Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Danton O'Day | -- | 2945 | 2023-08-15 14:27:23 | | | |
| 2 | Jessie Wu | Meta information modification | 2945 | 2023-08-16 03:16:38 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
O’day, D.H. Proteins of the Nucleolus of Dictyostelium discoideum. Encyclopedia. Available online: https://encyclopedia.pub/entry/48087 (accessed on 07 February 2026).
O’day DH. Proteins of the Nucleolus of Dictyostelium discoideum. Encyclopedia. Available at: https://encyclopedia.pub/entry/48087. Accessed February 07, 2026.
O’day, Danton H.. "Proteins of the Nucleolus of Dictyostelium discoideum" Encyclopedia, https://encyclopedia.pub/entry/48087 (accessed February 07, 2026).
O’day, D.H. (2023, August 15). Proteins of the Nucleolus of Dictyostelium discoideum. In Encyclopedia. https://encyclopedia.pub/entry/48087
O’day, Danton H.. "Proteins of the Nucleolus of Dictyostelium discoideum." Encyclopedia. Web. 15 August, 2023.
Copy Citation
The nucleolus is a multifunctional subnuclear compartment that has been studied for more than 200 years. The nucleoli of Dictyostelium discoideum have a comparatively unique, non-canonical, localization adjacent to the inner nuclear membrane. The verified nucleolar proteins of this eukaryotic microbe are detailed while other potential proteins are introduced. Heat shock protein 32 (Hsp32), eukaryotic translation initiation factor 6 (eIF6), and tumour necrosis factor receptor-associated protein 1 (TRAP1) are essential for cell survival. NumA1, a breast cancer type 1 susceptibility protein-C Terminus domain-containing protein linked to cell cycle, functions in the regulation of nuclear number.
nucleolus
Dictyostelium
gene
nucleomorphin
calmodulin
calcium binding protein
mitosis
1. The Dictyostelium Nucleolus
As in other eukaryotes, the multiple nucleoli are the largest intranuclear bodies in Dictyostelium discoideum. Early research revealed that the structural features of this social amoebozoan’s nucleoli differ from the classic nucleolar organization. Rather than localizing within the nucleoplasm, they exist as two to four dense patches that are tightly adhered to the inner nuclear envelope (Figure 1) [1][2][3][4][5]. What’s more, they are neither bipartite nor tripartite. Instead, ultrastructurally they present as a more-or-less homogenous structure consisting of continuous fibrous matrices within which different-sized ribosome-like granules (10–50 kDa) are distributed [6]. Fitting with the absence of defined FC regions, the rDNA instead forms a beaded ring-like structure (15–20 beads/ring) around the periphery of each nucleolus [7]. This rDNA is predominantly extrachromosomal with some being telomeric [8][9]. Other related amoebozoan species, including D. mucoroides, D. minutum, and Polysphondylium pallidum, appear to share the same nucleolar structure (e.g., [10][11]). In keeping with its role in rRNA synthesis in all species, treatment with actinomycin-D, an inhibitor of RNA polymerase I (i.e., rDNA transcription), leads to nucleolar breakdown in Dictyostelium [1][12].
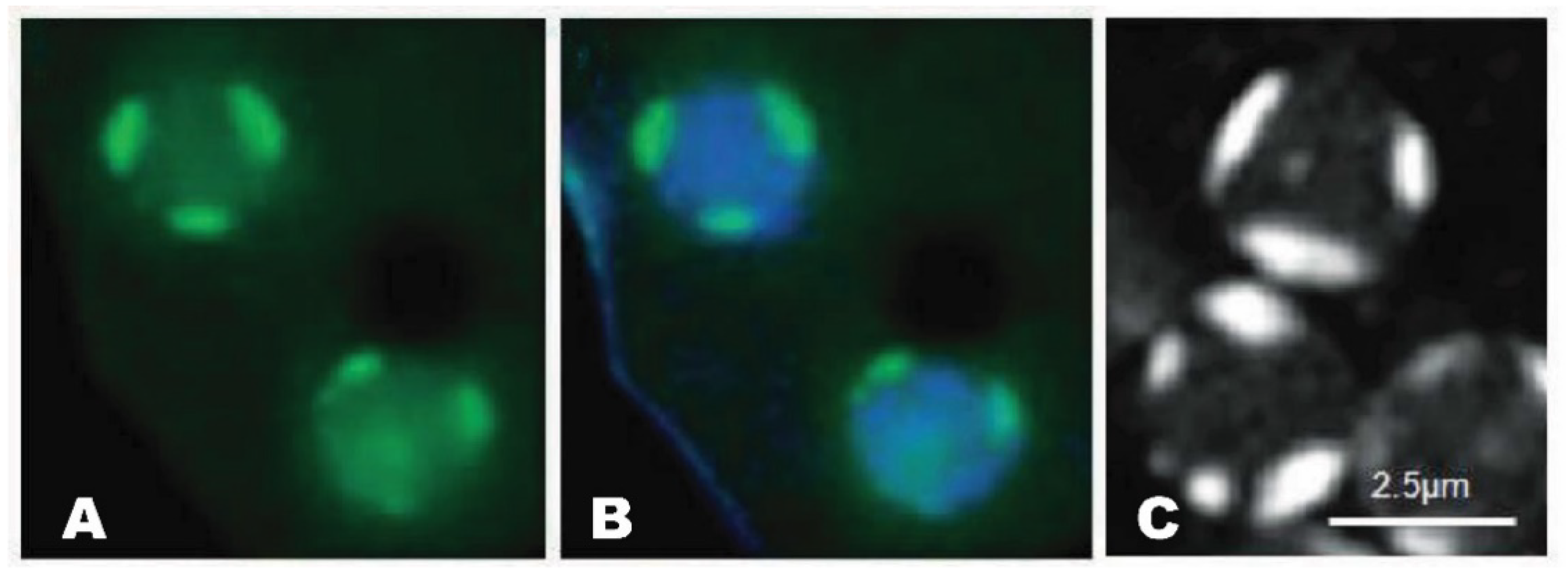
Figure 1. Nucleolar patches of Dictyostelium. A. GFP-NumA1 (green) localization. B. GFP-NumA1 localization with DNA (Hoescht, blue staining) stained. C. FITC-NumA1-NLS1 localization (white patches).
