the characterization and functional analysis of polysaccharides, providing a promising avenue for future research in this field. An example of interest in this field is the investigation of heparan sulfate (HS), a linear polysaccharide found in all animal cell plasma membranes. Guo et al. [37] investigated the interaction between heparan sulfate (HS) and antithrombin (AT) on the surface of a single endothelial cell under near-physiological conditions to understand the role of critical sulfates responsible for AT binding. The specific interaction between HS and a protein ligand is primarily determined by the sulfation patterns on the HS chain. The research revealed that AT interacts with endothelial HS through multiple binding sites, and the presence of N-, 2-O-, and/or 6-O-sulfates on HS is essential for this interaction.
2.4. Peptide
Peptides are short chains of amino acids that can have a wide range of functions in biological systems, including signaling molecules, neurotransmitters, cell regulation and homeostasis, etc. Li et al. [38] conducted a study focusing on peptide-assembled hydrogels, investigating the morphology and mechanical properties of individual nanofibrils during the gelation and degradation processes of these hydrogels. With the help of topographic imaging, they were able to observe distinct assembly behaviors of peptide-formed nanofibrils throughout the gelation and degradation stages, revealing a correlation between these behaviors and changes in nanofibrillar mechanics. Gaspar et al. [39] conducted a study investigating the impact of a peptide called APN-1 on the mechanical characteristics of cancer cells. Their findings revealed that APN-1 can induce alterations in the mechanical properties of cancer cells, resulting in cell death and suppressed tumor growth. Specifically, APN-1 was observed to enhance the stiffness of cancer cells and disrupt their cytoskeleton—a network of protein fibers responsible for cellular structural support. These changes in mechanical properties were associated with increased cell death and diminished tumor growth. These findings suggest the potential of APN-1 as an antitumor agent, with its effects on cellular biomechanics playing a crucial role in its therapeutic activity.
2.5. Enzymes
Enzymes help in speeding up chemical reactions in physiological processes such as digestion, metabolism, and DNA replication. Enzymes have been extensively studied due to their significance and diverse applications. Zhang et al. [40] investigated the enzymatic hydrolysis of pretreated biomass and observed real-time changes in the cellulose structure of plant cell walls during enzymatic hydrolysis (Figure 2a,b). The enzymes’ action depended on the size and width of the cracks in the cellulose. Smaller cracks led to progressive degradation, while larger cracks caused the peeling of glycan chains. The combination of CBH I and β-G enzymes effectively hydrolyzed the biomass, emphasizing the role of crack size in the depolymerization and peeling of cellulose microfibrils. Cellulolytic enzymes, such as CBM3a, play a crucial role in binding specifically to crystalline cellulose. Zhang et al. [41] utilized CBM3a-functionalized gold nanoparticles (GNPs) to monitor the binding activity of CBM3a to poplar cell-wall cellulose. The GNPs-CBM3a complexes showed specific binding to the cellulose surface, aligning with the cellulose fibril axis. This work advances the comprehension of biomass–enzyme interactions and facilitates the development of efficient cellulolytic enzymes for biofuel production. In their research, Zhang and Wang et al. [42] investigated the affinity interactions between the carbohydrate-binding module (CBM) and plant cell-wall cellulose at the single-molecule level. They used AFM tips functionalized with CBM3a and CBM2a to determine the binding efficiencies of CBMs to cellulose. Recognition imaging (Figure 2c) revealed that CBM3a exhibited slightly higher binding efficiency and affinity than CBM2a on both natural and extracted cellulose surfaces. Moreover, both CBMs showed higher affinities towards natural cell-wall cellulose microfibrils compared to extracted single cellulose microfibrils. This study provides valuable insights into the binding properties of CBMs to cellulose, highlighting the differences between CBM3a and CBM2a and their interactions with cellulose surfaces. They [43] also further examined the CBM–cellulose binding process on extracted crystalline cellulose. CBM3a molecules were utilized for both functionalizing the AFM tip and binding as free CBM molecules. The in situ AFM imaging revealed the efficient and regular binding of CBM molecules to cellulose, particularly within the initial 60–120 min. This research significantly contributes to in-depth understanding of the binding mechanism between CBM and crystalline cellulose at the single-molecule level.