This breakdown occurs in one of two ways: the progressive disappearance of protein localization (e.g., NumA1) or the formation of nucleolar buds—containing specific proteins (e.g., Cbp4a, Snf12, and FhkA)—that are released intact into the cytoplasm [13].
2. The Nucleolar Proteins of Dictyostelium
2.1. Heat Shock Protein 32 (Hsp32)
The first “resident” nucleolar protein to be identified in Dictyostelium was heat shock protein 32 (Hsp32) [14]. Colocalizing with rDNA as beads on a string around the periphery of the nucleolus in unstressed cells, during heat shock it redistributes throughout the nucleolus and nucleoplasm [7][14]. Extended periods of heat shock produce a nucleolus with more pronounced rDNA beads revealing that the structure of the nucleolus in Dictyostelium responds to stress, as it does in other organisms [14][15]. In keeping with this idea, heat shock treatment of Dictyostelium induces a redistribution of Hsp32 from the nucleolar periphery and Snf12 from the nucleoplasm to nucleolus, as discussed below. Attempts to knock out the Hsp32 gene have failed suggesting it may be a critical protein [14].
Due to the presence of a highly acidic region rich in aspartic acid (asp) and glutamic acid (glu) residues, Hsp32 shares sequence similarity to nucleophosmin (NPM1) and nucleolin, both highly conserved nucleolar proteins in mammals [16]. The highly acidic regions are a common feature thought to be responsible for binding to basic ribosomal proteins and to NLSs of other proteins [17][18]. The role of the acid rich glu/asp region is discussed again for NumA1 below. Hsp32 also binds with high affinity to DNA but this association is not involved in its localization [14].
Hsp32 possesses a monopartite and bipartite NLS, thus sharing similarity with nucleolar proteins from other species [14][19][20]. However, an NoLS has not been identified and the means by which Hsp32 localizes to the nucleolus remains to be elucidated.
2.2. Eukaryotic Translation Initiation Factor 6 (eIF6)
Eukaryotic translation initiation factor 6, eIF6, was originally identified as a nucleolar protein based on its localization as peripheral patches in DAPI-stained nuclei plus its sensitivity of actinomycin D treatment). [21]. This highly conserved protein is essential to the production of 60S ribosomal subunits serving as a rate-limiting step in the cell cycle [22][23][24][25]. The pathogenesis of two forms of leukemia—inherited Shwachman-Diamond syndrome (SDS) and sporadic SDS—involve a common pathway in 60S-subunit maturation and the functional activation of ribosomes [26][27]. eIF6 is involved in 60S-subunit maturation and thus could play a central role in the disease process. Since eIF6 is shared by eukaryotes and archaea, Dictyostelium serves as a model to detail its function. For example, Weiss et al. [28] used single-particle cryo-EM to dissect the mechanism by which eIF6 gets released from nascent 60S ribosomal subunits in Dictyostelium. As in other species, eIF6 prevents 60S maturation by blocking the binding of essential maturation factors and, thus, must be removed for functional ribosomal formation to occur. This conserved mechanism involving eIF6 release is impaired in both inherited and sporadic leukemias.
The N-terminal region of eIF6 contains both an NLS and NoLS with a second potential NLS in the C-term [21]. Scott et al. [29] published an “Experimentally determined NoLS” and a predicted NoLS for eIF6, the latter generated from a program they compiled. However, based on their information summarized in Table 2 of their publication, they erroneously analyzed the sequence of NumA1 not eIf6, albeit with additional errors. Using their program to analyze eIF6 in fact detects no NoLS for this nucleolar protein (O’Day, unpublished results). So, although the NoLS in eIF6 has been mapped to a subdomain, the precise location of the NoLS for eIF6 remains to be revealed. Deletion of the eIF6 gene is lethal in Dictyostelium as it is in other species [23][24][25]. Examination of the data from Sillo et al. [30], from their study of genes linked to phagocytosis, revealed that eIF6 is upregulated by factors that induce phagocytosis in Dictyostelium.
2.3. Tumor Necrosis Factor Receptor-Associated Protein 1 (TRAP1)
Tumor necrosis factor receptor-associated protein 1 (TRAP1) is a member of the Hsp90 family [31][32]. It is a multifunctional protein linked to cell cycle progression, cell differentiation, and apoptosis. Found in the outermost layer of the spore coat it is believed to protect these dormant structures from physicochemical stresses [33][34]. While TRAP1 is a questionable nucleolar protein, there is some evidence it localizes there as well as in other cellular locations including mitochondria to where it translocates during early differentiation [34]. It was first observed in intranuclear patches reminiscent of nucleoli but not verified through actinomycin D or other treatments. As expected, TRAP1expression is induced by heat shock and like other members of the Hsp90 family null mutations are lethal [31][32]. More recently, TRAP1 has gained attention as a protein that can help with understanding mitochondrial diseases [35].
2.4. Nucleomorphin A1 (NumA1)
The nucleolar localization of a group of well-established calmodulin-binding proteins from other organisms such as calcineurin, CaM kinase II, and myosin light chain kinase supports a role for calmodulin in the nucleolus [36][37]. In keeping with this, this calcium-sensor and -effector has also been shown to bind and localize in mammalian nucleoli [38][39]. Nucleomorphin isoform NumA1 represents the only verified nucleolar calmodulin-binding protein in Dictyostelium. Acting as a regulator of nuclear number and interacting with calmodulin in a Ca2+-dependent manner, it is predominantly a nucleolar protein with secondary nucleoplasmic localization [40][41][42]. This pattern of localization is a common feature of nucleolar proteins (e.g., nucleophosmin, adenosine deaminases, and murine double minute 2) that shuttle between the nucleolus and nucleoplasm [43][44].
Full length nucleomorphin contains a breast cancer carboxy-terminus domain (BRCT) that is found in cell cycle checkpoint proteins in other organisms [40][45]. The presence of a highly acidic, glu/asp domain is a common feature of nucleolar proteins including nucleophosmin, nucleoplasmin, nucleolin, and Hsp32 [14][16]. Overexpression of GFP-NumA1 lacking its palindromic glu/asp or DEED domain results in multinuclearity fitting with NumA1’s involvement in cell cycle regulation [40]. In contrast to the regulation of nuclear number in human cells where over 100 proteins appear to be involved, this is the only protein so far linked to this function in Dictyostelium [46].
A large number of attributes of NumA1 suggest it is a functional equivalent of the mammalian nucleolar protein nucleophosmin (NPM1) [40]. NPM1 has diverse functions including a role in DNA repair, centrosome duplication and cell proliferation. Mutations in NPM1 are a major cause of acute myeloid leukemia (AML) being present in 20–30% of the cases [47]. The acidic glu/asp domain of NPM1 is involved in histone binding but this function has not been studied in NumA1. The DEED domain of NumA1 is sufficient to target FITC to the nucleus thus acting as an unconventional NLS [40]. Furthermore, NPM1 can act as both a proto-oncogene and as a tumor suppressor [48]. The study of NumA1 thus has the possibility of offering additional insight into the mode of action of NPM1. For example, as covered in the next section, its DEED domain binds Cbp4a in a calcium-dependent manner suggesting NumA1 could be involved in recruiting other proteins to the nucleolus, a mechanism apparently not yet studied for NPM1.
Yeast two hybrid studies revealed that NumA1 interacts with the calcium-binding protein Cbp4a and Zn2+-metallopeptidase puromycin-sensitive aminopeptidase A (PsaA) which, in other species, is associated with cell cycle progression and several human diseases including Huntington’s and Alzheimer’s disease [49]. Dictyostelium PsaA is similar to PSA from Drosophila, mouse, and human. In contrast to Cbp4a (see below), PsaA does not localize to the nucleolus but colocalizes with NumA1 in the nucleoplasm independent of Ca2+/Calmodulin [50]. The functional relationship between Cbp4a and NumA1 is strengthened by their apparent co-regulation by developmental morphogens where differentiation factor-1 (DIF-1) upregulates them while cyclic AMP and ammonia leads to their downregulation [51].
An attribute of some nucleolar proteins is the presence of multiple NLSs. NumA1 contains four identified NLSs [40][52]. Three of them (NLS-1, -2, and -4) reside within N-terminal residues 1-120: NLS1, 31PKSKKKF37 and NLS-1 and -4 within 48KKSYQDPEIIAHSRPRK64 (NLS-4 is underlined). NLS-3 is found in the C-terminus (246PTKKRSL252). Multiple NLSs may function to modulate the amount of the nuclear-localized protein, as seen for human TBX5, nucleolar ribosomal protein L7a and transcription factor Nrf2 [53][54][55]. FITC-peptide constructs showed each of these sequences, including the terminal RPRK sequence of the bipartite NLS (48–64), localize to nucleoli revealing the peptide sequences are all NoLS/NLS (e.g., Figure 1) [50]. As with the localization of GFP-NumA1 the nucleolar localization was abolished in the presence of actinomycin D but unaffected by treatments with calcium chelators or calmodulin antagonists. Hence these sequences serve as joint NLS/NoLS for the targeting of NumA1. NoLSs, like NLSs, are typically rich in basic residues but, in the absence of a nucleolar envelope, rather than serving as transport signals they appear to function more as retention signals [56]. Human proteins containing NLS/NoLSs include human NF-κB-inducing kinase, the novel human nucleolar protein phosphatidylinositol 4-kinase and others [56][57].
In addition to NumA1, mitotic translocations have only been studied for four other proteins—Cbp4a, Src1, Snf12 and FhkA—as discussed in the following sections (Figure 2). During prophase nucleoli become indistinct with NumA1 appearing in smaller accumulations adjacent to the inner nuclear envelope as well as being associated with the centrosome. By metaphase these inner membrane accumulations disappear and, for the rest of mitosis, NumA1 appears throughout the cytoplasm with a border of protein adjacent the outer nuclear envelope plus continued centrosomal localization.