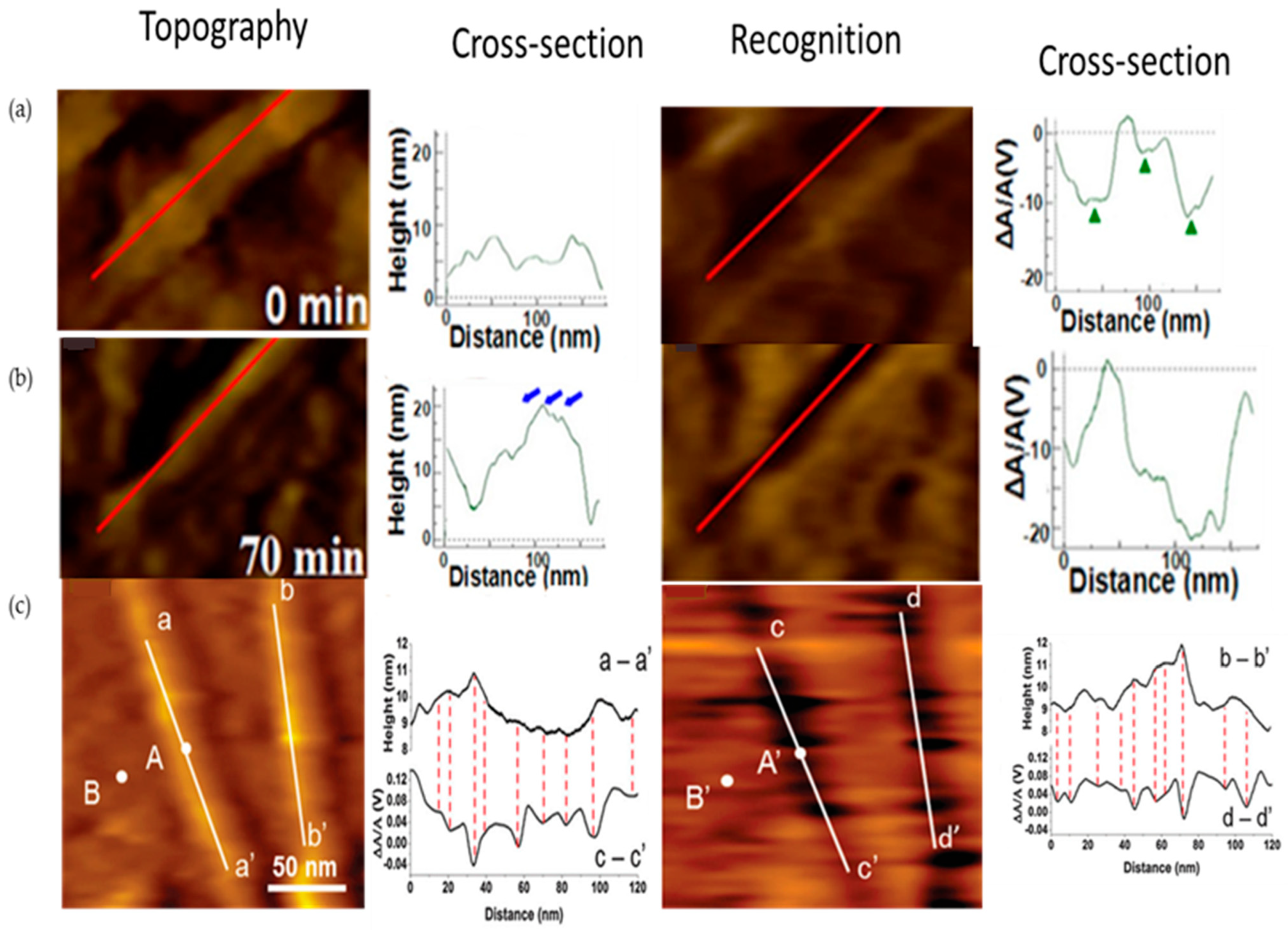
Figure 2. (a,b) Topography, recognition images, and cross-sectional analysis of pretreated poplar cellulose incubated with EG at 0 and 70 min, respectively. (c) Extracted single cellulose microfibrils.
References
- Steffens, C.; Leite, F.L.; Bueno, C.C.; Manzoli, A.; De Paula Herrmann, P.S. Atomic force microscopy as a tool applied to nano/biosensors. Sensors 2012, 12, 8278–8300.
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic force microscope. Phys. Rev. Lett. 1986, 56, 930.
- Zemła, J.; Danilkiewicz, J.; Orzechowska, B.; Pabijan, J.; Seweryn, S.; Lekka, M. Atomic force microscopy as a tool for assessing the cellular elasticity and adhesiveness to identify cancer cells and tissues. Semin. Cell Dev. Biol. 2018, 73, 115–124.
- Lyubchenko, Y.L.; Shlyakhtenko, L.S. AFM for analysis of structure and dynamics of DNA and protein–DNA complexes. Methods 2009, 47, 206–213.
- Schön, P. Imaging and force probing RNA by atomic force microscopy. Methods 2016, 103, 25–33.