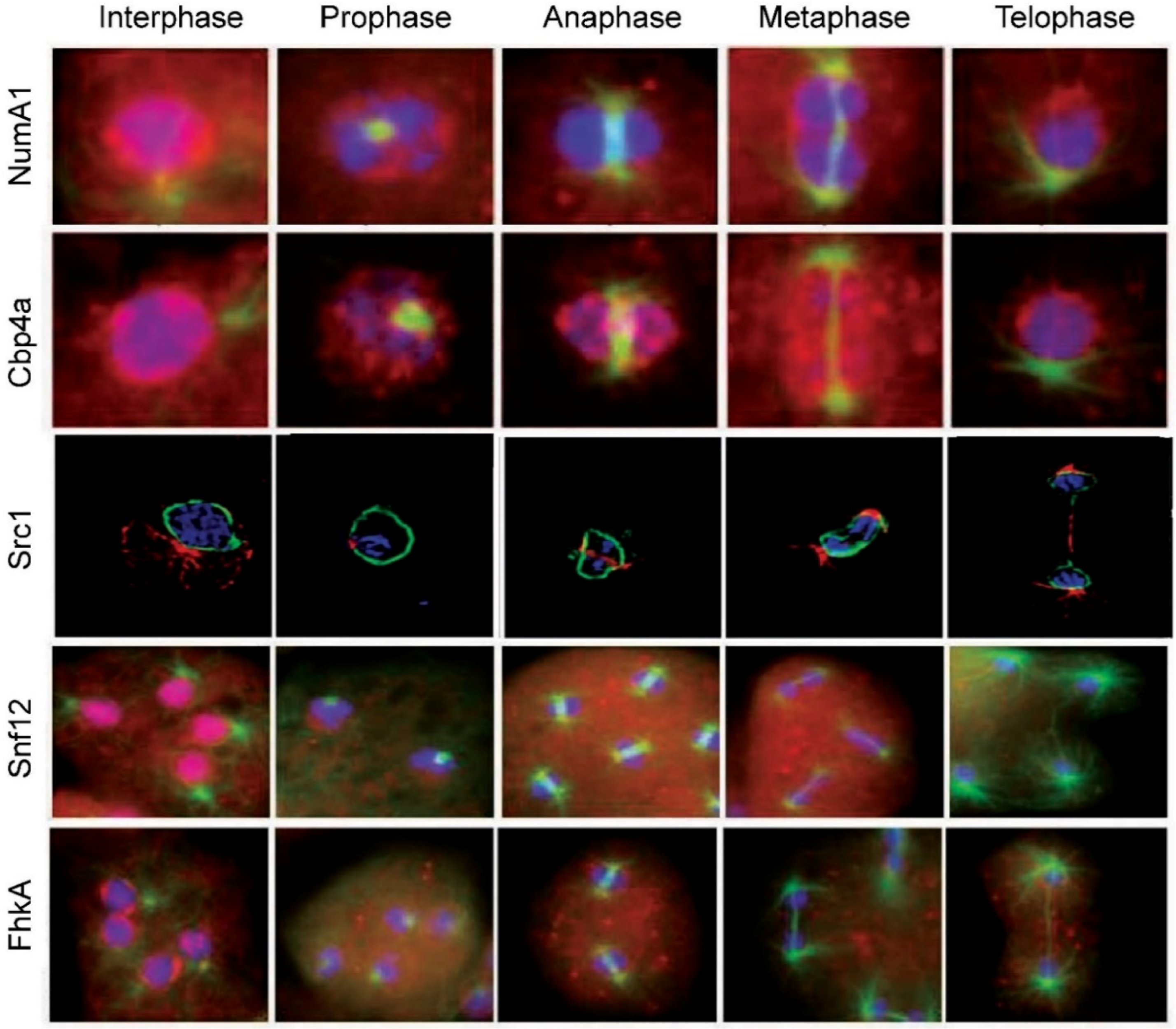
Figure 2. Nucleolar protein translocations during mitosis in Dictyostelium discoideum. NumA1, nucleomorphin A1, a cell cycle protein; Cbp4a, calcium-binding protein 4a, a NumaA1 binding protein; Src1, helix-extension-helix family homolog; Snf12, a nucleosome remodeling complex component; FhkA, a Rad53 (Chk2 in humans) tumor suppressor homolog. Note: the images for Src1 are from [58] with modifications
2.5. Calcium Binding Protein 4a (Cbp4a)
Studies using yeast two hybrid and coimmunoprecipitation, identified calcium binding protein 4a, (Cbp4a) as a nucleolar Ca2+-dependent, NumA1-binding partner [41]. This binding occurs via the glu/asp or DEED domain. Addition of actinomycin D leads to a loss of nucleolar CBP4a as does calcium-chelation with BAPTA-AM, supporting Cbp4a as a nucleolar protein that requires calcium for its localization. Fitting with its association with the cell cycle protein NumA1, CBP4a has a putative forkhead-associated domain that is present in numerous cell cycle proteins. Dictyostelium possesses 13 calcium-binding proteins (CBPs) including CaM, a major Ca2+ effector in all eukaryotes [59][60][61]. In contrast to CaM, calfumirin, and CBP3, the function of the other CBPs remains to be researched [42].
Cbp4a residues 40KKCK43 have been verified as a true NLS but not an NoLS since FITC-bound peptides show nuclear but not nucleolar localization. In total the results indicate that Cbp4a localizes to the nucleolus not via an NoLS but via the calcium-dependent binding to the DEED domain of NumA [50]. This would suggest that Cbp4a diffuses into the nucleolus to be held there through NumA1-binding. The relationship and behavioral differences between these two proteins was revealed during the events of mitosis where they were the first two nucleolar proteins to be studied during mitosis in Dictyostelium.
During mitosis CBP4a reveals a unique distribution that suggests the presence of previously undetected intranuclear subdomains that persist throughout the mitotic stages (Figure 2) [62]. During prophase, nucleolar dissolution is accompanied by the accumulation of CBP4a as multiple, discrete nucleoplasmic accumulations called “CBP4a islands”. In addition to these multiple smaller accumulations, during metaphase two larger islands localize to the metaphase plate region. Through anaphase and telophase, these accumulations migrate to the inner membrane as if in anticipation of reforming post-mitotic nucleoli. To date, no other nucleolar or nuclear protein has shown this sequence of events during mitosis. If the nucleolar binding of Cbp4a is dependent on diffusion, then retaining this protein within the nucleus may be essential to ensuring there is enough protein present when the nucleolus reforms during telophase.
2.6. SWI/SNF Complex Component SNF12 Homolog (Snf12)
SWI/SNF is a nucleosome remodeling complex, composed of 9–12 proteins called BAFs (Barrier-to-autointegration factor), highly conserve proteins that regulate gene transcription [63]. The complex mediates multiple other processes including cell proliferation, differentiation and DNA repair. It serves as a tumor suppressor by regulating the p53-mediated transcription of cell cycle genes. BAF60a (Snf12 in yeast) mediates its interaction with p53. Dictyostelium Snf12 is a predominately nucleoplasmic protein that localizes to nucleoli in ~20% of cells, as seen in mice [64][65]. Appropriately, it possesses conserved SWIB and COG domains found in BAF proteins, but these are not involved in nucleolar positioning. Instead an experimentally defined NLS/NoLS (372KRKR375) defines both its nuclear and nucleolar localization.
Unexpectedly, treatment of cells with actinomycin D increases the levels of nucleolar Snf12 which leads to an outward bulging of nucleoli followed by the cytoplasmic accumulation of Snf12-rich vesicles. Heat shock treatment also leads to a major increase in the nucleolar localization of Snf12. The rapid increases in nucleolar localization after heat shock and actinomycin D treatment, suggests Snf12 may function in the stress response. As mentioned above, heat shock treatment of Dictyostelium induces a redistribution of Hsp32 from the nucleolar periphery [14]. This is not surprising since the nucleolus is the central hub for coordinating the response to cell stress in other species where the composition of the nucleolus is stress-dependent [66]. In keeping with the results summarized here, heat shock and AM-D treatment both cause nucleolar accumulation of specific human proteins [67].
Snf12 undergoes several translocations during mitosis (Figure 2). With nucleolar dissolution during prometaphase, it first shifts from its nucleolar locale to take up a predominantly nucleoplasmic location with some localization in the cytoplasm. During metaphase and through anaphase it then exhibits a relatively uniform cellular distribution before reacquiring its nuclear/nucleolar localization during telophase.
2.7. Forkhead-Associated Kinase Protein A (FhkA)
Rad53 (CHK2 in humans) is a tumor suppressor protein involved in DNA damage (genotoxic) stress response [68]. It is recognized as a nuclear protein that possesses a C-terminal bipartite NLS [69]. The Dictyostelium Rad53 homologue forkhead-associated kinase protein A, FhkA, is a nucleolar protein. Immunolocalization shows is resides at the periphery of the nucleolar patches (i.e., NoSC3) being more concentrated adjacent to the nuclear envelope. Like Snf12, actinomycin D treatment leads to its nuclear expulsion as nucleolar protein-containing vesicles that end up in the cytoplasm. Its mitotic dynamics are also fitting for a nucleolar protein, yet its specific function there remains to be elucidated. During mitosis FhkA redistributes throughout the cell with an enhanced level of localization evident adjacent to the nuclear envelope from prometaphase through telophase (Figure 2). Like NumA1, FhkA also localizes within the spindle fiber region.
2.8. Bud31
As one of the last nucleolar proteins to be identified so far, less is known about Bud31 than any of the others. A comparative study of spliceosomal genes in Dictyostelium discoideum identified Bud31 but no further analysis of the protein was carried out [70]. In yeast where the protein was first identified, Bud31 is involved in cell cycle regulation, specifically functioning at the G1/S regulatory or start point [71]. A search of dictyBase.org indicates Bud31 is a putative RNA splicing factor or transcription factor. While the gene has been identified in humans and other species, its function in Dictyostelium has not been studied. Selected as a nuclear protein for comparison, it was shown that Bud31 localized throughout the Dictyostelium nucleolus along with NumA1 and eIF6 [13]. However, its location during mitosis and the presence of an NoLS have not been assessed.
2.9. Src1
Dictyostelium Src1 is homolog of the helix-extension-helix family that localizes adjacent to the inner nuclear membrane [58]. Because of its interaction with the major nuclear lamina protein NE81, its juxtaposition to the inner nuclear membrane and its unchanging location during the cell cycle, Src1 has attributes of an inner nuclear membrane protein that is involved in nuclear lamina formation. In other species, Src1 is also implicated in nucleolar organization through its ability to stabilize repetitive rDNA sequences [72]. GFP-Src1 and immune-transmission electron microscopy reveal that Src1 is an inner nuclear membrane protein that is tightly linked to the positioning of nucleoli in Dictyostelium [58]. Future work will have to be done to verify the significance of this relationship and whether Src1 fits the description of a true nucleolar protein. Since Src1 retains its localization throughout mitosis, this puts it in a position to serve as part of the reformation points for nucleolar reassembly during telophase (Figure 2).