- Sajja, S.; Chandler, M.; Fedorov, D.; Kasprzak, W.K.; Lushnikov, A.; Viard, M.; Shah, A.; Dang, D.; Dahl, J.; Worku, B. Dynamic behavior of RNA nanoparticles analyzed by AFM on a mica/air interface. Langmuir 2018, 34, 15099–15108.
- Connell, S.D.; Smith, D.A. The atomic force microscope as a tool for studying phase separation in lipid membranes. Mol. Membr. Biol. 2006, 23, 17–28.
- Puech, P.-H.; Poole, K.; Knebel, D.; Muller, D.J. A new technical approach to quantify cell–cell adhesion forces by AFM. Ultramicroscopy 2006, 106, 637–644.
- Allison, D.P.; Mortensen, N.P.; Sullivan, C.J.; Doktycz, M.J. Atomic force microscopy of biological samples. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 618–634.
- Dufrêne, Y.F. Sticky microbes: Forces in microbial cell adhesion. Trends Microbiol. 2015, 23, 376–382.
- Wagh, A.A.; Roan, E.; Chapman, K.E.; Desai, L.P.; Rendon, D.A.; Eckstein, E.C.; Waters, C.M. Localized elasticity measured in epithelial cells migrating at a wound edge using atomic force microscopy. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2008, 295, L54–L60.
- Luo, Q.; Kuang, D.; Zhang, B.; Song, G. Cell stiffness determined by atomic force microscopy and its correlation with cell motility. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2016, 1860, 1953–1960.
- Fang, T.; Alvelid, J.; Spratt, J.; Ambrosetti, E.; Testa, I.; Teixeira, A.I. Spatial regulation of T-cell signaling by programmed death-ligand 1 on wireframe DNA origami flat sheets. ACS Nano 2021, 15, 3441–3452.
- Shi, X.; Zhang, X.; Xia, T.; Fang, X. Living cell study at the single-molecule and single-cell levels by atomic force microscopy. Nanomedicine 2012, 7, 1625–1637.
- Zhang, Q.; Yang, J.; Tillieux, S.; Guo, Z.; Natividade, R.d.S.; Koehler, M.; Petitjean, S.; Cui, Z.; Alsteens, D. Stepwise Enzymatic-Dependent Mechanism of Ebola Virus Binding to Cell Surface Receptors Monitored by AFM. Nano Lett. 2022, 22, 1641–1648.
- Neaves, K.J.; Cooper, L.P.; White, J.H.; Carnally, S.M.; Dryden, D.T.; Edwardson, J.M.; Henderson, R.M. Atomic force microscopy of the EcoKI Type I DNA restriction enzyme bound to DNA shows enzyme dimerization and DNA looping. Nucleic Acids Res. 2009, 37, 2053–2063.
- Clark, C.G.; Sun, Z.; Meininger, G.A.; Potts, J.T. Atomic force microscopy to characterize binding properties of α7-containing nicotinic acetylcholine receptors on neurokinin-1 receptor-expressing medullary respiratory neurons. Exp. Physiol. 2013, 98, 415–424.
- Bergler, F.; Fuentes, C.; Kadir, M.F.; Navarrete, C.; Supple, J.; Barrera, N.P.; Edwardson, J.M. Activation of P2X4 receptors induces an increase in the area of the extracellular region and a decrease in receptor mobility. FEBS Lett. 2020, 594, 4381–4389.
- Murrough, J.W.; Huang, Y.; Hu, J.; Henry, S.; Williams, W.; Gallezot, J.-D.; Bailey, C.R.; Krystal, J.H.; Carson, R.E.; Neumeister, A. Reduced amygdala serotonin transporter binding in posttraumatic stress disorder. Biol. Psychiatry 2011, 70, 1033–1038.
- Ruozi, B.; Tosi, G.; Leo, E.; Vandelli, M.A. Application of atomic force microscopy to characterize liposomes as drug and gene carriers. Talanta 2007, 73, 12–22.
- Cardoso, F.L.; Brites, D.; Brito, M.A. Looking at the blood–brain barrier: Molecular anatomy and possible investigation approaches. Brain Res. Rev. 2010, 64, 328–363.
- Zhou, Y.; Peng, Z.; Seven, E.S.; Leblanc, R.M. Crossing the blood-brain barrier with nanoparticles. J. Control. Release 2018, 270, 290–303.
- Fotiadis, D. Atomic force microscopy for the study of membrane proteins. Curr. Opin. Biotechnol. 2012, 23, 510–515.
- Dhar-Chowdhury, P.; Malester, B.; Rajacic, P.; Coetzee, W. The regulation of ion channels and transporters by glycolytically derived ATP. Cell. Mol. Life Sci. 2007, 64, 3069–3083.