References
- Benichou, J.C.; Quiviger, B.; Ryter, A. Cytochemical study of the nucleolus of the slime-mold Dictyostelium discoideum. J. Ultrastruct. Res. 1983, 84, 60–66.
- Maclean, N.; Garside, K.; Bradley, M.C.; Wood, C. The nucleus of axenically grown Dictyostelium discoideum: Studies on its division cycle isolation and conformation. Experientia 1984, 40, 1207–1214.
- Roos, U.P.; Bottini, F.; Jenni, V. Morphology of the nucleolus in undifferentiated amebas of Dictyostelium discoideum. Eur. J. Protistol. 1992, 28, 94–101.
- Sameshima, M.; Fujimoto, H.; Imai, Y.; Tsukita, S.; Hashimoto, Y. Relation of nucleolar structure and position to the cytoplasmic microtubule system in Dictyostelium. Cell Motil. Cytoskel. 1991, 18, 293–303.
- Sameshima, M. The orientation of nucleus, nucleus-associated body and protruding nucleolus in aggregating Dictyostelium discoideum. Exp. Cell Res. 1985, 156, 341–350.
- Maeda, Y.; Takeuchi, I. Cell differentiation and fine structures in the development of the cellular slime molds. Dev. Growth Differ. 1969, 11, 232–245.
- Simon, I.; Olins, D.E. Higher-order association of extrachromosomal ribosomal RNA genes in Dictyostelium discodeum. Cell Biol. Int. 1994, 18, 1091–1094.
- Williams, J.G. Dictyostelium finds new roles to model. Genetics 2010, 185, 717–726.
- Cockburn, A.F.; Taylor, W.C.; Firtel, R.A. Dictyostelium rDNA consists of non-chromosomal palindromic dimers containing 5S and 36S coding regions. Chromosoma 1978, 70, 19–29.
- Hohl, H.R.; Miura-Santo, L.Y.; Cotter, D.A. Ultrastructural changes during formation and germination of microcysts in Polysphondylium pallidum, a cellular slime mould. J. Cell Sci. 1970, 7, 285–305.
- Schaap, P.; Vandermolen, L.; Konijn, T.M. The vacuolar apparatus of the simple cellular slime mold Dictyostelium minutum. Biol. Cell. 1981, 41, 133–142.
- Kawashima, K.; Sameshima, M.; Izawa, M. Isolation of nucleoli from the cellular slime mold Dictyostelium discoideum, strain A-3. Cell Struct. Funct. 1979, 4, 183–191.
- Catalano, A.; O’Day, D.H. Evidence for nucleolar subcompartments in Dictyostelium. Biochem. Biophys. Res. Commun. 2015, 456, 901–907.
- Moerman, A.M.; Klein, C. Dictyostelium discoideum Hsp32 is a resident nucleolar heat- shock protein. Chromosoma 1998, 107, 145–154.
- Shaw, P.J.; Highett, M.I.; Beven, A.F.; Jordan, E.G. The nucleolar architecture of polymerase I transcription and processing. EMBO J. 1995, 14, 2896–2906.
- Grisendi, S.; Mecucci, G.; Falini, B.; Pandolfi, P. Nucleophosmin and cancer. Nat. Rev. Cancer 2006, 6, 493–505.
- Xue, Z.; Melese, T. Nucleolar proteins that bind NLSs: A role in nuclear import or ribosome biogenesis? Trends Cell Biol. 1994, 4, 414–417.
- Szebeni, A.; Herrera, J.E.; Olson, M.O.J. Interaction of nucleolar protein B23 with peptides related to nuclear localization signals. Biochemistry 1995, 34, 8037–8042.
- Creancier, L.; Prats, H.; Zanibellato, C.; Amalric, F.; Bugler, B. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol. Biol. Cell 1993, 4, 1239–1250.
- Dingwall, C.; Laskey, R.A. Nuclear targeting sequences—A consensus. Trends Biochem. Sci. 1991, 16, 478–481.
- Balbo, A.; Bozzaro, S. Cloning of Dictyostelium eIF6 (p27BBP) and mapping its nucle(ol)ar localization subdomains. Eur. J. Cell Biol. 2006, 85, 1069–1078.
- Villacís, L.N.; Wong, M.S.; Ferguson, L.L.; Hein, N.; George, A.J.; Hannan, K.M. New roles for the nucleolus in health and disease. BioEssays 2018, 40, 1700233.
- Sanvito, F.; Piatti, S.; Villa, A.; Bossi, M.; Lucchini, G.; Marchisio, P.C.; Biffo, S. The β4 integrin interactor p27BBP/eIF6 is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J. Cell Biol. 1999, 144, 823–837.
- Si, K.; Maitra, U. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell Biol. 1999, 19, 1416–1426.
- Wood, L.C.; Ashby, M.N.; Grunfeld, C.; Feingold, K.R. Cloning of murine translation initiation factor 6 and functional analysis of the homologous sequence YPR016c in Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 11653–11659.
- Shimarmura, A. Shwachman-Diamond Syndrome. Semin. Hematol. 2006, 43, 178–188.
- Ng, C.L.; Waterman, D.G.; Koonin, E.V.; Walters, A.D.; Chong, J.P.J.; Isupov, M.N.; Lebedev, A.A.; Bunka, D.H.J.; Stockley, P.G.; Ortiz-Lombardia, M.; et al. Conformational flexibility and molecular interactions of an archaeal homologue of the Shwachman-Bodian-Diamond syndrome protein. BMC Struct. Biol. 2009, 9, 32.
- Weiss, F.; Guidice, E.; Churcher, M.; Jin, L.; Hilcenko, C.; Wong, C.C.; Traynor, D.; Kay, R.R.; Warren, A.J. Mechanism of eIF6 release from the nascent 60s ribosomal subunit. Nature Struct. Molec. Biol. 2016, 22, 914–919.
- Scott, M.S.; Troshin, P.V.; Barton, G.J. NoD: A nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinform. 2011, 12, 317.
- Sillo, A.; Bloomfield, G.; Balest, A.; Balbo, A.; Pergolizzi, B.; Peracino, B.; Skelton, J.; Ivens, A.; Bozzaro, S. Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC Genom. 2008, 9, 291.
- Morita, T.H.; Amagai, A.; Maeda, Y. Unique behavior of a Dictyostelium homologue of TRAP-1, coupling with differentiation of D. discoideum cells. Exp. Cell Res. 2002, 280, 45–54.
- Morita, T.H.; Amagai, A.; Maeda, Y. Translocation of the Dictyostelium TRAP1 homologue to mitochondria induces a novel prestarvation response. J. Cell Sci. 2004, 117, 5759–5770.
- Morita, T.; Yamaguchi, H.; Amagai, A.; Maeda, Y. Involvement of the TRAP-1 homologue, Dd-TRAP1, in spore differentiation during Dictyostelium development. Exp. Cell Res. 2005, 303, 425–431.
- Yamaguchi, H.; Morita, T.; Amagai, A.; Maeda, Y. Changes in spatial and temporal localization of Dictyostelium homologues of TRAP1 and GRP94 revealed by immunoelectron microscopy. Exp. Cell Res. 2005, 303, 415–424.
- Francione, L.M.; Annesley, S.J.; Carilla-Latorre, S.; Escalante, R.; Fisher, P.R. The Dictyostelium model for mitochondrial disease. Semin. Cell. Dev. Biol. 2011, 22, 120–130.
- Ohta, Y.; Ohba, T.; Miyamoto, E. Ca2+/calmodulin-dependent protein kinase II: Localization in the interphase nucleus and the mitotic apparatus of mammalian cells. Proc. Natl. Acad. Sci. USA 1990, 87, 5341–5345.
- Torgan, C.E.; Daniels, M.P. Calcineurin localization in skeletal muscle offers insights into potential new targets. J. Histochem. Cytochem. 2006, 54, 119–128.
- Wong, E.C.; Saffitz, J.E.; McDonald, J.M. Association of calmodulin with isolated nuclei from rat hepatocytes. Biochem. Biophys. Res. Commun. 1991, 181, 1548–1556.
- Thorogate, R.; Torok, K. Role of Ca2+activation and bilobal structure of calmodulin in nuclear and nucleolar localization. Biochem. J. 2007, 402, 71–80.
- Myre, M.A.; O’Day, D.H. Nucleomorphin. A novel, acidic, nuclear calmodulin-binding protein from Dictyostelium that regulates nuclear number. J. Biol. Chem. 1974, 277, 19735–19744.
- Myre, M.A.; O’Day, D.H. Dictyostelium calcium-binding protein 4a interacts with nucleomorphin, a BRCT-domain protein that regulates nuclear number. Biochem. Biophys. Res. Commun. 2004, 322, 665–671.
- Catalano, A.; O’Day, D.H. Calmodulin-binding proteins in the model organism Dictyostelium: A complete and critical review. Cell. Signal. 2008, 20, 277–291.
- Sansam, C.L.; Wells, K.S.; Emeson, R.B. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl. Acad. Sci. USA 2003, 100, 14018–14023.
- Lim, M.J.; Wang, X.W. Nucleophosmin and human cancer. Cancer Detect. Prev. 2006, 30, 481–490.
- Myre, M.A.; O’Day, D.H. Dictyostelium nucleomorphin is a member of the BRCT- domain family of cell cycle checkpoint proteins. Biochim. Biophys. Acta 2004, 1675, 192–197.
- Farley-Barnes, K.; McCann, K.L.; Ogawa, L.M.; Merkel, J.; Surovtseva, Y.V.; Baserga, S.J. Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep. 2018, 22, 1923–1934.
- Heath, E.M.; Chan, S.M.; Minden, M.D.; Murphy, T.; Shlush, L.I.; Schimmer, A.D. Biological and clinical consequences of NPM1 in mutations in AML. Leukemia 2017, 31, 798–807.
- Box, J.K.; Paquet, N.; Adams, M.N.; Boucher, D.; Bolderson, E.; O’Byrne, K.J.; Richard, D.J. Nucleophosmin: From structure and function to disease development. BMC Mol. Biol. 2016, 17, 19.
- Bhutani, N.; Venkatraman, P.; Goldberg, A.L. Puromycin-sensitive aminopeptidase is the major peptidase responsible for digesting polyglutamine sequences released by proteasomes during protein degradation. EMBO J. 2007, 26, 1385–1396.
- Catalano, A.; Poloz, Y.; O’Day, D.H. Dictyostelium puromycin-sensitive aminopeptidase A is a nucleoplasmic nucleomorphin-binding protein that relocates to the cytoplasm during mitosis. Histochem. Cell. Biol. 2011, 136, 677–688.
- O’Day, D.H.; Poloz, Y.; Myre, M.A. Differentiation inducing factor-1 (DIF-1) induces gene and protein expression of the Dictyostelium nuclear calmodulin-binding protein nucleomorphin. Cell. Signal. 2009, 21, 317–323.