- Alessandrini, A.; Facci, P. Phase transitions in supported lipid bilayers studied by AFM. Soft Matter 2014, 10, 7145–7164.
- Viles, J.H. Imaging Amyloid-β Membrane Interactions; Ion-channel pores and Lipid-Bilayer Permeability in Alzheimer’s Disease. Angew. Chem. 2023, e202215785.
- Creasey, R.; Sharma, S.; Gibson, C.T.; Craig, J.E.; Ebner, A.; Becker, T.; Hinterdorfer, P.; Voelcker, N.H. Atomic force microscopy-based antibody recognition imaging of proteins in the pathological deposits in pseudoexfoliation syndrome. Ultramicroscopy 2011, 111, 1055–1061.
- Best, R.B.; Li, B.; Steward, A.; Daggett, V.; Clarke, J. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 2001, 81, 2344–2356.
- Kawakami, M.; Smith, D. A new atomic force microscope force ramp technique using digital force feedback control reveals mechanically weak protein unfolding events. Nanotechnology 2008, 19, 495704.
- Peng, Q.; Li, H. Atomic force microscopy reveals parallel mechanical unfolding pathways of T4 lysozyme: Evidence for a kinetic partitioning mechanism. Proc. Natl. Acad. Sci. USA 2008, 105, 1885–1890.
- Mahmood, I.A.; Moheimani, S.R.; Bhikkaji, B. A new scanning method for fast atomic force microscopy. IEEE Trans. Nanotechnol. 2009, 10, 203–216.
- Hansma, H.G.; Bezanilla, M.; Zenhausern, F.; Adrian, M.; Sinsheimer, R.L. Atomic force microscopy of DNA in aqueous solutions. Nucleic Acids Res. 1993, 21, 505–512.
- Zhu, R.; Howorka, S.; Pröll, J.; Kienberger, F.; Preiner, J.; Hesse, J.; Ebner, A.; Pastushenko, V.P.; Gruber, H.J.; Hinterdorfer, P. Nanomechanical recognition measurements of individual DNA molecules reveal epigenetic methylation patterns. Nat. Nanotechnol. 2010, 5, 788–791.
- Shim, W.C.; Woo, S.; Park, J.W. Nanoscale Force-Mapping-Based Quantification of Low-Abundance Methylated DNA. Nano Lett. 2022, 22, 1324–1330.
- Strobl, K.; Mateu, M.; de Pablo, P.J. Exploring nucleic acid condensation and release from individual parvovirus particles with different physicochemical cues. Virology 2023, 581, 1–7.
- Abu-Lail, N.; Camesano, T. Polysaccharide properties probed with atomic force microscopy. J. Microsc. 2003, 212, 217–238.
- Guo, C.; Fan, X.; Qiu, H.; Xiao, W.; Wang, L.; Xu, B. High-resolution probing heparan sulfate–antithrombin interaction on a single endothelial cell surface: Single-molecule AFM studies. Phys. Chem. Chem. Phys. 2015, 17, 13301–13306.
- Li, M.; Xi, N.; Wang, Y.; Liu, L. Nanoscale multiparametric imaging of peptide-assembled nanofibrillar hydrogels by atomic force microscopy. IEEE Trans. Nanotechnol. 2019, 18, 315–328.
- Gaspar, D.; Freire, J.M.; Pacheco, T.R.; Barata, J.T.; Castanho, M.A. Apoptotic human neutrophil peptide-1 anti-tumor activity revealed by cellular biomechanics. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2015, 1853, 308–316.
- Zhang, Y.; Zhang, M.; Alexander Reese, R.; Zhang, H.; Xu, B. Real-time single molecular study of a pretreated cellulose hydrolysis mode and individual enzyme movement. Biotechnol. Biofuels 2016, 9, 1–12.
- Zhang, M.; Wu, S.-C.; Zhou, W.; Xu, B. Imaging and measuring single-molecule interaction between a carbohydrate-binding module and natural plant cell wall cellulose. J. Phys. Chem. B 2012, 116, 9949–9956.
- Zhang, M.; Wang, B.; Xu, B. Measurements of single molecular affinity interactions between carbohydrate-binding modules and crystalline cellulose fibrils. Phys. Chem. Chem. Phys. 2013, 15, 6508–6515.
- Zhang, M.; Wang, B.; Xu, B. Mapping single molecular binding kinetics of carbohydrate-binding module with crystalline cellulose by atomic force microscopy recognition imaging. J. Phys. Chem. B 2014, 118, 6714–6720.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
683
Revisions:
2 times
(View History)
Update Date:
23 Aug 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