- Myre, M.A.; O’Day, D.H. An N-terminal nuclear localization sequence but not the calmodulin-binding domain mediates nuclear localization of nucleomorphin, a protein that regulates nuclear number in Dictyostelium. Biochem. Biophys. Res. Commum. 2005, 332, 157–166.
- Russo, G.; Ricciardelli, G.; Pietropaolo, C. Different domains cooperate to target the human ribosomal L7a protein to the nucleus and to the nucleoli. J. Biol. Chem. 1997, 272, 5229–5235.
- Collavoli, A.; Hatcher, C.J.; He, J.; Okin, D.; Deo, R.; Basson, C.T. TBX5 nuclear localization is mediated by dual cooperative intramolecular signals. J. Mol. Cell. Cardiol. 2003, 35, 1191–1195.
- Theodore, M.; Kawai, Y.; Yang, J.; Kleshchenko, Y.; Reddy, S.P.; Villalta, F.; Arinze, I.J. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J. Biol. Chem. 2008, 283, 14176.
- Birbach, A.; Bailey, S.T.; Ghosh, S.; Schmid, J.A. Cytosolic, nuclear and nucleolar localization signals determine subcellular distribution and activity of the NF-κB-inducing kinase NIK. J. Cell Sci. 2004, 117, 3615–3624.
- Kakuk, A.; Friedlander, E.; Vereb Jr, G.; Lisboa, D.; Bagossi, P.; Toth, G.; Gergely, P.; Vereb, G. Nuclear and nucleolar localization signals and their targeting function in phosphatidylinositol 4-kinase PI4K230. Exp. Cell Res. 2008, 314, 2376–2388.
- Batsios, P.; Ren, X.; Baumann, O.; Larochelle, D.A.; Graf, R. Src1 is a protein of the inner nuclear membrane interacting with the Dictyostelium. Lamin NE81. Cells 2016, 5, 13.
- Dharamsi, A.; Tessarolo, D.; Coukell, B.; Pun, J. CBP1 associates with the Dictyostelium cytoskeleton and is important for normal cell aggregation under certain developmental conditions. Exp. Cell Res. 2000, 258, 298–309.
- Dorywalska, M.; Coukell, B.; Dharamsi, A. Characterization and hetereologous expression of cDNAs encoding two novel closely related Ca2+-binding proteins in Dictyostelium discoideum. Biochim. Biophys. Acta 2000, 356–361.
- Sakamoto, H.; Nishio, K.; Tomisako, M.; Kuwayama, H.; Yoshimasa, T.; Suetake, I.; Tajima, S.; Ogihara, S.; Coukell, B.; Maeda, M. Identification and characterization of novel calcium-binding proteins of Dictyostelium and their spatial expression patterns during development. Develop. Growth Differ. 2003, 45, 507–514.
- Catalano, A.C.; O’Day, D.H. Rad53 homologue forkhead-associated kinase A (FhkA) and Ca2+-binding protein 4a (CBP4a) are nucleolar proteins that differentially redistribute during mitosis in Dictyostelium. Cell Division 2013, 8, 4.
- Reisman, D.; Glaros, S.; Thompson, E.A. The SWI/SNF complex and cancer. Oncogene 2009, 28, 1653–1668.
- Li, S.; Liu, C.; Li, N.; Hao, T.; Han, T.; Hill, D.E.; Vidal, M.; Lin, J.D. Genome-wide coactivation analysis of PGC-1α identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008, 8, 105–117.
- Catalano, A.C.; O’Day, D.H. Nucleoplasmic/nucleolar translocation and identification of a nuclear localization signal (NLS) in Dictyostelium BAF60a/SMARCD1 homologue Snf12. Histochem. Cell Biol. 2012, 138, 515–530.
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.-M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227.
- Kodiha, M.; Chu, A.; Lazrak, O.; Stochaj, U. Stress inhibits nucleocytoplasmic shuttling of heat shock protein hsc70. Am. J. Physiol. Cell. Physiol. 2005, 289, C1034–C1041.
- Stracker, T.H.; Usui, T.; Petrini, J.H.J. Taking the time to make important decisions: The checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair 2009, 8, 1047–1054.
- Smolka, M.B.; Albuquerque, C.P.; Chen, S.-H.; Schmidt, K.H.; Wei, X.X.; Kolodner, R.D.; Zhou, H. Dynamic changes in protein-protein interaction and protein phosphorylation probed with amine-reactive isotope tag. Mol. Cell Proteom. 2005, 4, 1358–1369.
- Yu, B.; Fey, P.; Kestin-Pilcher, K.E.; Federov, A.; Prakash, A.; Chisholm, R.L.; Wu, J.Y. Spliceosomal genes in the Dictyostelium genome: A comparison with those in H. sapiens, D. melanogaster, A. thaliana and S. cerevisiae. Protein Cell 2011, 2, 395–409.
- Saha, D.; Banerjee, S.; Bashir, S.; Vijayraghavan, U. Context dependent splicing functions of Bud31/Ycr063w define its role in budding and cell cycle progression. Biochem. Biophys. Res. Commun. 2012, 424, 579–585.
- Mekhail, K.; Seebacher, J.; Gygi, S.P.; Moazed, D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 2008, 456, 667–670.
More
Information
Subjects:
Cell Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
627
Revisions:
2 times
(View History)
Update Date:
16 Aug 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




